Abstract
This paper reports studies of two patients proven by a variety of studies to have mitochondrial depletion syndromes due to mutations in either their MPV17 or DGUOK genes. Each was initially investigated metabolically because of plasma methionine concentrations as high as 15–21-fold above the upper limit of the reference range, then found also to have plasma levels of S-adenosylmethionine (AdoMet) 4.4–8.6-fold above the upper limit of the reference range. Assays of S-adenosylhomocysteine, total homocysteine, cystathionine, sarcosine, and other relevant metabolites and studies of their gene encoding glycine N-methyltransferase produced evidence suggesting they had none of the known causes of elevated methionine with or without elevated AdoMet. Patient 1 grew slowly and intermittently, but was cognitively normal. At age 7 years he was found to have hepatocellular carcinoma, underwent a liver transplant and died of progressive liver and renal failure at age almost 9 years. Patient 2 had a clinical course typical of DGUOK deficiency and died at age 8 ½ months. Although each patient had liver abnormalities, evidence is presented that such abnormalities are very unlikely to explain their elevations of AdoMet or the extent of their hypermethioninemias. A working hypothesis is presented suggesting that with mitochondrial depletion the normal usage of AdoMet by mitochondria is impaired, AdoMet accumulates in the cytoplasm of affected cells poor in glycine N-methyltransferase activity, the accumulated AdoMet causes methionine to accumulate by inhibiting activity of methionine adenosyltransferase II, and that both AdoMet and methionine consequently leak abnormally into the plasma.
Keywords: mitochondria, depletion, methionine, S-adenosylmethionine, MPV17, DGUOK
1 Introduction
Mitochondrial DNA depletion syndromes are autosomal recessive diseases characterized by a severe reduction in mitochondrial DNA content leading to dysfunction of the affected organs or tissues. This group of mitochondrial disorders is clinically and genetically heterogeneous, but can be classified into three major forms: the myopathic, encephalomyopathic, and hepatocerebral. The latter group has been associated with mutations in POLG1, TWINKLE, DGUOK, and MPV17 genes [1–6]. POLG is the mitochondrial DNA (mtDNA) polymerase gamma. POLG deficiency can cause autosomal recessive or dominant heterogeneous disorders with mtDNA depletion or mtDNA multiple deletions [5]. The most common POLG-related disorder is Alpers syndrome, characterized by the clinical triad of psychomotor retardation, intractable seizures, and liver failure [7]. Deficiency in deoxyguanosine kinase (E.C. 2.7.1.113) (DGUOK) is the most common hepatocerebral form of mtDNA depletion syndrome. Patients with mutations in the DGUOK gene usually present with liver dysfunction at birth, with or without neurological impairment, and most die within the first year of life due to liver failure.[8] In some POLG or DGUOK deficient cases, fulminant liver failure may be triggered by infection [9;10]. MPV17 encodes a mitochondrial inner membrane protein and plays an as yet poorly understood role in the maintenance of mitochondrial DNA integrity. Mutations in the MPV17 gene have been reported in patients who came to medical attention during infancy with liver failure, hypoglycemia, failure to thrive and neurological symptoms [3;4]. More recently, severe liver failure was found to be associated with autosomal recessive mutations in the TWINKLE gene, encoding the DNA helicase [6].
The two patients reported in this paper each had elevations of plasma methionine (as high as 15–22-fold above the upper limit of the reference range), and plasma S-adenosylmethionine (AdoMet) (4.4–8.6-fold elevated). As detailed in the Results and Discussion sections, both these elevations were shown not to be due to any of the known causes of such abnormalities. We present a working hypothesis that relates the underlying mitochondrial disorders to these metabolite abnormalities, as well as to the 8.2-fold elevation of plasma cystathionine found in one of the patients. Based on the available results, we suggest that, further studies of AdoMet, methionine, and cystathionine in mitochondrial disorders are needed, and that, pending the results of such studies, mitochondrial disorders be added to the differential diagnosis of combined elevations of AdoMet and methionine.
2. Materials and Methods
2.1 Patients and DNA samples
This research study was conducted according to IRB approved protocols. Tissue or blood samples from the two patients described were submitted to the Mitochondrial Diagnostics Laboratory at the Baylor College of Medicine for molecular and/or biochemical diagnosis. Total genomic (nuclear and mitochondrial) DNA was extracted from peripheral blood leukocytes, liver, or skin fibroblasts using a commercially available DNA isolation kit (Gentra Systems Inc., Minneapolis, MN) according to the manufacturer’s protocols.
2.2 Metabolite assays
AdoMet and S-adenosylhomocysteine (AdoHcy) were assayed as described [11]. Other metabolites were assayed by capillary gas chromatography-mass spectrometry [12–14]. GNMT activity was assayed as described [15].
2.3 Molecular analyses
Sequence-specific oligonucleotide primers linked to M13 universal primers were designed to amplify all coding exons and the flanking intronic 50 nucleotides of the genes of interest. Sequencing analyses were performed using the BigDye Terminator cycle sequencing kit (version 3.1) on an ABI3730XL automated DNA sequencer (Applied Biosystems) as previously described [1;2;4]. DNA sequences were aligned with the corresponding GenBank reference sequences using Mutation Surveyor version 3.23 (SoftGenetics®, State College, PA).
The mtDNA copy number in liver or blood was determined in triplicate by the real-time quantitative polymerase chain reaction (RT-qPCR) using primers specific for the mitochondrial transfer RNALeu.(UUR) gene and nuclear single-copy gene, β-2-microglobulin [16]. Depletion of mtDNA was determined by comparing patient’s mtDNA copy number with that of tissue- and age-matched controls [17].
Intragenic deletions in the nuclear genes responsible for mtDNA depletion syndrome were investigated by using a custom-designed oligonucleotide Comparative Genomic Hybridization (CGH) array (MitoMet®). The MitoMet® (version 2.8) is a clinically validated 60 K oligonucleotide array with complete coverage of the mitochondrial genome and 351 nuclear genes related to mitochondrial structure/function and metabolic diseases, including the DGUOK and MPV17 genes [18].
For the analysis of transcripts, total RNA was isolated from skin fibroblast culture of patient 1 using RiboPure-Blood (Applied Biosystem, Foster City, CA) and cDNA was made with the iScript cDNA Synthesis Kit (Bio-Rad Laboratories, Hercules, CA) primed with random hexamers according to the manufacturer’s protocol. The cDNAs were cloned and 28 clones were picked for sequencing. The primers used for MPV17 RT-PCR and cDNA sequencing were MPV17-E1FcDNA: 5’-GCGGAAGTTCCTAGGCCA-3’ and MPV17-E7RcDNA: 5’-GTACACTGACTGCTTTAGA-3’.
2.4 Respiratory chain enzyme analysis
Spectrophotometric analysis of the respiratory chain complexes was performed on the liver specimen of patient 1 and on cultured skin fibroblasts of patients 1 and 2 according to published procedures [3].
3. Case reports
3.1 Patient 1
This boy was born at full term with normal birth weight and stature (Table 1). Although he grew relatively slowly during his first year, he smiled at 9 weeks, held up his head or rolled over at 4 months, sat alone at 8 months, stood without help at 10 months, and walked without help at 18 months. At about 9 months of age, his appetite diminished and he lost almost a kilogram of body weight during the next two months. At age 14 months he was seen by a metabolic physician and found to have a very elevated methionine of 947 µM (Table 1). (Methionine had not been measured during newborn screening). Because his plasma total homocysteine (tHcy) was minimally above the reference range for his age, suggesting possible CBS deficiency, he was started on pyridoxine, 150 mg/day. At age 16 months the physician judged him to be “mildly hypotonic”. He began to gain weight and grow in stature so that by age 3–5 years these parameters were within normal limits. More detailed metabolic studies at age 17 months found that his hypermethioninemia persisted while on B6, plasma AdoMet was very elevated, tHcy was virtually normal, cystathionine slightly above the reference range, and plasma AdoHcy, sarcosine, and tyrosine were normal. Serum transaminases were slightly elevated. At age approximately 2½ years his physician found him to be a “cute boy with mild dysmorphic features, very personable, verbal and interactive with mild motor delay”. Between ages 2 and 5 to 6 years the levels of both methionine and AdoMet gradually diminished, staying low after discontinuation of the supplement of B6 at 5 years. During a routine visit to his local physician at age 6½ years he was found to have slight splenomegaly, a heart murmur, and slightly elevated serum transaminases, but was otherwise clinically normal and continuing to gain weight and height within the lower areas of the references ranges. At age 7 years he was about to start 3rd grade when, he had an episode of hematemesis, a CT scan showed multiple masses in the liver, and he had a liver transplant. The pathology report for the liver described a 1.7 cm hepatocellular carcinoma, multifocal hepatocellular carcinomas, and hepatic microadenomas on a background of cirrhotic liver. The cytoplasm of the hepatocytes was granular. Periodic acid Schiff staining showed a diastase-digestible material indicative of a storage disease. Subsequently the boy deteriorated clinically with intermittent periods of fatigue, extreme tiredness and sleepiness during the day. His mother described him as behaving like a 90-year-old. However, he remained cognitively perfectly normal. At age 8½ he fell and sustained a fracture of his skull. He had a seizure and was found to have renal insufficiency, loss of muscle mass, and poor musculoskelatal tone. Molecular studies (detailed below) found the boy to have MPV17 deficiency and mtDNA depletion, but while the latter studies were in progress the boy continued to have progressive liver failure, renal failure, stiffness, muscle wasting, bone demineralization, and he died at age almost 9 years.
Table 1.
Height and weights and plasma metabolite concentrations in Patient 1.
| Age, months | 0.25 | 6 | 9 | 12 | 14 | 17 | 22 | 38 | 64 | 707 275 |
83 | 102 | ||
| B6 supplement, mg/d | 0 | 0 | 0 | 0 | 150 | 100 | 100 | 100 | 100 2 |
2 | 2 | 2 | 2 | |
| —— | —— | —— | —— | —— | —— | —— | —— | —— | —— | —— | —— | —— | —— | |
| Height, percentile* | 50–75 | 5–10 | <3 | 3–5 | <3 | <3 | 3–5 | 5–10 | 5–10 | 3–5 | <3 | |||
| Weight, percentile | 5–10 | <3 | <3 | <3 | <3 | <3 | 10–25 | 10–25 | <3 | <3 | ||||
| Methionine (13–43 µM) | ( | 705)** | 947 | 877 | 249 | 33 | 71 | 52 | 70 | |||||
| AdoMet (92.8 ± 16.2 nM) | 936 | 891 | 475 | 144 | 143 | 172 | ||||||||
| AdoHcy (27.8 ± 7.9 nM) | 19 | 16 | 27 | 11 | 13 | |||||||||
| tHcy (5.1–13.9 µM) | (13.5) | 10.7 | 12.2 | 6.7 | 4.3 | 4.5 | 4.1 | |||||||
| Cystathionine (44–342 nM) | 446 | 498 | 625 | 647 | 464 | 565 | ||||||||
| tCys (203–369 µM | 202 | 213 | 214 | 219 | 209 | 222 | ||||||||
| Sarcosine (0.6–2.7 µM) | 1.8 | 1.7 | 3.4 | 2.3 | 3.9 | 4.3 | ||||||||
| Dimethyglycine (1. 4–5.3 µM) | 4.9 | 4.2 | 8.5 | 8.5 | 7.1 | 8.9 | ||||||||
| Citrulline (16–55 µM) | 7 | 64 | ||||||||||||
| Tyrosine (30–90M) | (normal) | (29) | (36) (150) |
(224) | ||||||||||
| Serine | (97–267 µM) | 141 100 162 120 169 |
165 210 |
|||||||||||
| Threonine (92–240 µM) | 148 | |||||||||||||
| ALT (10–60 U/L) | 101 | 111 | 96 71 |
(117) | ||||||||||
| AST (10–42 U/L) | 147 | 228 | 129 88 |
(91) | ||||||||||
| Alkalinephosphatase (29–144 U/L) | 305 |
Percentiles of height and weight were estimated by comparison with the values for males listed by the National Center for Health Statistics (http://www.cdc.gov/growthcharts/html_charts.).
Values in parentheses were obtained in laboratories which may have somewhat different reference ranges than those shown.
3.2. Patient 2
This baby girl, was delivered by caesarian section at gestation week 41 due to late fetal heart rate decelerations. Her weight was 2.7 kg. In an expanded newborn screen her methionine was not elevated, and, although tyrosine was above the reference range at 342 µM, succinylacetone was normal (Table 2). She was hypotonic and floppy from birth, had loose stools, decreasing with age in frequency from 10/day down to 2–3/day, and did not track with her eyes. On formula feeding she gained weight only slowly. At 2 months of age her mother reported hypotonia and concern about vision. On evaluation at age 4 months she weighed 3.83 kg, was markedly hypotonic and either areflexic or hyporeflexic, had limited movement for her age, impaired visual interactions, and some nystagmus. Plasma methionine was now elevated to 344 µM, and six weeks later was extremely high at 975 µM (Table 2). Plasma tHcy was virtually normal for her age, AdoMet and cystathionine were elevated, AdoHcy normal, plasma sarcosine low normal, and serum transaminases slightly elevated. Molecular studies (detailed below) showed the patient to be a homozygote for a deletion in the gene encoding DGUOK. However, before this diagnosis became available the baby went into liver failure and renal shutdown and died at age 6½ months.
Table 2.
Plasma metabolite concentrations in Patient 2.
| Age, months | NBS |
4 |
5½ |
6 |
6½ |
|---|---|---|---|---|---|
| Methionine (8–75 µM) | (44) | 344 | 908 | 975 | 1184 |
| AdoMet (92.8 ± 16.2 nM) | 475 | ||||
| AdoHcy (27.8 ± 7.9 nM) | 16 | ||||
| tHcy (5.1–13.9 µM) | 7.0 | 10.9 | |||
| Cystathionine (44–342 nM) | 3160 | ||||
| tCys (203–369 µM) | 191 | ||||
| Sarcosine(0.6–2.7 µM) | 0.6 | ||||
| Dimethyglycine (1.4–5.3 µM) | 1.6 | ||||
| Citrulline (6–54 µM) | 38 | 36 | 40 | 21 | |
| Tyrosine (<180 µM) | 342 | 103 | 288 | 334 | 376 |
| Succinylacetone | (normal) | (normal) | |||
| Alanine (152–734 µM) | 907 | 964 | 980 | 1401 | |
| Lactate (0.5–1.6 mM) | 4.3;4.4; | 6.1 | 23 | ||
| Guanidinoacetate (0.8–3 µM) | 0.6 | ||||
| Creatine (10–100 µM) | 84 | ||||
| Creatine kinase (15–110 U/L) | 31 | 106 | |||
| ALT (10–60 U/L) | 262 | 161 | 205 | ||
| AST (10–42 U/L | 188 | 206 | 485 | ||
| Ammonia | 79 | 518 | |||
| Bilirubin, total (0.2–1.3 mg/dl) | 4.4 | 4.9 | 11.1 | ||
| Bilirubin, durect (0.1–0.3 mg/dl | 6.4 | ||||
| Prothrombin time (12.6–15.2 seconds | 50.8 | 79.1 | |||
| Activated partial prothrombin time (23.3–35.7 seconds) | 94.0 | 124.6 | |||
4. Results
4.1. Molecular studies
4.1.1 Patient 1: mitochondrial studies
Due to the suspicion of mitochondrial involvement, the CTLN2 (citrin), DGUOK, MPV17, and TK2 genes were sequenced. The only mutation detected was a heterozygous c.22insC, in the MPV17 gene. A search for an intragenic deletion using targeted array CGH (MitoMet®) with dense oligonucleotide probe coverage of the MPV17 gene, failed to detect any deletion. However, mitochondrial depletion was shown by the facts that the mtDNA content in the liver of the patient was found to be drastically reduced to 12% of the mean of three control liver specimens (Table 3), and that electron transport chain complexes I and IV activities in the liver specimen were reduced compared to control means, but all complex activities in cultured skin fibroblast cells were normal (Table 4).
Table 3.
mtDNA copy numbers.
| Patient | Tissue type | Copy number |
% of control | Method | |
|---|---|---|---|---|---|
| Patient | Control | ||||
| 1 | liver | 450 | 3784 | 12 | qPCR |
| 1 | skin fibroblasts | 863 | 637 | 135 | qPCR |
| 2 | blood | 70 | 295 | 24 | qPCR |
| 2 | skin fibroblasts | 593 | 718 | 83 | qPCR |
| 2 | blood | 25 | aCGH | ||
| Mother of 2 | blood | 71 | aCGH | ||
| Father of 2 | blood | 93 | aCGH | ||
Table 4.
Electron transport chain (ETC) results.
| Patient 1 Liver |
Patient 1 SFC* |
Patient 2 SFC* |
|
|---|---|---|---|
| NADH ferricyanide reductase (complex I) | 19 | 93 | 80 |
| NADH-cytochrome c reductase (complex I + III) | 62 | 160 | 75 |
| Succinate cytochrome c reductase (complex II + III) | 46 | 137 | 99 |
| Succinate dehydrogenase (complex II) | 62 | 114 | 129 |
| Cytochrome c oxidase (complex IV) | 26 | 74 | 62 |
| Citrate synthase | 60 | 86 | 96 |
ETC activities are expressed in % of normal mean.;
SFC : skin fibroblast cells
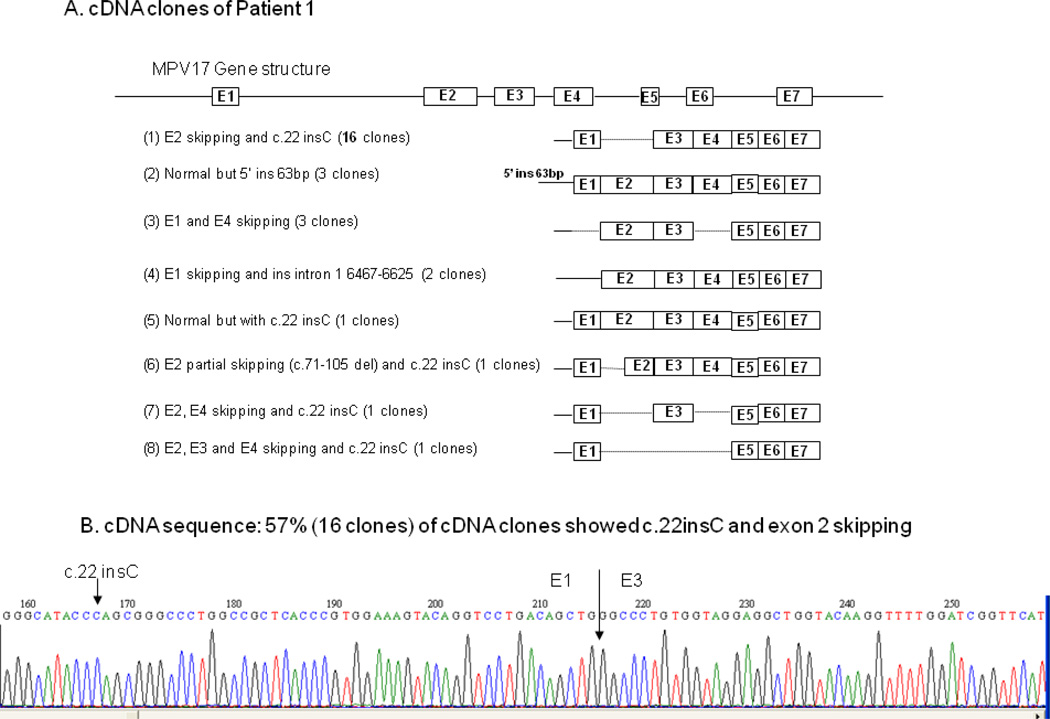
Given the proven hepatic mtDNA depletion, the combined complex deficiency, and the identification of one mutation in the MPV17 gene, the functionality of this gene was further investigated by sequence analysis of cDNA isolated from the patient’s cultured skin fibroblasts. A total of 28 MPV17 cDNA clones were sequenced (Fig. 1A and 1B), and eight different types of transcripts were detected, none of them normal (Fig. 1A). (1) Sixteen clones (the majority, 57%), had c.22insC in exon 1 with skipping of exon 2 (Fig. 1B); (2) three clones had an insertion of 63 bp in the 5’ promoter region; (3) three clones had skipping of exons 1 and 4; (4) two clones had skipping of exon 1 and an insertion of 158 bp in the intron 1 region; (5) one clone was normal except for c.22 insC, which results in frame-shift; (6) one clone had c. 22insC and partial skipping of exon 2 (7) one clone had c.22insC and skipping of exons 2 and 4; (8) one clone had C.22insC and skipping of exons 2, 3, and 4. Thus, although a second mutant allele was not identified, there were no normal transcripts, a finding consistent with the presence of MPV17 deficiency and mtDNA depletion.
Figure 1.
cDNA clones and sequence from patient 1.
1A. Total of 28 TA clones c.22insC were constructed and sequenced. Eight different types of MPV17 cDNA were detected. The majority, 16/28 clones (57%) had c.22insC with E2 skipping. The remaining 12 clones were distributed as follows: three had insertion of 63 bp at 5’ UTR which is part of promoter region; three had E1 and E4 skipping; two had E1 skipping and insertion of 158 bp from intron 1 region; one had c.22insC only, with no exon skipping; one.had c. 22insC with skipping of partial E2; one had c. 22insC with skipping of E2 and E4, one had c.22 insC with skipping of E2, E3 and E4.
1B. cDNA sequence of clone type (1) shows both c.22insC and E2 skipping.
4.1.2 Patient 2: mitochondrial studies
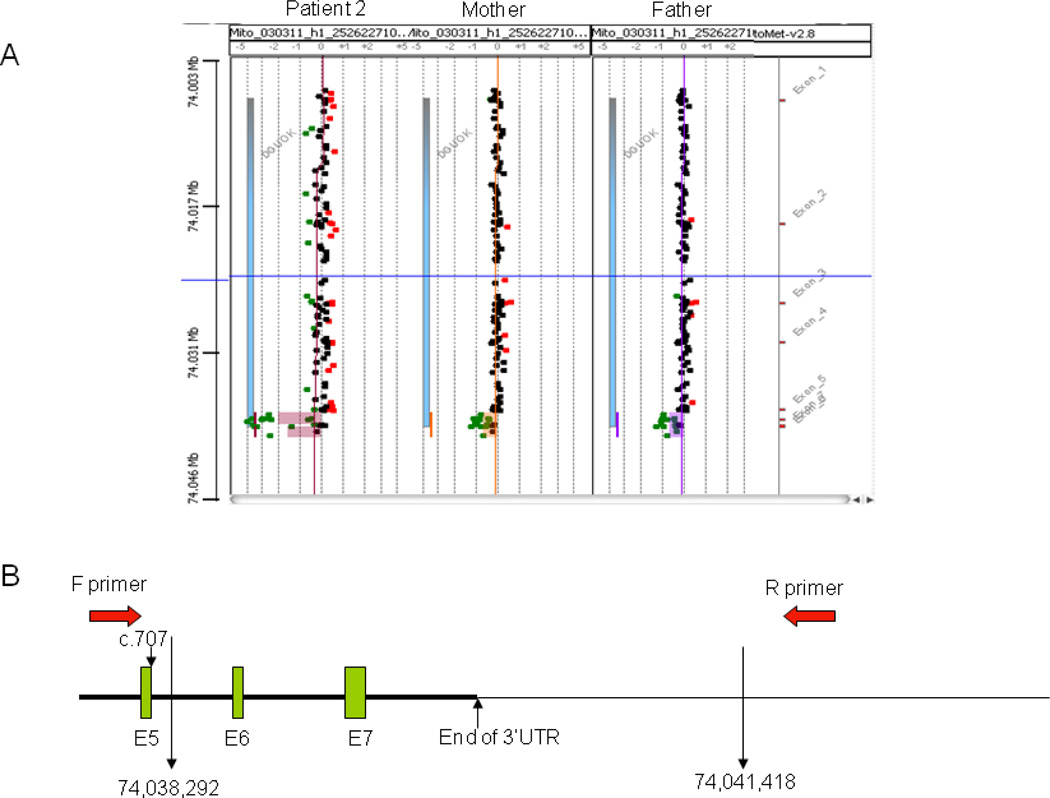
Patient 2 had a clinical presentation typical of the DGUOK deficient hepatocerebral form of mtDNA depletion syndrome in the newborn period. Sequence analysis of her DGUOK gene detected no mutations in exons 1–5. However, PCR failed to amplify exons 6 and 7, suggesting homozygous deletion of these two exons. Oligonucleotide aCGH (MitoMet®) detected a homozygous deletion of 3.3 kb encompassing exons 6 and 7 of the DGUOK gene (Fig. 2A). Both parents are heterozygous carriers of the 3.3 kb deletion (Fig. 2A). Subsequent PCR/sequence analysis confirmed the size of deletion to be 3,127 bp with the breakpoints at c.707+417 (intron 5) and c.834 (end of 3’UTR)+ 3416, containing the last two exons of the DGUOK gene (Fig. 2B). The mitochondrial DNA profile on the same array showed mtDNA reduction (about 25% of age-matched control mean) in a blood specimen of the patient, consistent with the mtDNA content evaluated by qPCR (24%) (Table 3).
Figure 2.
A. A homozygous large deletion encompassing exons 6 and 7 of DGUOK gene was present in patient 2. Both parents are heterozygous for the same deletion.
B. Schematic diagram showing the deletion breakpoints confirmed by PCR. The exact deletion size is 3126bp (c.707+417_c.834+3416).
4.1.3 Studies of glycine methyltransferase in patients 1 and 2
For patient 1 the GNMT gene was sequenced and no mutations were found. GNMT activity in extracts of the explanted liver was low when normalized to total protein (0.031 U/mg protein compared to 0.29 and 0.38 U/mg protein in two normal livers); but not low when normalized to GNMT protein calculated from Western blots (1.9 U/mg GNMT protein compared to 2.2 and 1.2 U/mg GNMT protein in the normal livers).
Sequencing of the GNMT gene of patient 2 revealed a heterozygous mutation, c.469C>T (p.Arg157Trp) [NM_018960], but no additional changes. Expressed in E. coli, this variant, located on the surface of the molecule, had Km and Vmax values close to those of the wild-type enzyme. and size-exclusion chromatography showed it to be a tetramer, as is wild-type GNMT [19].
5. Discussion
This paper reports two children with different mitochondrial DNA depletion syndromes and hepatic abnormalities. For patient 1, mtDNA was shown to be drastically reduced in the liver, and no normal MPV17 transcripts were found. There was a heterozygous mutation, c22insC in MPV17 that caused not only frameshift, but frequently also exon skipping. Although a second mutant allele was not identified, the remaining aberrantly spliced transcripts suggest there was such a mutation, located perhaps at the promoter or 5’UTR region or intron 1, affecting mostly exon 1 or exon 2 skipping. For patient 2, the identification of a homozygous deletion of exons 6 and 7 of the DGUOK gene directly confirms the hepatocerebral form of the mtDNA depletion syndrome.
5.1. Evidence as to the cause of the elevated methionine and AdoMet
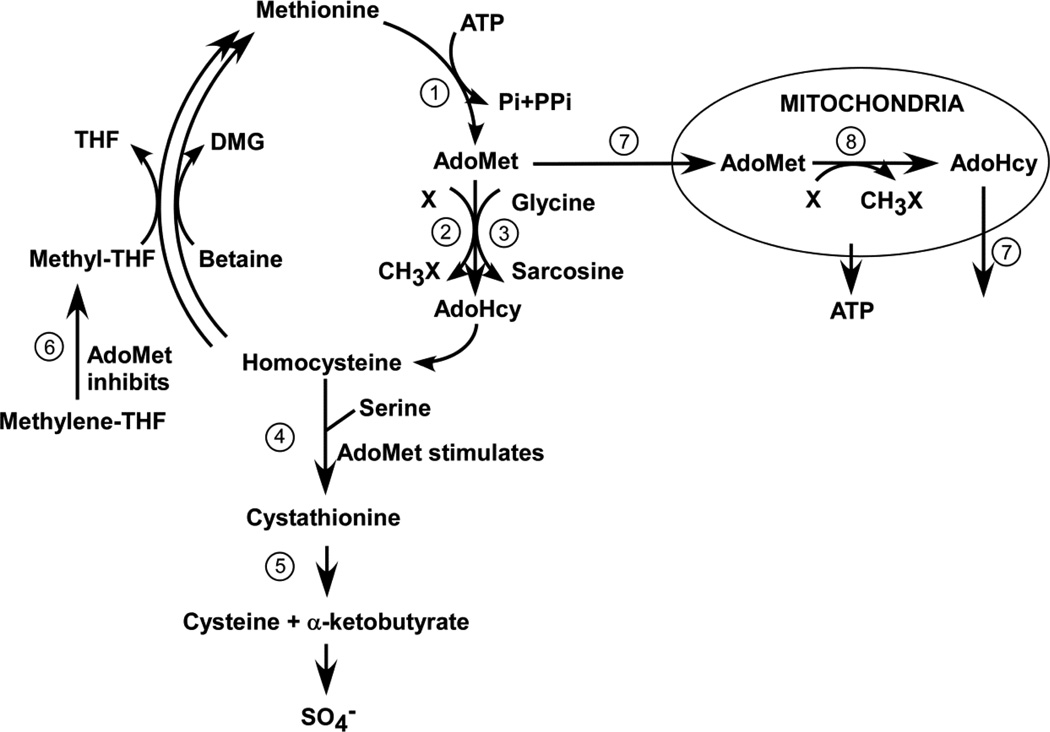
Genetic defects directly involved in the methionine/homocysteine cycle that are presently known to cause hypermethioninemia and/or elevations of AdoMet are the following (see Fig. 3): deficient activity of either methionine adenosyltransferase (E.C. 2.5.1.6) (MAT) I/III, cystathionine beta-synthase (E.C. 4.2.1.22) (CBS), glycine N-methyltranserase (E.C. 2.1.1.20) (GNMT),or S-adenosylhomocysteine hydrolase (E.C. 3.3.1.1) [20]. For both patients 1 and 2, the virtually normal values for tHcy and the cystathionine values above the reference range, ruled out CBS deficiency. The very elevated plasma AdoMet concentrations ruled out deficiency of MAT I/III. The normal concentrations of plasma AdoHcy indicated that S-adenosylhomocysteine hydrolase deficiency was not the cause [21]. Although the elevated AdoMet, together with normal sarcosine and the slightly elevated serum transaminases were consistent with deficiency of GNMT activity [22;23], as described above, sequencing of the GNMT genes from both patients revealed no inactivating mutations.
Figure 3.
Methionine metabolism and its relationship with mitochondrial function. Activities are identified numerically as follows: 1. Methionine adenosyltransferase (MAT (chiefly isozymes MAT I and MAT III in liver, Mat II in non-hepatic tissues; 2. A variety of AdoMet-dependent methyltransferases; 3 Glycine N-methyltransferase (GNMT) (separated in this figure from the other methyltransferases because of its unique role in AdoMet disposal); 4. Cystathionine beta synthase (CBS); 5. Cystathioinine gamma-lyase (CGL). 6. Methylenetetrahydrofolate reductase (MTHFR); 7. The transporter (SAMC) that imports AdoMet into mitochondria in exchange for export of the the counter-substrate, AdoHcy.
Deficiency of citrin, a factor that brings about the exchange of mitochondrial aspartate for glutamate [24] is an additional cause of hypermethioninemia. A compilation of published reports found methionine elevations in 73/112 patients with childhood citrin deficiency [20] and more recently 10/10 elevations have been added [25] to bring the totals to 83/122). The methionine elevations are usually transient in childhood and hypermethioninemia has not been reported in adult citrin deficiency. The citrin genes of both patients 1 and 2 were sequenced, but no mutations were detected.
Deficient activity of fumarylacetoacetate hydrolase (E.C. 3.7.1.2) (tyrosinemia type I) may cause hypermethioninemia [20], but the normal tyrosine or succinylacetone levels in patients 1 and 2 are strong evidence against such a deficiency
Taken together, the above findings rule out any of the known genetic causes of elevation of methionine and/or AdoMet. Liver disease is well-established to be a non-genetic cause of elevated methionine. Because both patient 1 and 2 had elevations of serum transaminases and, in time, more serious liver abnormalities, the possibilities that their abnormal levels of both AdoMet and methionine were due to liver disease merit discussion:
5.1 1. AdoMet
Several lines of evidence indicate that ordinary liver disease is an unlikely explanation of abnormally high levels of this compound: (a) The rate of enzymatic conversion of methionine to AdoMet in livers of cirrhotics was found to be only 38–56% of control activity [26]. (b) Patients with cirrhosis have been reported to have hepatic concentrations of AdoMet of 17.3 ± 2.6 µM, not significantly different from the control level of 17.8 ± 3.1 µM [27]. (c) Plasma AdoMet’s in three cases of hepatitis were 16–77 nM. A single case, a two-week old child, had a slightly high concentration of 180 nM [22]. However we have recently found plasma AdoMet may be elevated to this extent in children without liver disease during the first month or two after birth (unpublished observations). (d) Median serum AdoMet level in 40 individuals with alcoholic liver disease was 120 nM (range 63–328) compared to the median in 28 healthy subjects of 89 nM (72–160) [28]. Thus none of these patients with liver disease had an AdoMet concentration as high as the highest observed in the two present patients with mtDNA mutations.
5.2 Hypermethioninemia
Elevation of methionine has previously been reported in patients with hepatic mtDNA depletion syndromes [2;3]. In published papers that explicitly mention amino acid assays for patients with DGUOK deficiency [2;29–37], 6/22 were found to have elevations of methionine, with plasma concentrations as high as 513 [2], 816 µM [35], or 26 x normal [37] [probably being equal to more than 1100 µM]. Less information is available for MPV17, but among the four such patients reported by Wong and coauthors, one was hypermethioninemic with a concentration 15-fold above the upper limit of the normal range [3], and elevated methionines have been present in the patient reported by Navarro-Sastre et al [38] and in at least three of the Navaho patients studied by S. Holve and his colleagues (personal communication, S. Holve).
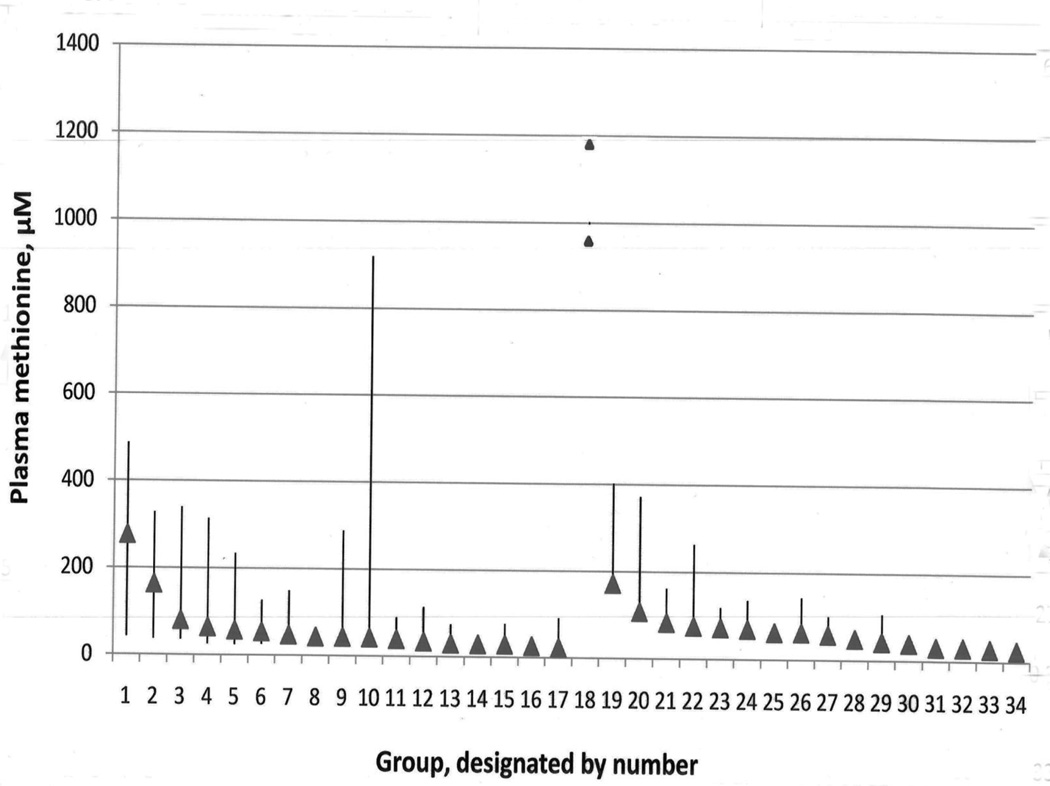
To provide some perspective on the methionine elevations in mitochondrial patients relative to those found in other liver disorders, a search was carried out for publications in which quantitative values had been reported for plasma or serum methionine in patients with liver abnormalities. The results are plotted in Fig. 4, which shows the means and indicates the upper ends of the ranges among 33 groups with hepatic problems ranging from deep hepatic coma to hepatitis or fatty liver. The highest methionine concentrations found in the two patients reported in this paper are shown at interval 18. It is apparent that the elevations of methionine in the 777 cases composing the groups with liver abnormalities were almost always far less than those for patients 1 or 2, and that at least some of the methionine concentrations previously found in mitochondrial patients are also relatively high in comparison.
Figure 4.
Plasma methionine concentrations in the two patients reported in this paper with proven mitochondrial disorders, and in 33 groups of patients with liver disease (taken from previous publications). Peak values for the two mitochondrial disorder patients are indicated by the triangles placed in position 18 between groups 17 and 19 because these patients are not members of any of the 33 groups with liver disease. The reference range for the laboratory in which plasma methionines for these two patients were assayed is 13.3 to 42.7 µM. For each group of liver disease patients the mean is indicated by a triangle. Groups 1–17 are arranged from left to right according to descending values of their means with the vertical bars indicating the overall range for each group. Patient groups 19–34 are similarly arranged by descending means, but the vertical bars represent means + 2 SD because overall ranges for these groups were not reported. A total of 777 patients are included in the combined groups with liver diseases. The patient groups used are the following: 1. Deep hepatic coma [59]; 2. Hepatic precoma [59]; 3. Cryptogenic cirrhosis [60]; 4. Cirrhosis [61]; 5. Chronic active hepatitis with progression to cirrhosis [60]; 6. Viral hepatitis [60]; 7. Primary biliary cirrhosis [60]; 8. Severe cirrhosis, Child-Pugh class B/C[62]; 9. Decompensated cirrhosis [63]; 10. Alcoholic hepatitis with or without cirrhosis [60]; 11. Hepatitis [22]; 12. Alcoholic fatty liver: [60]; 13. Alcoholic hepatitis [64]; 14. Mild cirrhosis, Child-Pugh class A [62]; 15. Alcoholic liver disease [28]; 16. Compensated cirrhosis [63]; 17. Chronic persistent/active hepatitis [63]; 18. Patients 1 and 2 with proven mitochondrial abnormalities. 19. Acute hepatic necrosis/encephalopathy [65]; 20. Extrahepatic biliary atresia and cirrhosis [66] 21. Cirrhosis with coma [67]; 22. Cirrhosis, Child-Pugh C. [67]; 23. Chronic cirrhosis/encephalopathy [65]; 24. Cirrhosis, Child-Pugh B or C [68]; 25. Micronodular alcoholic cirrhosis [69]; 26. Alcoholic cirrhosis [70]; 27. Alcoholic cirrhosis [71]; 28. Alcoholic cirrhosis [72]; 29. Cirrhosis, Child-Pugh B. [67]; 30. Chronic cirrhosis [65] 31. Hepatitis [67]; 32. Cirrhosis, Child-Pugh A [67]; 33. Inactive cirrhosis:[73]; 34. Fatty liver [67].
5.3 Working hypothesis
In summary, the evidence presented and discussed above indicates the elevations of AdoMet and the (sometimes extreme) hypermethioninemias of the mitochondrial patients reported here are not explained by any of the previously recognized causes of these abnormalities. We therefore suggest the following working hypothesis that relates these elevations to the underlying mitochonrial depletion syndromes: A transporter that takes AdoMet from the cytoplasm, where it is synthesized in an ATP-dependent reaction, into mitochondria has been identified [39]. The human protein (SAMC) functions almost exclusively by a counter-exchange mechanism, so that the uptake of AdoMet into mitochondria requires the efflux of a counter-substrate, usually AdoHcy [40] (Fig. 3). Mitochondrial AdoMet comprises about 30% of the total AdoMet in rat hepatocytes, and, although it turns over less rapidly than cytoplasmic AdoMet [41], within the mitochondria there are a variety of AdoMet-dependent methyltransferases that utilize AdoMet as a methyl donor [42]. We suggest that, if the mitochondria are depleted or otherwise unable to utilize the AdoMet they would normally, the AdoMet may accumulate in the cytoplasm and leak out of the cells, explaining the elevation in the plasma. Why is the AdoMet not disposed of by GNMT, as is the case in most situations when it tends to accumulate [19;43]? One possibility is that GNMT, located in the cytosol and strongly inhibited by 5-methyl-THF [19], may be inhibited by 5-methyl-THF not retained by abnormal mitochondria. That possibility is unlikely because practically all the cellular 5-methyl-THF is normally already located in the cytoplasm [44]. A more plausible possibility is based on the tissue distributions of MAT and GNMT expression. A tissue such as skeletal muscle has MAT activity [45] but no detected GNMT protein [46]. Thus, AdoMet may accumulate abnormally in muscle if the mitochondria of that tissue are deleted or non-functional, but will not be disposed of by GNMT, and may leak into the plasma. However such extracellular AdoMet will not have access to GNMT in liver, the tissue with the major amount of GNMT activity [46;47], because hepatic cells do not have a transport system for AdoMet uptake [39]. The plasma AdoMet will then be removed by urinary excretion [48]. Accumulation of AdoMet in muscle will tend also to cause elevation of methionine because, among the MAT isozymes, muscle expresses only MAT II, the form that is highly sensitive to inhibition by AdoMet [49;50]. Even though the renal clearance of AdoMet is close to that of creatinine [48], the leakage of AdoMet from non-hepatic tissues needed to sustain a plasma concentration of 500–1000 nM need be only 0.1–0.2 millimol/day, an amount small relative to the total bodily daily rate of synthesis of AdoMet that ranges in adults from 15.5–23.4 millimol/day [43], so the proposed working hypothesis does not require the normal flux of AdoMet into mitochondria to be a major component of total AdoMet utilization. In presenting the above working hypothesis, we do not mean to imply that methionine elevations in mitochondrial disorders are due solely to leakage from non-hepatic tissues. Liver dysfunction may also play a role, and, when plasma concentrations are not extremely high, may be contributing especially significant proportions.
5.4: Cystathionine elevations in the patients
Mild elevations of plasma cystathionine (as much as 1.9-fold above the upper limit of the reference range) were found in patient 1 (Table 1). Similar mild elevations of plasma cystathionine (up to as high as 1.7-fold) have been reported in patients with severely elevated methionine due to MAT I/III deficiency. Inhibition of cystathionine gamma-lyase (CGL), the enzyme that catabolizes cystathionine, was shown to be a plausible explanation of such elevations. [51]. However, in the sample from patient 2, cystathionine was far more elevated −9.2-fold (Table 2). A possible explanation for this quantitative difference between patients 1 and 2 may be that brain mitochondria were affected in patient 2 (who certainly had CNS abnormalities), but not in patient 1, who remained cognitively normal throughout his life. Human brain has a concentration of cystathionine of 23–57 mg per cent, far above the level of 0.8 mg per cent present in either liver or muscle [52]. This is attributable to the fact that, although CBS activity is present in brain of mice and rats at 3 [53] to 20 % [45] of the specific activities in liver [data for human brain not available], CGL activity is so low in rat, monkey, or mouse brain as to be only marginally measurable (perhaps 0.2 % of the liver activity) or to require especially sensitive methods to detect [45;54;55]. If the DGUOK deficiency in patient 2 led to abnormal elevation of AdoMet in her brain, the AdoMet would tend not only to increase the rate of homocysteine formation by increasing the fluxes through AdoMet-dependent methyltransferases, but also, by virtue both of its inhibition of MTHFR [56], and its stimulation of CBS [57], to cause available homocysteine to be converted to cystathionine rather than being reconverted to methionine (Fig. 3). As a result, cystathionine would accumulate abnormally in brain of patient 2 and leak into her plasma. A similar sequence would not happen in patient 1 if his MPV-17 abnormality did not cause AdoMet to accumulate in his brain tissue.
5.5. Why did the elevations of AdoMet and methionine diminish in patient 1 as he aged?
A remaining uncertainty is why the elevations of both methionine and AdoMet in patient 1 abated as he aged. For this patient, the chief source of AdoMet leakage from non-hepatic cells into the plasma was probably skeletal muscle, a tissue which, although it has a relatively low MAT activity per unit of protein [45], may well contribute significantly to overall AdoMet synthesis because muscle is a major total body component, during childhood amounting to between 22 and 37% of the body weight [58]. Because muscle has no GNMT activity, AdoMet accumulated due to mitochondrial depletion or malfunction will leak out, rather than being utilized by GNMT. Patient 1 was mildly myotonic as early as age 16 months, and toward the end of his life had gross loss of muscle mass and poor muscle tone, so clearly his muscle mitochondria were affected. Loss of muscle mass would perhaps lead to less leakage. An additional (but not mutually exclusive) possibility is that, as his muscle mitochondrial function became worse, their ability to produce ATP became progressively impaired, and the ATP-requiring synthesis of AdoMet diminished. Further studies of patients with progressive involvement of muscle mitochondria will help to resolve these uncertainties
5.6 Future directions
For the moment, the cases reported in this paper suggest that mitochondrial disease might be included in the differential diagnosis of hypermethioninemia, especially when there is an accompanying elevation of plasma AdoMet. Further studies that include assays of plasma AdoMet, methionine, and cystathionine concentrations are needed to ascertain which mitochondrial disorders are most likely to cause these metabolic abnormalities, how often they occur, their extent, and the timing of their appearance. Once further knowledge becomes available, it will be possible to judge whether, and in what situations, such assays will be useful additions to the study of mitochondrial abnormalities.
Highlights.
-
>
We describe two patients with different fatal mitochondrial depletion syndromes.
-
>
Each had marked elevations of plasma S-adenosylmethionine and methionine.
-
>
Evidence is provided that neither had any of the known causes of these elevations.
-
>
An hypothesis is presented relating these abnormalities to mitochondrial disorders.
-
>
Further studies of these metabolites in mitochondrial disorders are needed.
Acknowledgements
We thank the many physicians who participated in the management of these patients or in studies of their metabolic abnormalities, and the patient’s families for their care and consideration. We also thank Ms. Linya Tang for excellent technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Wong L-JC, Naviaux RK, Brunetti-Pierri N, Zhang Q, Schmitt ES, Truong C, Milone M, Cohen BH, Wical B, Ganesh J, Basinger AA, Burton BK, Swoboda K, Gilbert DL, Vanderver A, Saneto RP, Maranda B, Arnold G, Abdenur JE, Waters PJ, Copeland WC. Molecular and clinical genetics of mitochondrial diseases due to POLG mutations. Hum.Mutat. 2008;29:E150–E172. doi: 10.1002/humu.20824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dimmock DP, Zhang Q, Dionisi-Vici C, Carrozzo R, Shieh J, Tang L-Y, Truong C, Schmitt E, Sifry-Platt M, Lucioli S, Santorelli FM, Ficicioglu CH, Rodriguez M, Wierenga K, Enns GM, Longo N, Lipson MH, Vallance H, Craigen WJ, Scaglia F, Wong L-J. Clinical and molecular features of mitochondrial DNA depletion due to mutations in deoxyguanosine kinase. Hum.Mutat. 2007;29:330–345. doi: 10.1002/humu.9519. [DOI] [PubMed] [Google Scholar]
- 3.Wong LJ, Brunetti-Pierri N, Zhang Q, Yazigi N, Bove KE, Dahms BB, Puchowicz MA, Gonzalez-Gomez I, Schmitt ES, Truong CK, Hoppel CL, Chou PC, Wang J, Baldwin EE, Adams D, Leslie N, Boles RG, Kerr DS, Craigen WJ. Mutations in the MPV17 gene are responsible for rapidly progressive liver failure in infancy. Hepatology. 2007;46:1218–1227. doi: 10.1002/hep.21799. [DOI] [PubMed] [Google Scholar]
- 4.El-Hattab AW, Li F-Y, Schmitt E, Zhang S, Craigen WJ, Wong L-JC. MPV17-associated hepatocerebral mitochondrial DNA depletion syndrome: New patients with novel mutations. Mol.Genet.Metab. 2010;99:300–308. doi: 10.1016/j.ymgme.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 5.Tang S, Wang J, Lee N-C, Milone M, Li Y, Halberg M, Schmitt ES, Craigen WJ, Zhang W, Wong L-J. Mitochondrial DNA polymerase gamma mutations: an ever expanding molecular and clinical spectrum. J.Med.Genet. 2011;48:669–681. doi: 10.1136/jmedgenet-2011-100222. [DOI] [PubMed] [Google Scholar]
- 6.Goh V, Helbling D, Biank V, Jarzembowski J, Dimmock D. Next generation sequencing facilitates the diagnosis in a child with Twinkle mutations causing cholestatic liver failure. J.Pediatr Gastroenterol.Nutr. 2011 doi: 10.1097/MPG.0b013e318227e53c. DOI: 10.1097/MPG.0b013e318227e53e. [DOI] [PubMed] [Google Scholar]
- 7.Naviaux RK, Nguyen KV. POLG mutations associated with Alpers’ syndrome and mitochondrial DNA depletion. Ann.Neurol. 2004;55:706–712. doi: 10.1002/ana.20079. [DOI] [PubMed] [Google Scholar]
- 8.Dimmock DP, Dunn JK, Feigenbaum A, Rupar A, Horvath R, Freisinger P, Mousson de, Camaret B, Wong L-J, Scaglia F. Abnormal neurological features predict poor survival and should preclude liver transplantation in patients with deoxyguanosine kinase deficiency. Liver Transplantation. 2008;14:1480–1485. doi: 10.1002/lt.21556. [DOI] [PubMed] [Google Scholar]
- 9.Lutz RE, Dimmock D, Schmitt ES, Zhang Q, Tang L-Y, Reyes C, Truemper E, McComb RD, Hernandez A, Basinger A, Wong L-JC. De novo mutations in POLG presenting with acute liver failure or encephalopathy. J.Pediatr Gastroenterol.Nutr. 2009;49:125–129. doi: 10.1097/MPG.0b013e31817d9cad. [DOI] [PubMed] [Google Scholar]
- 10.Shieh JTC, Berquist WE, Zhang Q, Chou P-C, Wong L-JC, Enns GM. Novel deoxyguanosine kinase gene mutations and viral infection predispose apparently healthy children to fulminant liver failure. J.Pediatr.Gastroenterol.Nutr. 2009;49:130–132. doi: 10.1097/MPG.0b013e31819de7a6. [DOI] [PubMed] [Google Scholar]
- 11.Capdevila A, Wagner C. Measurement of plasma S-adenosylmethionine and S-adenosylhomocysteine as their fluorescent isoindoles. Analyt.Biochem. 1998;264:180–184. doi: 10.1006/abio.1998.2839. [DOI] [PubMed] [Google Scholar]
- 12.Allen RH, Stabler SP, Lindenbaum J. Serum betaine N,N-dimethylglycine and N-methylglycine levels in patients with cobalamin and folate deficiency and related inborn errors of metabolism. Metabolism. 1993;42:1448–1460. doi: 10.1016/0026-0495(93)90198-w. [DOI] [PubMed] [Google Scholar]
- 13.Stabler SP, Marcell PD, Podell ER, Allen RH. Quantitation of total homocysteine total cysteine and methionine in normal serum and urine using capillary gas chromatography-mass spectrometry. Analyt.Biochem. 1987;162:185–196. doi: 10.1016/0003-2697(87)90026-1. [DOI] [PubMed] [Google Scholar]
- 14.Stabler SP, Lindenbaum J, Savage DG, Allen RH. Elevation of serum cystathionine levels in patients with cobalamin and folate deficiency. Blood. 1993;81:3404–3413. [PubMed] [Google Scholar]
- 15.Cook RJ, Wagner C. Glycine N-methyltransferase is a folate-binding protein of rat liver cytosol. Proc.Natl.Acad.Sci.USA. 1984;81:3631–3634. doi: 10.1073/pnas.81.12.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Venegas V, Wang J, Dimmock D, Wong LJ. Real-time quantitative PCR analysis of mitochondrial DNA content. Curr.Protoc.Hum.Genet. 2011;68:19.7.1–19.7.12. doi: 10.1002/0471142905.hg1907s68. [DOI] [PubMed] [Google Scholar]
- 17.Dimmock D, Tang L-Y, Schmitt ES, Wong L-JC. Quantitative evaluation of the mitochondrial DNA depletion syndrome. Clin.Chem. 2010;56:1119–1127. doi: 10.1373/clinchem.2009.141549. [DOI] [PubMed] [Google Scholar]
- 18.Wong L-JC, Dimmock D, Geraghty MT, Quan R, Lichter-Konecki U, Wang J, Brundage EK, Scaglia F, Chinault AC. Utility of oligonucleotide array-based comparative genomic hybridization for detection of target gene deletions. Clin.Chem. 2008;54:1141–1148. doi: 10.1373/clinchem.2008.103721. [DOI] [PubMed] [Google Scholar]
- 19.Luka Z, Mudd SH, Wagner C. Glycine N-methyltransferase and regulation of S-adenosylmethionine levels. J.Biol.Chem. 2009;284:22507–22511. doi: 10.1074/jbc.R109.019273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mudd SH. Hypermethioninemias of genetic and non-genetic origin: a review. Am.J.Med.Genet.Part C. 2011;157:3–32. doi: 10.1002/ajmg.c.30293. [DOI] [PubMed] [Google Scholar]
- 21.Baric I, Fumic K, Glenn B, Cuk M, Schulze A, Finkelstein JD, James SJ, Mejaski-Bosnjak V, Pazanin L, Pogribny IP, Rados M, Sarnavka V, Scukanec-Spoljar M, Allen RH, Stabler S, Uzelac L, Vugrek O, Wagner C, Zeisel S, Mudd SH. S-Adenosylhomocysteine hydrolase deficiency in a human: A genetic disorder of methionine metabolism. Proc.Natl.Acad.Sci.USA. 2004;101:4234–4239. doi: 10.1073/pnas.0400658101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mudd SH, Cerone R, Schiaffino MC, Fantasia AR, Minniti G, Caruso U, Lorini R, Watkins D, Matiaszuk N, Rosenblatt D, Schwahn B, Rozen R, LeGros L, Kotb M, Capdevila A, Luka Z, Finkelstein JD, Tangerman A, Stabler SP, Allen RH, Wagner C. Glycine N-methyltransferase deficiency: a novel inborn error causing persistent isolated hypermethioninemia. J.Inherit.Metab.Dis. 2001;24:448–464. doi: 10.1023/a:1010577512912. [DOI] [PubMed] [Google Scholar]
- 23.Luka Z, Cerone R, Phillips JA, III, Mudd SH, Wagner C. Mutations in glycine N-methyltransferase give insight into its role in methionine metabolism. Hum.Genet. 2002;110:68–72. doi: 10.1007/s00439-001-0648-4. [DOI] [PubMed] [Google Scholar]
- 24.Palmieri L, Pardo B, Lasorsa FM, del Arco A, Kobayashi K, Iijima M, Runswick MJ, Walker JE, Saheki T, Satrustegui J, Palmieri F. Citrin and aralar1 are Ca(2+)-stimulated aspartate/glutamate transporters in mitochondria. EMBO J. 2001;20:5060–5069. doi: 10.1093/emboj/20.18.5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chew HB, Ngu LH, Zabedah MY, Keng WT, Balasubramaniam S, Hanifah MJM, Kobayashi K. Neonatal intrahepatic cholestasis associated with citrin deficiency (NICCD): a case series of 11 Malaysian patients. J.Inherit.Metab.Dis. 2010 doi: 10.1007/s10545-010-9248-6. DOI 10.1007/s10545-010-9248-6. [DOI] [PubMed] [Google Scholar]
- 26.Martin Duce A, Ortiz P, Cabrero C, Mato JM. S-Adenosyl-L-methionine synthetase and phospholipid methyltransferase are inhibited in human cirrhosis. Hepatology. 1988;8:65–68. doi: 10.1002/hep.1840080113. [DOI] [PubMed] [Google Scholar]
- 27.Cabrero C, Martin-Duce A, Ortiz P, Alemany S, Mato JM. Specific loss of the high-Mr form of S-adenosyl-L-methionine synthetase in human liver cirrhosis. Hepatology. 1988;8:1530–1534. doi: 10.1002/hep.1840080610. [DOI] [PubMed] [Google Scholar]
- 28.Medici V, Peerson JM, Stabler SP, French SW, Gregory JF, III, Virata MC, Albanese A, Bowlus CL, Devaraj S, Panacek EA, Rahim N, Richards JR, Rossaro L, Halsted CH. Impaired homocysteine transsulfuration is an indicator of alcoholic liver disease. J.Hepatol. 2010;53:551–557. doi: 10.1016/j.jhep.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Müller-Höcker J, Muntau A, Schäfer S, Jaksch M, Staudt F, Pongratz D, Taanman J-W. Depletion of mitochondrial DNA in the liver of an infant with neonatal giant cell hepatitis. Hum.Pathol. 2002;33:247–253. doi: 10.1053/hupa.2002.31477. [DOI] [PubMed] [Google Scholar]
- 30.Tadiboyina VT, Rupar A, Atkison P, Feigenbaum A, Kronick J, Wang J, Hegele RA. Novel mutation in DGUOK in hepatocerebral mitochondrial DNA depletion syndrome associated with cystathioninuria. Am.J.Med.Genet.A. 2005;135:289–291. doi: 10.1002/ajmg.a.30748. [DOI] [PubMed] [Google Scholar]
- 31.Freisinger P, Futterer N, Lankes E, Gempel K, Berger TM, Spalinger J, Hoerbe A, Schwantes C, Lindner M, Santer R, Burdelski M, Schaefer H, Setzer B, Walker UA, Horvath R. Hepatocerebral mitochondrial DNA depletion syndrome caused by deoxyguanosine kinase (DGUOK) mutations. Arch.Neurol. 2006;63:1129–1134. doi: 10.1001/archneur.63.8.1129. [DOI] [PubMed] [Google Scholar]
- 32.Lee N-C, Dimmock D, Hwu W-L, Tang L-Y, Huang W-C, Chinault AC, Wong L-JC. Simultaneous detection of mitochondrial DNA depletion and single-exon deletion in the deoxyguanosine gene using array-based comparative genomic hybridisation. Arch.Dis.Child. 2009;94:55–58. doi: 10.1136/adc.2008.139584. [DOI] [PubMed] [Google Scholar]
- 33.Ji JQ, Dimmock D, Tang L-Y, Descartes M, Gomez R, Rutledge SL, Schmitt ES, Wong LJ. A novel c.592-4_c.592-3delTT mutation in DGUOK gene causes exon skipping. Mitochondrion. 2010;10:188–191. doi: 10.1016/j.mito.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 34.Pronicka E, Weglewska-Jurkiewicz A, Taybert J, Pronicki M, Szymanska-Debinska T, Karkucinska-Wieckowska A, Jakóbkiewicz-Banecka J, Kowalski P, Piekutowska-Abramczuk D, Pajdowska M, Socha P, Sykut-Cegielska J, Wegrzyn G. Post mortem identification of deoxyguanosine kinase (DGUOK) gene mutations combined with impaired glucose homeostasis and iron overload features in four infants with severe progressive liver failure. J.Appl.Genetics. 2010 doi: 10.1007/s13353-010-0008-y. doi 10.1007/s13353-010-0008-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hanchard NA, Shchelochkov OA, Roy A, Wiszniewska J, Wang J, Popek EJ, Karpen S, Wong L-JC, Scaglia F. Deoxyguanosine kinase deficiency presenting as neonatal hemochromatosis. Mol.Genet.Metab. 2011;103:262–267. doi: 10.1016/j.ymgme.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 36.Kiliç M, Sivri HS, Dursun A, Tokatli A, De Meirleir L, Seneca S, Akçören Z, Yigit S, Topaloglu H, Coskun T. A novel mutation in the DGUOK gene in a Turkish newborn with mitochondrial depletion syndrome. Turkish J.Pediatr. 2011;53:79–82. [PubMed] [Google Scholar]
- 37.Brahimi N, Jambou M, Sarzi E, Serre V, Boddaert N, Romano S, de Lonlay P, Slama A, Munnich A, Rötig A, Bonnefont JP, Lebre AS. The first founder DGUOK mutation associated with hepatocerebral miotochondrial DNA depletion syndrome. Mol.Genet.Metab. 2011;97:221–226. doi: 10.1016/j.ymgme.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 38.Navarro-Sastre A, Martín-Hernández E, Campos Y, Quintana E, Medina E, Simón de las Heras R, Lluch M, Muñoz A, del Hoyo P, Martín R, Gort L, Briones P, Ribes A. Lethal hepatopathy and leukodystrophy caused by a novel mutation in MPV17 gene: Description of an alternative MPV17 spliced form. Mol.Genet.Metab. 2008;94:234–239. doi: 10.1016/j.ymgme.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 39.Horne DW, Holloway RS, Wagner C. Transport of S-adenosylmethionine in isolated rat liver mitochondria. Arch.Biochem.Biophys. 1997;343:201–206. doi: 10.1006/abbi.1997.0167. [DOI] [PubMed] [Google Scholar]
- 40.Agrimi G, Di Noia MA, Marobbio CMT, Fiermonte G, Lasorsa FM, Palmieri F. Identification of the human mitochondrial S-adenosylmethionine transporter: bacterial expression reconstitution, functional characterization and tissue distribution. Biochem.J. 2004;379:183–190. doi: 10.1042/BJ20031664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Farooqui JZ, Lee HW, Kim S, Paik WK. Studies on compartmentation of S-adenosyl-L-methionine in Saccharomyces cerevisiae and isolated rat hepatocytes. Biochim.Biophys.Acta. 1983;757:342–351. [PubMed] [Google Scholar]
- 42.Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong S-E, Walford GA, Sugiana C, Boneh A, Chen WK, Hill DE, Vidal M, Evans JG, Thorburn DR, Carr SA, Mootha VK. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2011;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mudd SH, Brosnan JT, Brosnan ME, Jacobs RL, Stabler SP, Vance DE, Wagner C. Methyl balance and transmethylation fluxes in humans. Am.J.Clin.Nutr. 2007;85:19–25. doi: 10.1093/ajcn/85.1.19. [DOI] [PubMed] [Google Scholar]
- 44.Shane B. Folate chemistry and metabolism. In: Bailey LB, editor. Folate in Health and Disease. New York: Marcel Dekker, Inc; 1995. pp. 1–22. [Google Scholar]
- 45.Mudd SH, Finkelstein JD, Irreverre F, Laster L. Transsulfuration in mammals: Microassays and tissue distributions of three enzymes of the pathway. J.Biol.Chem. 1965;240:4382–4392. [PubMed] [Google Scholar]
- 46.Cook RJ, Wagner C. Measurement of a folate binding protein from rat liver cytosol by radioimmunoassay. Arch.Biochem.Biophys. 1981;208:358–364. doi: 10.1016/0003-9861(81)90520-8. [DOI] [PubMed] [Google Scholar]
- 47.Kerr SJ. Competing methyltransferase systems. J.Biol.Chem. 1972;247:4248–4252. [PubMed] [Google Scholar]
- 48.Stabler SP, Allen RH. Quantification of serum and urinary S-adenosylmethionine and S-adenosylhomocysteine by stable-isotope-dilution liquid chromatography-mass spectrometry. Clin.Chem. 2004;50:365–372. doi: 10.1373/clinchem.2003.026252. [DOI] [PubMed] [Google Scholar]
- 49.Kotb M, Geller AM. Methionine adenosyltransferase: structure and function. Pharmacol.Ther. 1993;59:125–143. doi: 10.1016/0163-7258(93)90042-c. [DOI] [PubMed] [Google Scholar]
- 50.Kotb M, Mudd SH, Mato JM, Geller AM, Kredich NM, Chou JY, Cantoni GL. Consensus nomenclature for the mammalian methionine adenosyltransferase genes and gene products. Trends Genet. 1997;13:51–52. doi: 10.1016/s0168-9525(97)01013-5. [DOI] [PubMed] [Google Scholar]
- 51.Stabler SP, Steegborn C, Wahl MC, Oliveriusova J, Kraus JP, Allen RH, Wagner C, Mudd SH. Elevated plasma total homocysteine in severe MAT I/III deficiency. Metabolism. 2002;51:981–988. doi: 10.1053/meta.2002.34017. [DOI] [PubMed] [Google Scholar]
- 52.Tallan HH, Moore S, Stein WH. L-Cystathionine in human brain. J.Biol.Chem. 1958;230:707–716. [PubMed] [Google Scholar]
- 53.Vitvitsky V, Dayal S, Stabler S, Zhou Y, Wang H, Lentz SR, Banerjee R. Perturbations in homocysteine-linked redox homeostasis in a murine model for hyperhomocysteinemia. Am.J.Physiol.Regulatory Integrative Comp.Physiol. 2011;287:R39–R46. doi: 10.1152/ajpregu.00036.2004. [DOI] [PubMed] [Google Scholar]
- 54.Sturman JA, Gaull GE, Niemann WH. Cystathionine synthesis degradation in brainliver and kidney of the developing monkey. J.Neurochem. 2011;26:457–463. doi: 10.1111/j.1471-4159.1976.tb01496.x. [DOI] [PubMed] [Google Scholar]
- 55.Vitvitsky V, Thomas M, Ghorpade A, Gendelman HE, Banerjee R. A functional transsuulfuration pathway in the brain links to glutathione homeostasis. J.Biol.Chem. 2006;281:35785–35793. doi: 10.1074/jbc.M602799200. [DOI] [PubMed] [Google Scholar]
- 56.Kutzbach C, Stokstad ELR. Feedback inhibition of methylene-tetrahydrofolate reductase in rat liver by S-adenosylmethionine. Biochim.Biophys.Acta. 1967;139:217–220. doi: 10.1016/0005-2744(67)90140-4. [DOI] [PubMed] [Google Scholar]
- 57.Finkelstein JD, Kyle WE, Martin JJ, Pick A-M. Activation of cystathionine synthase by adenosylmethionine and adenosylethionine. Biochem.Biophys.Res.Commun. 1975;66:81–87. doi: 10.1016/s0006-291x(75)80297-x. [DOI] [PubMed] [Google Scholar]
- 58.Holliday MA. Body composition and energy needs during growth. In: Falkner F, Tanner JM, editors. Human Growth: a Comprehensive Treatise. Volume 2. New York: Plenum Press; 1986. pp. 101–117. [Google Scholar]
- 59.Wu C, Bollman JL, Butt HR. Changes in free amino acids in the plasma during hepatic coma. J.Clin.Invest. 1955;34:845–849. doi: 10.1172/JCI103139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Morgan MY, Marshall AW, Milsom JP, Sherlock S. Plasma amino-acid patterns in liver disease. Gut. 1982;23:362–370. doi: 10.1136/gut.23.5.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Horowitz JH, Rypins EB, Henderson JM, Heymsfield SB, Moffitt SD, Bain RP, Chawla RK, Bleier JC, Rudman D. Evidence for impairment of transsulfuration pathway in cirrhosis. Gastroenterology. 1981;81:668–675. [PubMed] [Google Scholar]
- 62.Look MP, Riezler R, Reichel C, Brensing KA, Rockstroh JK, Stabler SP, Spengler U, Berthold HK, Sauerbruch T. Is the increase in serum cystathionine levels in patients with liver cirrhosis a consequence of impaired homocysteine transsulfuration at the level of g-cystathionase? Scand.J.Gastroenterol. 2000;35:866–872. doi: 10.1080/003655200750023255. [DOI] [PubMed] [Google Scholar]
- 63.Almasio P, Bianchi G, Marchesini G, Luca A, Bugianesi E, Le Grazie C, Pagliaro L. Sulphur amino acid pattern in chronic liver disease. Ital.J.Gastroenterol. 1994;26:21–25. [PubMed] [Google Scholar]
- 64.Ning M, Lowenstein LM, Davidson CS. Serum amino acid concentrations in alcoholic hepatitis. J.Lab.Clin.Med. 1967;70:554–562. [PubMed] [Google Scholar]
- 65.Rosen HM, Yoshimura N, Hodgman JM, Fischer JE. Plasma amino acid patterns in hepatic encephalopathy of differing etiology. Gastroenterology. 1977;72:483–487. [PubMed] [Google Scholar]
- 66.Weisdorf SA, Freese DK, Fath JJ, Tsai MY, Cerra FB. Amino acid abnormalities in infants with extrahepatic biliary atresia and cirrhosis. J.Pediatr Gastroenterol.Nutr. 1987;6:860–864. doi: 10.1097/00005176-198711000-00007. [DOI] [PubMed] [Google Scholar]
- 67.Bosy-Westphal A, Ruschmeyer M, Czech N, Oehler G, Hinrichsen H, Plauth M, Lotterer E, Cascino A, Cangiano C, Calcaterra V, Rossi-Fanelli F, Capocaccia L. Plasma amino acids imbalance in patients with liver disease. Digestive Diseases. 1978;23:591–598. doi: 10.1007/BF01072593. [DOI] [PubMed] [Google Scholar]
- 68.Marchesini G, Bugianesi E, Bianchi G, Fabbri A, Marchi E, Zoli M, Pisi E. Defective methionine metabolism in cirrhosis: relation to severity of liver disease. Hepatology. 1992;16:149–155. doi: 10.1002/hep.1840160125. [DOI] [PubMed] [Google Scholar]
- 69.Tribble DL, Jones DP, Ardehali A, Feeley RM, Rudman D. Hypercysteinemia and delayed sulfur excretion in cirrhotics after oral cysteine loads. Am.J.Clin.Nutr. 1989;50:1401–1406. doi: 10.1093/ajcn/50.6.1401. [DOI] [PubMed] [Google Scholar]
- 70.Marchesini G, Bianchi GP, Vilstrup H, Checchia GA, Patrono D, Zoli M. Plasma clearances of branched-chain amino acids in control subjects and in patients with cirrhosis. J.Hepatol. 1987;4:108–117. doi: 10.1016/s0168-8278(87)80017-x. [DOI] [PubMed] [Google Scholar]
- 71.Chawla RK, Berry CJ, Kutner MH, Rudman D. Plasma concentrations of transsulfuration pathway products during nasoenteral and intravenous hyperalimentation of malnourished patients. Am.J.Clin.Nutr. 1985;42:577–584. doi: 10.1093/ajcn/42.4.577. [DOI] [PubMed] [Google Scholar]
- 72.Russmann S, Junker E, Lauterburg BH. Remethylation and transsulfuration of methionine in cirrhosis: studies with L-[2H3-methyl-1-13C]methionine. Hepatology. 2002;36:1190–1196. doi: 10.1053/jhep.2002.36499. [DOI] [PubMed] [Google Scholar]
- 73.Pisi E, Marchesini G. Mechanisms and consequences of the impaired trans-sulphuration pathway in liver disease: Part II. Clinical consequences and potential for pharmacological intervention in cirrhosis. Drugs. 1990;40 Suppl. 3:65–72. doi: 10.2165/00003495-199000403-00007. [DOI] [PubMed] [Google Scholar]