Abstract
Aberrations in IL-3, GM-CSF and G-CSF induced signaling are frequently reported in acute myeloid leukemia (AML). Herein, we utilized a unique human myeloid leukemic cell line, AML-193, which responds to all three cytokines to analyze the regulation at microRNA level. Using real-time PCR-based miRNA expression profiling, we investigated miRNA signatures regulated by IL-3, GM-CSF and G-CSF for n=704 miRNAs. We discovered that in addition to regulating specific miRNAs, these cytokines also regulate common set of miRNAs, which includes miR-590-5p, miR-219-5p, miR-15b and miR-628-5p. Taken together, we have identified novel candidate miRNAs that may be instructive during leukemic and normal hematopoiesis.
Keywords: AML-193, AML, miRNA, IL-3, GM-CSF, G-CSF
Introduction
Hematopoiesis is mainly regulated by signaling cascades that are activated by a variety of cytokines 1. IL-3, GM-CSF and G-CSF are predominant regulators for growth and differentiation of myeloid progenitors 2–4. Interestingly, they all signal via a common JAK2-STAT5 pathway in myeloid progenitor compartments 5–8. However, the mechanism through which STAT5 responds differentially to early-acting and lineage restricted cytokines, particularly in leukemic and stem/progenitor cells, is unknown. Previous studies investigated STAT5 response/activity to a mixture of factors, precluding analysis of pathways initiated by specific cytokines.
AML-193 is one of eight cell lines established from 50 patients with childhood acute leukemia 9,10. Of those 8 lines, AML-193, which was derived from a 13-year-old female with AML, was the only myeloid leukemic cell line that required conditioned media (from 5637 cell line) to grow and proliferate. Subsequently the active factors were identified as GM-CSF, G-CSF and Hematopoietin-I, which later came to be known as IL-19. IL-3, a cytokine that supports the growth of hematopoietic stem cells, is also known to support the growth of AML-193 cells11. In addition, response to IL-3 confirms the expression of functional CD123 (alpha subunit of IL-3 receptor), which is known to be strongly expressed on leukemic stem cells (LSCs) and leukemic blasts 12. Furthermore, AML-193 cells co-express both surface CD135 (FLT3 receptor) and FLT3 ligand 13, a well-established marker for leukemic and hematopoietic stem cells 14–16.
MicroRNAs (miRNAs), a class of naturally occurring noncoding RNAs of 17 to 30 nucleotides, are known to regulate gene expression by inducing translational inhibition and cleavage of their target mRNAs 17. miRNAs play several crucial roles during hematopoiesis that include lineage decisions, stem cell progenitor transitions, niche control and other cell functions 18–20.
Recent investigations have linked miRNA expression with leukemia. The first miR that was discovered to have a role in myelopoiesis was miR-223, which was regulated by CEBPa transcription factor and was shown to target MEF2c transcription factor, a known promoter of myeloid progenitor differentiation 21. Recently, miR-223 has been reported to target cell cycle regulator E2F1 and regulate cell cycle progression and myeloid proliferation 22. Interestingly, trancriptional repressor GFI1, required for normal granulopoiesis, has been shown to negatively regulate the expression of miR-21 and miR-196b. Retinoic acid (RA/ATRA) induces the expression of miR-15a/−16-1 and several let-7 family members which target oncogenes BCL2 and RAS; and regulate RA induced differentiation in acute promyelocytic leukemia (APL) cells 23,24. Baltimore and his colleagues showed that overexpression of human miR-125b caused a dose dependent myeloproliferative disorder that progressed to lethal myeloid leukemia in mice and also enhanced hematopoietic engraftment 18, which again reveals their potential to regulate hematopoietic output. A cluster of two miRNAs, miR-15a and miR-16-1, located at 13q14 was discovered to be deleted or down-regulated in approximately 60% of chronic lymphocytic leukemia (CLL) samples 25. Furthermore, a recent study evaluated miRNA expression in 122 untreated adult acute myeloid leukemia (AML) cases using a microarray platform. AML patients with high expression of miR-191 and miR-199a had significantly worse overall and event-free survival than AML patients with low expression 26. Taken together, miRNAs can either act as tumor suppressors or as oncogenes and their altered levels can have functional relevance in leukemogenesis. Either single miRNA or signatures have demonstrated prognostic utility and complements the diagnostic information derived from cytogenetics, gene mutations and altered gene expressions.
Despite increasing evidence demonstrating a critical role for JAK2-STAT5 in leukemic and normal hematopoiesis, the distinct nature of signaling response regulated by early-acting (IL-3) and lineage-restricted cytokines (GM-CSF and G-CSF) is largely unresolved. We hypothesized that a unique response of leukemic myeloid progenitors to IL-3, GM-CSF, and G-CSF are possibly mediated in part by distinct regulation at the miRNA level. Therefore by utilizing a unique leukemic myeloid cell line, AML-193, that responds to both early and late acting cytokines, we attempted to profile IL-3, GM-CSF and G-CSF regulated miRNA signatures in leukemic myeloid progenitors and identify novel candidate miRNAs that may be instructive during leukemic and normal hematopoiesis.
Materials and methods
Cell culture
AML-193 cells were cultured in Iscove’s Modified Dulbecco’s Medium (IMDM) with -CSF, 5ng/mL IL-3, or 5ng/mL G-CSF.
RT-qPCR for STAT5 response factors
In analyses of bona fide STAT5 response factors, AML-193 cells were cultured for 16 hours in IMDM starvation media and were then exposed to either IL-3 (5ng/mL), GM-CSF (10ng/mL), or G-CSF (5ng/mL) for 90 minutes. RNA was extracted using an RNeasy Mini Kit (Qiagen) and then reverse transcription (QScript cDNA Synthesis Kit, cat # 95047-100, Quanta) and real-time quantitative polymerase chain reactions (iQ SYBR Green, i-Cycler, Bio-Rad) were performed. qPCR primer pairs were obtained for Pim1 (NM_002648), Cish (NM_145071) and Gapdh (NM_002046) from SA Biosciences.
microRNA profiling and analyses
Post IL-3/GM-CSF/G-CSF treatment of AML-193 cells, miRNA was isolated using miRNeasy Mini Kit and RNeasy MinElute Cleanup Kit (cat # 217004 and 74204, Qiagen) to obtain high quality qPCR grade total RNA and microRNA. 400 ng of enriched miRNA per sample was used to perform reverse transcription to generate cDNAs (RT miRNA First Strand Kit, cat # 331401, SA Biosciences). The complete Human V2.0 miRNA Genome Array representing a total of n=704 miRNAs in 96-well format (MAH-100 and -200, SA Biosciences) was used to perform profiling. Four housekeeping small nuclear RNA (SNORD 48, 47, 44 and RNU6) were used for normalization. qPCR was performed using 2X SYBR Green master mix (SYBR Green/Fluorescein, cat # 330513, SA Biosciences) on an IQ5 cycler (BioRad). The raw Ct values obtained were loaded onto an online miRNA PCR Array Data Analysis web portal system, http://pcrdataanalysis.sabiosciences.com/pcr/arrayanalysis.php, developed by SA Biosciences to retrieve fold change analysis using delta delta Ct method. For filtering criteria, greater than (>) 35 cycle threshold (Ct) scores were considered to be nonspecific and miRNAs that showed a raw Ct score > 35 were excluded from the final data analysis. Fold change filters were applied to select the miRNAs that were regulated more than 2-fold.
Results and Discussion
The process of hematopoiesis that involves HSC self-renewal and lineage commitment decisions is tightly regulated by extrinsic (bone marrow microenvironment, cell adhesion molecules, growth factors and cytokines) and intrinsic factors (signaling networks and transcription factors) 1. miRNAs bring in another regulatory dimension to hematopoesis by selectively targeting signaling molecules, receptors, ligands and transcription factors and thus influence major outcomes related to HSC self renewal, differentiation, apoptosis and balance of myeloid/lymphoid compartments within the hematopoietic progenitor pool 27. Importantly, miRNAs also contribute to oncogenesis, where an miRNA that targets oncogenes acts as a tumor suppressor and therefore decreased expression of such miRNAs contribute towards promoting tumorogenesis and comparably, up regulation of miRNAs that target tumor suppressors will also lead to oncogenesis 28,29. Recently, dysregulation of miRNAs has been shown in leukemias where its genomic locations have been linked to breakpoint regions associated with chromosome aberrations and regions of amplification and loss of heterozygosity 30. Importantly, aberrations in IL-3, GM-CSF, and G-CSF induced signaling are frequently reported in acute myeloid leukemia (AML) 31,32. Here, we present outcomes from an miRNA profiling study that utilized a unique leukemic myeloid cell line, AML-193, which responds to both early and late acting cytokines, to investigate distinct miRNA signatures regulated by IL-3, GM-CSF and G-CSF in leukemic myeloid progenitors.
In order to assess the Jak2-STAT5 responsiveness of various cytokines, AML-193 cells were cultured in the absence of cytokines for 16 hours before being treated with either IL-3 (5ng/mL), GM-CSF (50ng/mL) or G-CSF (50ng/mL) for 90 minutes. RNA was isolated and qPCR was performed on two bona fide STAT5 response genes Pim1 and Cish 33–35. The data was normalized using GAPDH. All three cytokines induced significant increases in Pim1 and Cish transcripts (Figure 1B). Though the oncogenic role of JAK2-STAT5 is mediated through direct involvement in the up-regulation of genes such as cyclin D, bcl-2, mcl-1, bcl-XL, and myc 36, the specific mechanisms by which JAK2-STAT5 critically controls preleukemic expansion in the myeloid lineage have not been defined. Identifying cytokine specific Jak2-STAT5-regulated miRNAs that govern this transition will provide candidate regulators for functional testing. Further, study of the distinct IL-3/GM-CSF/G-CSF induced miRNA signatures accompanying progenitor and lineage restricted stages will provide important mechanistic cues regarding the complexity of the regulatory network.
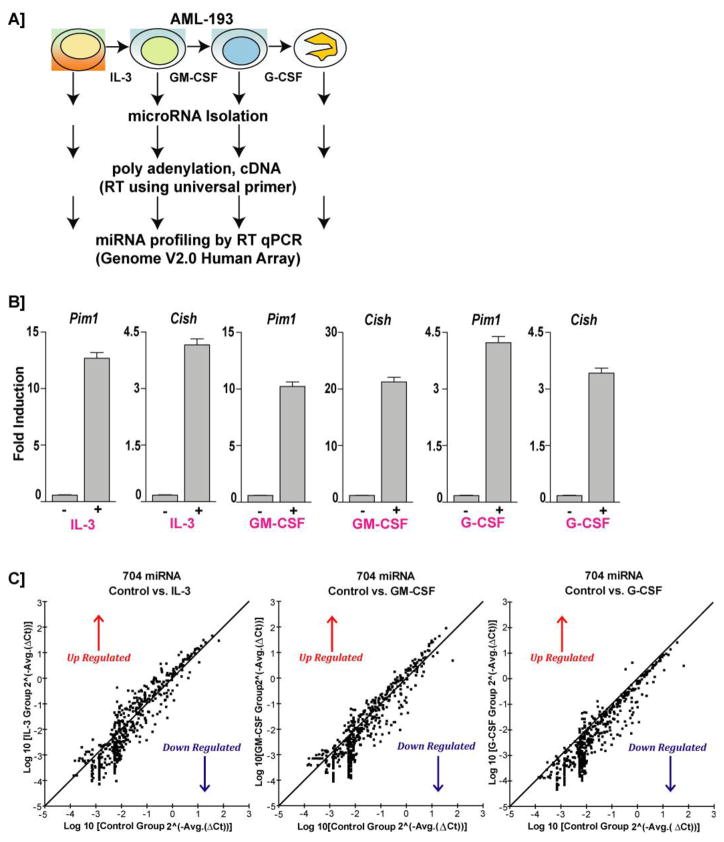
Figure 1. AML-193 as a model system for testing IL-3, GM-CSF, and G-CSF induced miRNA profiling via potential JAK2-STAT5 pathway.
A. Schematic diagram of overall experimental strategies for IL-3, GM-CSF and G-CSF induced miRNA profiling in AML-193 cells. B. In order to confirm the JAK2-STAT5 responsiveness to cytokines, AML-193 cells were cultured under growth factor deprivation conditions for 16 hours and were treated with either IL-3 (5ng/mL) or GM-CSF (50ng/mL) or G-CSF (50ng/mL) for 90 minutes. RNA was isolated and RTqPCR was performed on two bona fide STAT5 response genes Pim1 and Cish. The data was normalized using GAPDH. Data are representative of two independent experiments performed in triplicate (mean +/− SEM). C. Scatter plot showing the comparison of IL-3, GM-CSF and G-CSF induced miRNA expression profiling in AML-193 cells. Fold change filters were applied to select the miRNAs that were regulated more than 2-fold.
For miRNA profiling, AML-193 cells were initially exposed to IL-3 for 3 days followed by GM-CSF for 3 more days and subsequently to G-CSF for 3 days. We then profiled miRNA expression induced by IL-3, GM-CSF and G-CSF in AML-193 cells by treating the respective cohorts post growth factor deprivation with corresponding cytokines (Figure 1A). Using the complete Human V2.0 miRNA Genome Array platform representing a total of n=704 miRNAs, we analyzed the cytokine specific expression of miRNAs in AML-193 early myeloid leukemic cells. To further specify our search for miRNAs differentially induced by cytokines in AML-193, we added an additional filter. We only considered miRNAs that were at least 2-fold up- or down-regulated in their expression compared to untreated control. In addition, miRNAs having a Ct value of >35, which are likely due to background noise, were excluded from the analysis. Frequencies of false positives were avoided using these stringent filters. Using these empirically determined thresholds, we identified 90 miRNAs were up regulated by IL-3 and 339 miRNAs were down regulated (Figure 1C and 2B). GM-CSF elevated the expression of 65 miRNAs and decreased expression of 358 miRNAs (Figure 1C and 2B). G-CSF up regulated 10 miRNAs and down regulated 435 miRNAs (Figure 1C and 2B). Interestingly, this list of highly down regulated miRNAs includes Let-7a and miR-16. Tumor suppressors like Let-7a and miR-16 have been shown to target multiple oncogenes such as RAS, HMGA2 and BCL2, MCL1, CCND1, WNT3A respectively 25,37,38. Significantly, down regulation of Let-7a and miR-16 have been reported in chronic lymphocytic leukemia (CLL) 39 and in addition, decreased levels of miR-16 has been found in blasts isolated from high-risk MDS (myelodysplastic syndrome) patients 40. Therefore, it is possible that in AML associated with skewed IL-3 or GM-CSF signaling may result in the down regulation of these miRs and promote a proliferative phenotype for the blasts.
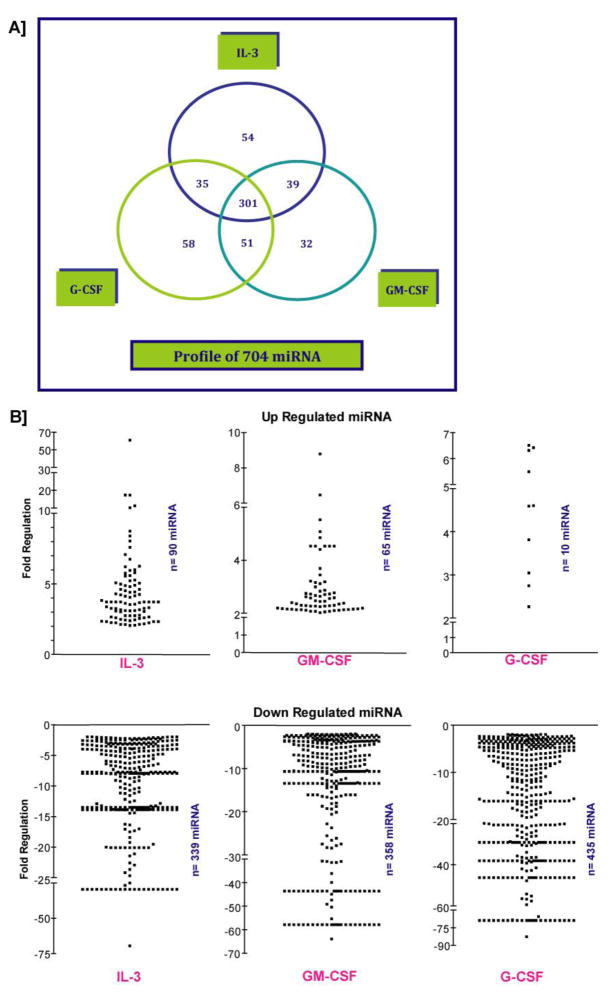
Figure 2. Profiling outcomes of IL-3, GM-CSF, and G-CSF regulated miRNA in AML-193 cells.
A. A three-way Venn diagram illustrating the comprehensive miRNA profiling outcome by three different cytokines. 301 miRNAs were commonly regulated by IL-3, GM-CSF and G-CSF. B. Individual scatter plots for up- and down- regulated miRNAs. 90 miRNAs were upregulated by IL-3 and 339 miRNAs were down regulated. GM-CSF elevated the expression of 65 miRNAs and decreased expressions of 358 miRNAs. G-CSF upregulated 10 miRNAs and downregulated 453 miRNAs. As previously mentioned, fold change filters were applied to select the miRNAs that were regulated more than 2-fold.
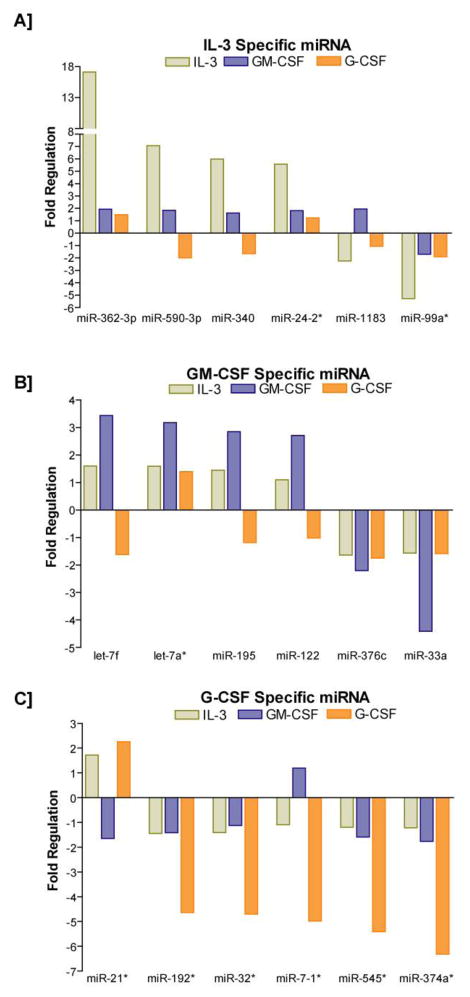
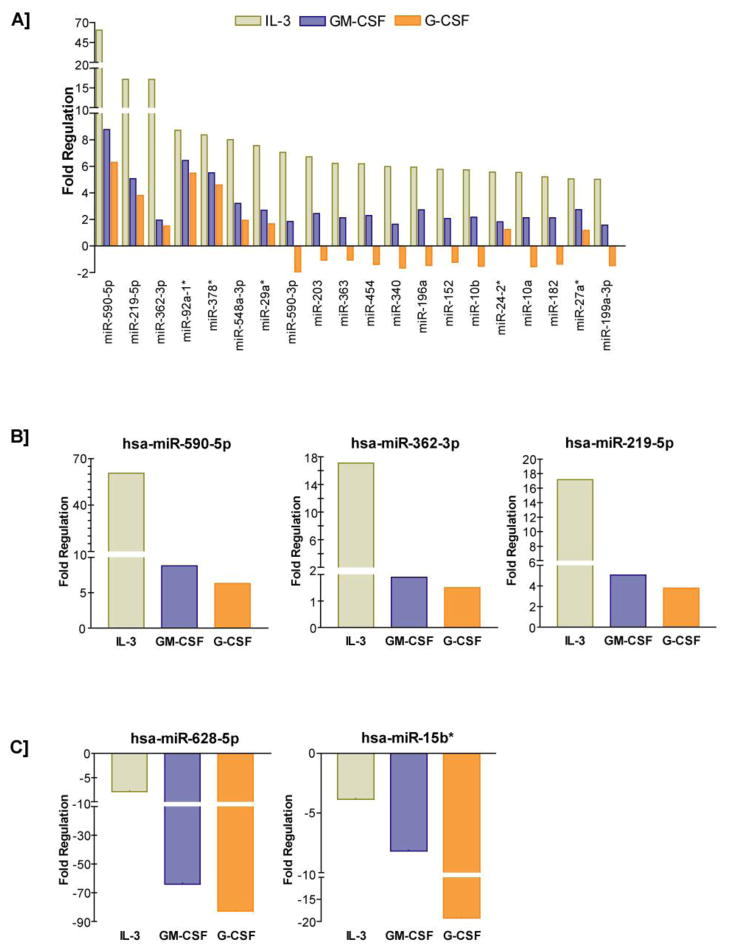
A detailed view of the comprehensive miRNA profiling outcome by three different cytokines is presented as three-way Venn diagram in Figure 2A. Interestingly, 301 miRNAs were commonly regulated by IL-3, GM-CSF and G-CSF. IL-3 specifically regulated 54 miRNAs and those miRNAs that were highly regulated included miR-362-39, miR-590-3p, miR-340, miR-24-2, miR-1183 and miR-99a (Figure 3A). GM-CSF specifically regulated miRNAs included let-7f, let-7a*, miR-195, miR-122, miR-376-c and miR-33a (Figure 3B). G-CSF specifically regulated set included miR-21*, miR-192*, miR-32*, miR-7-1*, miR-545* and miR-37-4a* (Figure 3C). Among the commonly regulated miRNAs, the ones that were subjected to high levels of regulation included miR-590-5p, miR-219-5p, miR-362-3p, miR-92-a1*, miR-378*, miR-548-3p, miR-29a*, miR-590-3p, miR-203, miR-363, miR-454, miR-340, miR-196a, miR-152, miR-10b, miR-24-2*, miR-10a, miR-182, miR-27a*, and miR-199a-3p (Figure 4A). Interestingly, the commonly regulated miRNAs demonstrated a directional regulation in the order of IL-3>GM-CF>G-CSF (Figure 4A-C). A complete list of all miRNAs distinctly regulated as well as commonly regulated by IL-3, GM-CSF and G-CSF in AML-193 cells is provided as Supplemental Table 1, 2, 3, and 4 respectively. miR-10b has been shown to be up regulated in cytogenetically normal AML (CN-AML) with FLT3-ITD and nucleophosmin (NPM) mutations 26,41. Likewise, in a previous Cancer and Leukemia Group B (CALGB) 19808 study, in AML patients with FLT3-ITD with wild type NPM1 or both, increased expression of miR-219-5p was associated with increased risk of an event 42. Similarly and significantly, increased expression of miR-199a has been identified in AML patients with isolated Trisomy 8 that is associated with poor outcome 26. Furthermore, increased expression of miR-199a is also part of a miRNA signature that is significantly up regulated in six solid tumors 43. The currently observed up regulation of these select miRNAs by IL-3, GM-CSF and G-CSF in AML-193 cells provides an insight into the possible deviant regulatory mechanism responsible for the high expressions of these miRNAs in AML. Further analyses of the potential targets of these significantly regulated miRNAs revealed important functional roles for these putative targets in myeloid cell development and differentiation. For example, a few of the highly conserved targets of miR-590-5p included BMPR2 and PCBP2. Interestingly, comparative analysis of gene expression on Agilent 22K oligonucleotide microarrays in purified CD34+ cells from the blood of myeloid metaplasia patients showed decreased expressions of BMPR2 44. BMPR2 is also a conserved target of miR-125b and significantly, Baltimore and his colleagues showed that overexpression of human miR-125b caused a dose dependent myeloproliferative disorder that progressed to lethal myeloid leukemia in mice and also enhanced hematopoietic engraftment 18. Furthermore, BMPR2 has been recently shown to possess tumor suppressive roles in mammary epithelia, and disruption of BMPR2 expression results in accelerated metastases of mammary carcinoma 45. Therefore, down regulation of BMPR2 by miR-590-5p may promote the IL-3 induced proliferation of early myeloid leukemic progenitors. Further, miR-590-5p is also predicted to target poly(C) binding protein 2 (PCBP2), which has been recently shown to be highly expressed in hematopoietic stem cells 46. Importantly, deficiency of PCBP2 in K562 cells results in p53 independent induction of cyclin-dependent kinase inhibitor (CDKN1A) and G1 arrest 47. Therefore, up regulation of miR-590-5p may attenuate onset of myeloid differentiation program, nonetheless, the direct effect of miR-590-5p on BMPR2 and PCBP2 in myelopoiesis will be explored in our future studies. Another directionally regulated miRNA that was discovered in the present studies was miR-219-5p. A highly conserved 8-mer target of miR-219-5p is a TGFβ/BMP induced signaling protein Smad4 48. Importantly, Smad4 has been showed to bind directly to Hoxa9 and inhibit Hoxa9-Nup98 induced AML 49. Most significantly, a recent study employing Smad4 −/− hematopoietic stem and progenitor cells (HSPCs) revealed a major negative regulatory role for Smad4 in Hoxa9 or Hoxa9-NUP98 induced AML 50. Disruptions of Smad4-Hoxa9 interaction in the cytoplasm leads to activation of the TGF-β pathway and apoptosis; deletion of Smad4 in HSPCs leads to the acceleration of or Hoxa9-NUP98 induced AML 50. One of miR-219-5p’s highly conserved target genes is Growth arrest and DNA damage 45 beta (Gadd45beta), one of the primary response genes induced during myeloid differentiation along with MyD88, Gadd45a, Gadd45g and Gadd34 51. Interestingly, Gadd45beta and Gadd45a knockout mice exhibit deficiencies in coping with hematological stresses like acute cytokine stimulation, myeloablation and inflammation 52,53. Significantly, a recent study discovered that miR-219 is one of the eight up regulated miRNAs in AML patients with 11q23 balanced translocation 26. The third most highly induced miRNA in our studies was miR-362-3p. miR-362-3p is predicted to target Sox17, an inhibitor of Wnt signaling pathway. The promoter of Sox17 has been shown to be hypermethylated in AML patients 54. Interestingly, the Wnt/beta-catenin pathway has recently been shown to be required for the development of leukemic stem cells (LSCs) in AML 55. Therefore, silencing of Sox17 by elevated expression of miR-362-3p may result in a similar up regulation of Wnt signaling pathway and promote growth of LSCs in AML.
Figure 3. Specific miRNAs independently regulated by IL-3, GM-CSF, and G-CSF in AML-193 cells.
A. IL-3 specifically regulated 54 miRNAs and highly regulated miRs included miR-362-39, miR-590-3p, miR-340, miR-24-2, miR-1183 and miR-99a. B. miRNAs specifically regulated by GM-CSF included let-7f, let-7a*, miR-195, miR-122, miR-376-c and miR-33a. C. miRNAs specifically regulated by G-CSF included miR-21*, miR-192*, miR-32*, miR-7-1*, miR-545* and miR-37-4a*.
Figure 4. IL-3, GM-CSF, and G-CSF directionally regulate specific miRNAs in AML-193 cells.
A. 301 miRNAs were commonly regulated by IL-3, GM-CSF and G-CSF and miRNAs that were subjected to high levels of regulation included miR-590-5p, miR-219-5p, miR-362-3p, miR-92-a1*, miR-378*, miR-548-3p, miR-29a*, miR-590-3p, miR-203, miR-363, miR-454, miR-340, miR-196a, miR-152, miR-10b, miR-24-2*, miR-10a, miR-182, miR-27a*, miR-199a-3p. B. Top three miRNAs (miR-590-5p, miR-362-3p and miR-219-5p) that were predominantly upregulated byIL-3, GM-CSF and G-CSFare individually graphed. C. Top two miRNAs (miR-628-5p and miR-15b*) that were predominantly down-regulated byIL-3, GM-CSF and G-CSF are individually graphed.
One of the most down regulated miRNA in our data set was a known STAT5 target miR-15b* 56. miR-15 has been shown to target Bcl-2 and inhibit apoptosis and promote cell survival 56. Importantly, miR-15a and miR-16-1, located at 13q14 were discovered to be deleted or down-regulated in approximately 60% of chronic lymphocytic leukemia (CLL) samples 25. Further, a recent study demonstrated that miR-15 and -16 target Cyclin E and conversely, inhibition of miR-15 and -16 enhanced E2F1-induced G(1) S transition 57. More importantly, a new study found that levels of miR-15 and -16 were significantly down regulated in a majority of the patients with prostate tumor 58. Therefore, it is tempting to speculate that GM-CSF mediated down regulation of miR-15 in leukemic cells may promote survival of blasts or even earlier leukemic myeloid progenitor pool or LSCs. Another highly down-regulated miR was miR-628-5p. One of the highly conserved predicted targets of miR-628-5p is Foxo3/Foxo3a 59. Recent clinical studies have implicated an important prognostic role for Foxo3 expression in AML. Elevated Foxo3 expression was shown to be associated with a poorer prognosis in AML with normal cytogenetics 60. Further, highly phosphorylated Foxo3 has been predicted to be an independent adverse prognostic factor in AML 61. In the light of these reports that reveal high expressions of Foxo3 in AML, it is possible that decreased levels of miR-628-5p caused by IL-3 signaling in leukemic progenitors may be playing a major role in promoting increased Foxo3a expressions in AML.
In summary, we have discovered for the first time novel miRNA profiles regulated by IL-3, GM-CSF and G-CSF in an acute myeloid leukemia progenitor cell model. Importantly, we have established a cytokine regulated miRNome for leukemic myeloid progenitors and set the stage for future investigations in leukemic stem cells to delineate the pathological roles of dysregulated miRNAs in AML. It will be interesting to examine how many of these miRNAs are directly regulated by STAT5 as the outcomes from such investigation may provide important leads to develop combinatorial therapeutic strategy for AML subtypes with STAT5 dysregulation via use of STAT5 specific inhibitors 62. Furthermore, these novel miRNA signatures may have therapeutic implications for targeting dysregulated miRNAs by antagomir strategy or miRNA replacement therapy, paving the way for the development of novel miRNA-based therapeutic interventions in AML.
Supplementary Material
Acknowledgments
This Research is supported in part through an American Cancer Society Research Grant, # IRG-82-003-26 and National Institute of Health National Center for Resaerch Resources COBRE (P20RR018789, Project #7). Additional support was provided by Maine Medical Center Research Institute Core facilities in Genomics and Bioinformatics; and Flow Cytometry and Progenitor Cell Analysis (as supported by NIH P20RR018789, DM Wojchowski, PI).
Footnotes
Authorship Contributions
All authors contributed to bench investigations and paper construction; A.F. performed miRNA profiling and analyses; P.S designed and directed all investigations.
Conflict of Interest
The Authors declare no conflict of interest related to this work.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132:631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spivak JL, Smith RR, Ihle JN. Interleukin 3 promotes the in vitro proliferation of murine pluripotent hematopoietic stem cells. J Clin Invest. 1985;76:1613–1621. doi: 10.1172/JCI112145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Broxmeyer HE, Williams DE. The production of myeloid blood cells and their regulation during health and disease. Crit Rev Oncol Hematol. 1988;8:173–226. doi: 10.1016/s1040-8428(88)80016-7. [DOI] [PubMed] [Google Scholar]
- 4.Broxmeyer HE, Williams DE, Hangoc G, et al. Synergistic myelopoietic actions in vivo after administration to mice of combinations of purified natural murine colony-stimulating factor 1, recombinant murine interleukin 3, and recombinant murine granulocyte/macrophage colony-stimulating factor. Proc Natl Acad Sci U S A. 1987;84:3871–3875. doi: 10.1073/pnas.84.11.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feldman GM, Rosenthal LA, Liu X, et al. STAT5A-deficient mice demonstrate a defect in granulocyte-macrophage colony-stimulating factor-induced proliferation and gene expression. Blood. 1997;90:1768–1776. [PubMed] [Google Scholar]
- 6.Mui AL, Wakao H, Harada N, O’Farrell AM, Miyajima A. Interleukin-3, granulocyte-macrophage colony-stimulating factor, and interleukin-5 transduce signals through two forms of STAT5. J Leukoc Biol. 1995;57:799–803. doi: 10.1002/jlb.57.5.799. [DOI] [PubMed] [Google Scholar]
- 7.Rosen RL, Winestock KD, Chen G, Liu X, Hennighausen L, Finbloom DS. Granulocyte-macrophage colony-stimulating factor preferentially activates the 94-kD STAT5A and an 80-kD STAT5A isoform in human peripheral blood monocytes. Blood. 1996;88:1206–1214. [PubMed] [Google Scholar]
- 8.Tian SS, Tapley P, Sincich C, Stein RB, Rosen J, Lamb P. Multiple signaling pathways induced by granulocyte colony-stimulating factor involving activation of JAKs, STAT5, and/or STAT3 are required for regulation of three distinct classes of immediate early genes. Blood. 1996;88:4435–4444. [PubMed] [Google Scholar]
- 9.Lange B, Valtieri M, Santoli D, et al. Growth factor requirements of childhood acute leukemia: establishment of GM-CSF-dependent cell lines. Blood. 1987;70:192–199. [PubMed] [Google Scholar]
- 10.Valtieri M, Boccoli G, Testa U, Barletta C, Peschle C. Two-step differentiation of AML-193 leukemic line: terminal maturation is induced by positive interaction of retinoic acid with granulocyte colony-stimulating factor (CSF) and vitamin D3 with monocyte CSF. Blood. 1991;77:1804–1812. [PubMed] [Google Scholar]
- 11.Tsubokawa M, Tohyama Y, Tohyama K, et al. Interleukin-3 activates Syk in a human myeloblastic leukemia cell line, AML193. Eur J Biochem. 1997;249:792–796. doi: 10.1111/j.1432-1033.1997.t01-2-00792.x. [DOI] [PubMed] [Google Scholar]
- 12.Du X, Ho M, Pastan I. New immunotoxins targeting CD123, a stem cell antigen on acute myeloid leukemia cells. J Immunother. 2007;30:607–613. doi: 10.1097/CJI.0b013e318053ed8e. [DOI] [PubMed] [Google Scholar]
- 13.Brasel K, Escobar S, Anderberg R, de Vries P, Gruss HJ, Lyman SD. Expression of the flt3 receptor and its ligand on hematopoietic cells. Leukemia. 1995;9:1212–1218. [PubMed] [Google Scholar]
- 14.Lyman SD, Jacobsen SE. c-kit ligand and Flt3 ligand: stem/progenitor cell factors with overlapping yet distinct activities. Blood. 1998;91:1101–1134. [PubMed] [Google Scholar]
- 15.Carow CE, Levenstein M, Kaufmann SH, et al. Expression of the hematopoietic growth factor receptor FLT3 (STK-1/Flk2) in human leukemias. Blood. 1996;87:1089–1096. [PubMed] [Google Scholar]
- 16.Sitnicka E, Buza-Vidas N, Larsson S, Nygren JM, Liuba K, Jacobsen SE. Human CD34+ hematopoietic stem cells capable of multilineage engrafting NOD/SCID mice express flt3: distinct flt3 and c-kit expression and response patterns on mouse and candidate human hematopoietic stem cells. Blood. 2003;102:881–886. doi: 10.1182/blood-2002-06-1694. [DOI] [PubMed] [Google Scholar]
- 17.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Connell RM, Chaudhuri AA, Rao DS, Gibson WS, Balazs AB, Baltimore D. MicroRNAs enriched in hematopoietic stem cells differentially regulate long-term hematopoietic output. Proc Natl Acad Sci U S A. 107:14235–14240. doi: 10.1073/pnas.1009798107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuchen S, Resch W, Yamane A, et al. Regulation of microRNA expression and abundance during lymphopoiesis. Immunity. 32:828–839. doi: 10.1016/j.immuni.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ooi AG, Sahoo D, Adorno M, Wang Y, Weissman IL, Park CY. MicroRNA-125b expands hematopoietic stem cells and enriches for the lymphoid-balanced and lymphoid-biased subsets. Proc Natl Acad Sci U S A. doi: 10.1073/pnas.1016218107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fazi F, Rosa A, Fatica A, et al. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 2005;123:819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 22.Pulikkan JA, Dengler V, Peramangalam PS, et al. Cell-cycle regulator E2F1 and microRNA-223 comprise an autoregulatory negative feedback loop in acute myeloid leukemia. Blood. 115:1768–1778. doi: 10.1182/blood-2009-08-240101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garzon R, Pichiorri F, Palumbo T, et al. MicroRNA gene expression during retinoic acid-induced differentiation of human acute promyelocytic leukemia. Oncogene. 2007;26:4148–4157. doi: 10.1038/sj.onc.1210186. [DOI] [PubMed] [Google Scholar]
- 24.Careccia S, Mainardi S, Pelosi A, et al. A restricted signature of miRNAs distinguishes APL blasts from normal promyelocytes. Oncogene. 2009;28:4034–4040. doi: 10.1038/onc.2009.255. [DOI] [PubMed] [Google Scholar]
- 25.Calin GA, Cimmino A, Fabbri M, et al. MiR-15a and miR-16–1 cluster functions in human leukemia. Proc Natl Acad Sci U S A. 2008;105:5166–5171. doi: 10.1073/pnas.0800121105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garzon R, Volinia S, Liu CG, et al. MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood. 2008;111:3183–3189. doi: 10.1182/blood-2007-07-098749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 28.Calin GA, Croce CM. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006;66:7390–7394. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- 29.Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 30.Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Testa U, Riccioni R, Militi S, et al. Elevated expression of IL-3Ralpha in acute myelogenous leukemia is associated with enhanced blast proliferation, increased cellularity, and poor prognosis. Blood. 2002;100:2980–2988. doi: 10.1182/blood-2002-03-0852. [DOI] [PubMed] [Google Scholar]
- 32.Marvin J, Swaminathan S, Kraker G, Chadburn A, Jacobberger J, Goolsby C. Normal bone marrow signal-transduction profiles: a requisite for enhanced detection of signaling dysregulations in AML. Blood. 117:e120–130. doi: 10.1182/blood-2010-10-316026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nosaka T, Kawashima T, Misawa K, Ikuta K, Mui AL, Kitamura T. STAT5 as a molecular regulator of proliferation, differentiation and apoptosis in hematopoietic cells. EMBO J. 1999;18:4754–4765. doi: 10.1093/emboj/18.17.4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsumoto A, Masuhara M, Mitsui K, et al. CIS, a cytokine inducible SH2 protein, is a target of the JAK-STAT5 pathway and modulates STAT5 activation. Blood. 1997;89:3148–3154. [PubMed] [Google Scholar]
- 35.Lehtonen A, Matikainen S, Miettinen M, Julkunen I. Granulocyte-macrophage colony-stimulating factor (GM-CSF)-induced STAT5 activation and target-gene expression during human monocyte/macrophage differentiation. J Leukoc Biol. 2002;71:511–519. [PubMed] [Google Scholar]
- 36.Ferbeyre G, Moriggl R. The role of Stat5 transcription factors as tumor suppressors or oncogenes. Biochim Biophys Acta. 1815:104–114. doi: 10.1016/j.bbcan.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 37.Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 38.Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21:1025–1030. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pons A, Nomdedeu B, Navarro A, et al. Hematopoiesis-related microRNA expression in myelodysplastic syndromes. Leuk Lymphoma. 2009;50:1854–1859. doi: 10.3109/10428190903147645. [DOI] [PubMed] [Google Scholar]
- 41.Garzon R, Garofalo M, Martelli MP, et al. Distinctive microRNA signature of acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin. Proc Natl Acad Sci U S A. 2008;105:3945–3950. doi: 10.1073/pnas.0800135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marcucci G, Radmacher MD, Maharry K, et al. MicroRNA expression in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358:1919–1928. doi: 10.1056/NEJMoa074256. [DOI] [PubMed] [Google Scholar]
- 43.Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andrieux J, Roche-Lestienne C, Geffroy S, et al. Bone morphogenetic protein antagonist gene NOG is involved in myeloproliferative disease associated with myelofibrosis. Cancer Genet Cytogenet. 2007;178:11–16. doi: 10.1016/j.cancergencyto.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 45.Owens P, Pickup MW, Novitskiy SV, et al. Breast Cancer Special Feature: Disruption of bone morphogenetic protein receptor 2 (BMPR2) in mammary tumors promotes metastases through cell autonomous and paracrine mediators. Proc Natl Acad Sci U S A. doi: 10.1073/pnas.1101139108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu XL, Yuan JY, Zhang JW, Zhang XH, Wang RX. Differential gene expression in human hematopoietic stem cells specified toward erythroid, megakaryocytic, and granulocytic lineage. J Leukoc Biol. 2007;82:986–1002. doi: 10.1189/jlb.0107014. [DOI] [PubMed] [Google Scholar]
- 47.Waggoner SA, Johannes GJ, Liebhaber SA. Depletion of the poly(C)-binding proteins alphaCP1 and alphaCP2 from K562 cells leads to p53-independent induction of cyclin-dependent kinase inhibitor (CDKN1A) and G1 arrest. J Biol Chem. 2009;284:9039–9049. doi: 10.1074/jbc.M806986200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blank U, Karlsson S. The role of Smad signaling in hematopoiesis and translational hematology. Leukemia. doi: 10.1038/leu.2011.95. [DOI] [PubMed] [Google Scholar]
- 49.Wang N, Kim H-G, Cotta CV, et al. TGF[beta]/BMP inhibits the bone marrow transformation capability of Hoxa9 by repressing its DNA-binding ability. EMBO J. 2006;25:1469–1480. doi: 10.1038/sj.emboj.7601037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Quere R, Karlsson G, Hertwig F, et al. Smad4 binds Hoxa9 in the cytoplasm and protects primitive hematopoietic cells against nuclear activation by Hoxa9 and leukemia transformation. Blood. 117:5918–5930. doi: 10.1182/blood-2010-08-301879. [DOI] [PubMed] [Google Scholar]
- 51.Liebermann DA, Hoffman B. Myeloid differentiation (MyD) primary response genes in hematopoiesis. Blood Cells Mol Dis. 2003;31:213–228. doi: 10.1016/s1079-9796(03)00160-8. [DOI] [PubMed] [Google Scholar]
- 52.Gupta SK, Gupta M, Hoffman B, Liebermann DA. Hematopoietic cells from gadd45a-deficient and gadd45b-deficient mice exhibit impaired stress responses to acute stimulation with cytokines, myeloablation and inflammation. Oncogene. 2006;25:5537–5546. doi: 10.1038/sj.onc.1209555. [DOI] [PubMed] [Google Scholar]
- 53.Hoffman B, Liebermann DA. Gadd45 modulation of intrinsic and extrinsic stress responses in myeloid cells. J Cell Physiol. 2009;218:26–31. doi: 10.1002/jcp.21582. [DOI] [PubMed] [Google Scholar]
- 54.Griffiths EA, Gore SD, Hooker C, et al. Acute myeloid leukemia is characterized by Wnt pathway inhibitor promoter hypermethylation. Leuk Lymphoma. 51:1711–1719. doi: 10.3109/10428194.2010.496505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Y, Krivtsov AV, Sinha AU, et al. The Wnt/beta-catenin pathway is required for the development of leukemia stem cells in AML. Science. 327:1650–1653. doi: 10.1126/science.1186624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li G, Miskimen KL, Wang Z, et al. STAT5 requires the N-domain for suppression of miR15/16, induction of bcl-2, and survival signaling in myeloproliferative disease. Blood. 115:1416–1424. doi: 10.1182/blood-2009-07-234963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ofir M, Hacohen D, Ginsberg D. miR-15 and miR-16 Are direct transcriptional targets of E2F1 that limit E2F-induced proliferation by targeting cyclin E. Mol Cancer Res. 9:440–447. doi: 10.1158/1541-7786.MCR-10-0344. [DOI] [PubMed] [Google Scholar]
- 58.Bonci D, Coppola V, Musumeci M, et al. The miR-15a-miR-16–1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. 2008;14:1271–1277. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- 59.Anderson MJ, Viars CS, Czekay S, Cavenee WK, Arden KC. Cloning and characterization of three human forkhead genes that comprise an FKHR-like gene subfamily. Genomics. 1998;47:187–199. doi: 10.1006/geno.1997.5122. [DOI] [PubMed] [Google Scholar]
- 60.Santamaria CM, Chillon MC, Garcia-Sanz R, et al. High FOXO3a expression is associated with a poorer prognosis in AML with normal cytogenetics. Leuk Res. 2009;33:1706–1709. doi: 10.1016/j.leukres.2009.04.024. [DOI] [PubMed] [Google Scholar]
- 61.Kornblau SM, Singh N, Qiu Y, Chen W, Zhang N, Coombes KR. Highly phosphorylated FOXO3A is an adverse prognostic factor in acute myeloid leukemia. Clin Cancer Res. 16:1865–1874. doi: 10.1158/1078-0432.CCR-09-2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nelson EA, Walker SR, Weisberg E, et al. The STAT5 inhibitor pimozide decreases survival of chronic myelogenous leukemia cells resistant to kinase inhibitors. Blood. 117:3421–3429. doi: 10.1182/blood-2009-11-255232. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.