Abstract
Facioscapulohumeral dystrophy (FSHD) is one of the most common inherited muscular dystrophies. The causative gene remains controversial and the mechanism of pathophysiology unknown. Here we identify genes associated with germline and early stem cell development as targets of the DUX4 transcription factor, a leading candidate gene for FSHD. The genes regulated by DUX4 are reliably detected in FSHD muscle but not in controls, providing direct support for the model that misexpression of DUX4 is a causal factor for FSHD. Additionally, we show that DUX4 binds and activates LTR elements from a class of MaLR endogenous primate retrotransposons and suppresses the innate immune response to viral infection, at least in part through the activation of DEFB103, a human defensin that can inhibit muscle differentiation. These findings suggest specific mechanisms of FSHD pathology and identify candidate biomarkers for disease diagnosis and progression.
Facioscapulohumeral dystrophy (FSHD) is the third most common muscular dystrophy. The mutation that causes FSHD was identified nearly 20 years ago (Wijmenga et al., 1992), yet the molecular mechanism(s) of the disease remains elusive. The most prevalent form of FSHD (FSHD1) is caused by the deletion of a subset of D4Z4 macrosatellite repeats in the subtelomeric region of chromosome 4q. Unaffected individuals have 11-100 of the 3.3kb D4Z4 repeat units, whereas FSHD1 individuals have 10 or fewer repeats. At least one repeat unit appears necessary for FSHD because no case has been identified with a complete deletion of D4Z4 repeats (Tupler et al., 1996). Each repeat unit contains a copy of the double homeobox retrogene DUX4 (Clapp et al., 2007; Gabriels et al., 1999; Lyle et al., 1995), and inappropriate expression of DUX4 was initially proposed as a possible cause of FSHD. This was supported by the observations that repeat contraction is associated with decreased repressive epigenetic marks in the remaining D4Z4 units (van Overveld et al., 2003; Zeng et al., 2009) and that overexpression of the DUX4 protein in a variety of cells, including skeletal muscle, causes apoptotic cell death (Kowaljow et al., 2007; Wallace et al., 2011; Wuebbles et al., 2010). However, initial attempts to identify DUX4 mRNA transcripts in FSHD muscle were unsuccessful, leading to the suggestion that other genes in the region were causative for FSHD (Gabellini et al., 2002; Klooster et al., 2009; Laoudj-Chenivesse et al., 2005; Reed et al., 2007).
Recent progress has returned the focus to the DUX4 retrogene as a leading candidate for FSHD. First, a subset of individuals with clinical features of FSHD do not have contracted D4Z4 repeats on chromosome 4 but do have decreased repressive heterochromatin at the D4Z4 repeats (de Greef et al., 2009) (FSHD2), indicating that loss of repressive chromatin at D4Z4 is the primary cause of FSHD. Second, genetic studies identified polymorphisms that create a DUX4 polyadenylation site as necessary for a D4Z4 contraction to cause FSHD (Lemmers et al., 2010). Third, high sensitivity RT-PCR assays detect DUX4 mRNA specifically in FSHD muscle (Dixit et al., 2007; Snider et al., 2010). Still, a major problem with the hypothesis that DUX4 expression causes FSHD has been the extremely low abundance of the mRNA and inability to reliably detect the protein in FSHD biopsy samples. Our prior work demonstrated that the low abundance of DUX4 in FSHD muscle cells represents a relatively high expression in a small subset of nuclei (Snider et al., 2010). However, it remained unclear whether the expression of DUX4 in FSHD muscle has a biological consequence that might drive the pathophysiology of FSHD.
The coding sequence of the DUX4 retrogene has been conserved in primates (Clapp et al., 2007), but whether this retrogene has a normal physiological function is unknown. Previously we found that DUX4 is normally expressed at high levels in germ cells of human testes and is epigenetically repressed in somatic tissues (Snider et al., 2010), whereas the epigenetic repression of the DUX4 locus in somatic tissues is less efficient in both FSHD1 and FSHD2, resulting in DUX4 expression in FSHD muscle cell nuclei. The germline-specific expression pattern of DUX4 is similar to that of other double homeodomain proteins (Booth and Holland, 2007; Wu et al., 2010). The function of this distinct family of DNA-binding proteins is unknown, but their shared tissue expression pattern may indicate a possible role for double homeodomain transcription factors in reproductive biology.
Here we report that DUX4 regulates the expression of genes involved in germline and early stem cell development. These DUX4 target genes are aberrantly expressed in FSHD skeletal muscle but not in control muscle biopsies. Therefore, the low level of DUX4 expression in FSHD is sufficient to effect numerous downstream changes and activate genes of germ cell and early development in postmitotic skeletal muscle. Additionally, we show that DUX4 binds and activates LTR elements from a class of MaLR endogenous primate retrotransposons and at the same time suppresses the innate immune response to retroviral infection, at least in part through transcriptional activation of DEFB103, a human defensin that can inhibit muscle differentiation. These findings suggest specific mechanisms of FSHD pathology and identify candidate biomarkers for disease diagnosis and progression.
RESULTS
Identification of genes regulated by DUX4 in human primary myoblasts
Previously, we identified two different DUX4 mRNA transcripts in human skeletal muscle, both at extremely low abundance: a full-length open reading frame mRNA (DUX4-fl) only detected in FSHD muscle and an internally spliced form of DUX4 mRNA (DUX4-s) that maintains the N-terminal double-homeobox domains but deletes the C-terminal domain and is detected in both control and FSHD muscle (Snider et al., 2010). Forced over-expression of DUX4-fl is toxic to cells, inducing apoptotic cell death (Kowaljow et al., 2007; Wallace et al., 2011), whereas forced over-expression of DUX4-s is not toxic to cultured human skeletal muscle cells (Geng et al., 2011). To determine whether gene expression is regulated by DUX4-fl and/or DUX4-s in human muscle cells, we transduced primary myoblasts from a control individual (unaffected by muscle disease) with a lentiviral vector expressing either DUX4-fl or DUX4-s and performed expression microarrays. At 24 hours after transduction, DUX4-fl increased the expression of 1071 genes and decreased the expression of 837 genes compared to a control myoblast population similarly infected with a GFP expressing lentivirus (2-fold change and FDR<0.01); whereas DUX4-s increased the expression of 159 genes and decreased expression of 45 genes (Figure 1A and see Table S1 for the complete list of genes regulated by DUX4-fl and/or DUX4-s). Using a slightly more stringent 3-fold criteria (> 1.584 log2-fold change and FDR<0.01), 466 genes were increased and 244 decreased by DUX4-fl; and 37 were increased and one decreased by DUX4-s. Only two annotated genes were increased 3-fold or more by both (CCNA1, MAP2), and none were decreased 3-fold or more by both. A representative sample of genes activated by DUX4-fl is shown in Table 1 and the full set of genes regulated by DUX4-fl or DUX4-s is in Table S1.
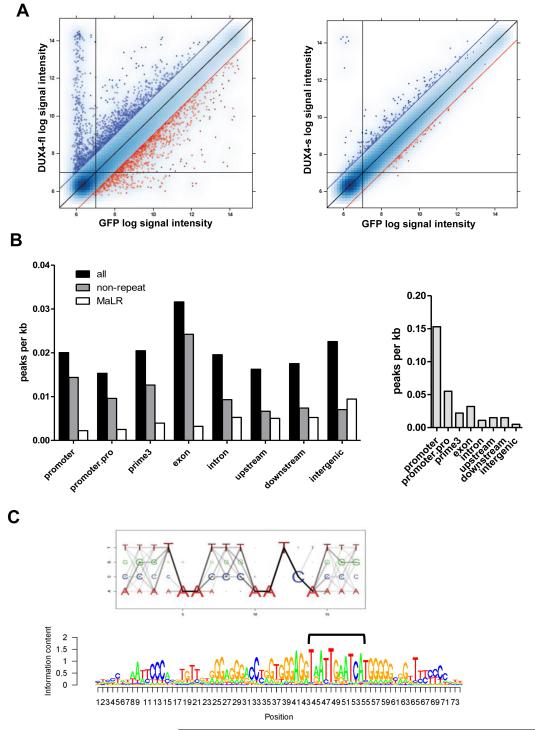
Figure 1. DUX4-fl activates the expression of germline genes and binds a double-homeobox motif.
(A) Pairwise comparison of normalized array data from DUX4-fl vs GFP (left), and DUX4-s vs GFP (right). Blue dots: upregulated genes; red dots: downregulated genes; blue and red diagonal lines represent 2-fold change. Vertical and horizontal lines represent signal thresholds for calling genes present or absent. (B) Gene context of DUX4-fl binding sites (left) or MyoD binding sites (right) represented by peak density, adjusting for prevalence of gene context category in the genome. Promoter: +/−500 bp from the transcription start site (TSS); promoter.pro: +/−2 kb from the TSS; prime3: +/−500 nt from the end of the transcript; upstream: −2 kb to −10 kb upstream of the TSS; downstream: +2 kb to +10 kb from the end of the transcript; intergenic: >10 kb from any annotated gene. (C) Top: DUX4-fl motif Logo. The size of each nucleotide at a given position is proportional to the frequency of the nucleotide at that position, and the darkness of the line connecting two adjacent nucleotides represents corresponding dinucleotide frequency. Bottom: DUX4 binding motif matches MaLR repeat consensus sequence. We identified the best DUX4 binding sites (bracket) within the MaLR repeats annotated in the RepeatMasker track provided by the UCSC genome browser (hg18) and extended the motif in the flanking regions to reflect general MaLR repeat consensus.
Table 1.
Representative genes induced by DUX4-fl
| Category | Log2 DUX4-fl Fc* | Log2 DUX4-s Fc* | Comments |
|---|---|---|---|
| Germline and Stem Cells | |||
| ZSCAN4 | 8.3 | 0.0 | Genome stability, telomere length |
| PRAMEF1 | 8.1 | 0.1 | Melanoma antigen family |
| SPRYD5 | 8.0 | −0.1 | Expressed in oocyte |
| KHDC1L | 8.0 | −0.1 | KH RNA binding domain |
| MBD3L2 | 7.6 | 0.0 | Methyl-CpG-binding protein |
| TRIM43 | 5.8 | 0.0 | Preimplantation embryo |
| TPRX1 | 4.5 | −0.1 | Homeobox protein |
| ZNF217 | 4.1 | −0.3 | Expressed in cancer stem cells |
| HSPA2 | 3.7 | −0.3 | Chaperone, heat shock 70 kd |
| JUP | 3.2 | −0.1 | expressed in germline and testicular cancers |
| FGFR3 | 3.1 | 0.0 | Expressed in spermatogonia |
| SLC2A14 | 2.4 | 0.2 | Spermatogenesis |
| ID2 | 2.3 | 0.3 | Negative regulator of cell differentiation |
| PVRL3 | 2.2 | 0.4 | Spermatid-sertoli junction |
| RNA Processing | |||
| SFRS2B | 4.2 | −0.3 | Splicing |
| THOC4 | 4.0 | −0.2 | Splicing, RNA transport |
| ZNHIT6 | 3.5 | 0.3 | sno-RNA processing |
| DBR1 | 3.4 | 0.2 | RNA lariat debranching enzyme |
| TFIP11 | 3.2 | 0.1 | Splicesome assembly |
| CWC15 | 2.6 | 0.1 | Spliceosome-associated |
| ARS2 | 2.6 | −0.2 | miRNA processing |
| PABPN1 | 2.6 | −0.3 | PolyA binding |
| RMRP | 2.3 | 0.1 | Mitochondrial RNA processing |
| Ubiquitin Pathway | |||
| SIAH1 | 3.7 | −0.1 | Targets TRF2 telomere maintenance |
| FBXO33 | 3.2 | 0.2 | E3 ubiquitin-protein ligase complex |
| PELI1 | 2.9 | 0.1 | E3 ligases involved in innate immunity |
| USP29 | 2.6 | −0.1 | Ubiquitin-specific peptidase |
| ARIH1 | 2.2 | 0.8 | Ubiquitin-conjugating enzyme E2 binding protein |
| TRIM23 | 2.2 | 0.6 | E3 ubiquitin ligase involved in immunity |
| Immunity and Innate Defense | |||
| DEFB103B | 6.4 | 0.1 | Innate defense |
| IFRD1 | 3.0 | −0.2 | Interferon-related developmental regulator |
| CXADR | 2.5 | −0.1 | Leukocyte migration |
| CBARA1 | 2.1 | −0.2 | T-helper 1-mediated autoreactivity |
| CXCR4 | 2.0 | −0.1 | Chemotaxis |
| General Transcription | |||
| GTF2F1 | 3.2 | 0.3 | General transcription factor IIF |
| MED26 | 2.1 | 0.1 | RNA Pol II mediator complex |
| RRN3 | 2.1 | 0.1 | RNA Pol I preinitiation complex |
| Cancer Expressed | |||
| CSAG3 | 5.9 | 0.1 | Chondrosarcoma-associated gene |
| SLC34A2 | 5.5 | 0.0 | Breast cancer biomarker |
| PNMA6B | 3.6 | −0.2 | Paraneoplastic antigen |
| Other | |||
| FLJ45337 | 3.7 | −0.2 | Endogenous retrovirus |
| HNRNPCL1 | 3.5 | −0.1 | Nucleosome assembly |
Fc = fold change
The Gene Ontology (GO) terms significantly enriched in 3-fold up-regulated genes by DUX4-fl included categories such as RNA polymerase II mediator complexes, RNA splicing and processing, and gamete/spermatogenesis (Table S2A); whereas down-regulated genes were enriched in immune response pathways (Table S2B). The up-regulation of a large number of transcription-related and RNA processing factors suggests that DUX4-fl might be a central component of a complex gene regulatory network, and the large number of germline associated genes suggests a possible role in reproductive biology.
In primary human myoblasts, DUX4-fl robustly induced a large number of genes not normally detected in skeletal muscle (see the contrail of genes along the Y-axis in the left panel of Figure 1A). These genes are good candidate biomarkers of DUX4 activity in skeletal muscle, since there is little to no background expression in control muscle. GO analysis for these highly induced genes showed enrichment for gamete generation and spermatogenesis categories (Table S2A). In many cases, DUX4-fl activated multiple members of gene families involved in germ cell biology and early development, including some primate-specific genes (Table 2). We validated the differential expression of 15 of the DUX4-fl regulated genes by RT-PCR (Figure S1).
Table 2.
DUX4 highly activates gene families involved in germ cell and early development
| Gene Family | Members | Biological Context | Fc range |
|---|---|---|---|
| Preferentially expressed in melanoma family |
PRAMEF1 | Cancer-testis antigen (Chang et al., 2011) | 9 - 269 |
| PRAMEF2 | |||
| PRAMEF4- 15 |
|||
| PRAMEF17 | |||
| PRAMEF20 | |||
| Tripartite motif- containing |
TRIM43 | Testis-expressed, preimplantation embryos (Stanghellini et al., 2009) |
27-235 |
| TRIM48 | |||
| TRIM49 | |||
| TRIM53 | |||
| TRIM64 | |||
| Methyl-binding protein- like |
MBD3L2 | Spermatids & germ cell tumors (Jiang et al., 2002; Jin et al., 2008) | 197-310 |
| MBD3L3 | |||
| MBD3L5 | |||
| Zinc finger and SCAN domain containing |
ZSCAN4 | Telomere maintenance in embryonic stem cells (Zalzman et al., 2010) |
13-320 |
| ZSCAN5B | |||
| ZSCAN5D | |||
| Ret-finger Protein-like | RFPL1 | Primate neocortex development (Bonnefont et al., 2008) |
20-336 |
| RFPL1S | |||
| RFPL2 | |||
| RFPL3 | |||
| RFPL4A | |||
| RFPL4B | |||
| KH homology domain containing |
KHDC1 | Oocyte- and embryo-expressed (Pierre et al., 2007) | 108-258 |
| KHDC1L | |||
| Family with sequence similarity 90 |
FAM90A1 | Primate-specific gene family with unknown function (Bosch et al., 2007) |
9-19 |
| FAM90A2P | |||
| FAM90A6P | |||
| FAM90A7 |
Identification of DUX4-fl binding sites and a consensus binding sequence motif
Double homeodomain proteins comprise a distinct group of DNA-binding proteins (Holland et al., 2007), but their consensus recognition sites and genomic targets are unknown. Therefore, we performed chromatin immunoprecipitation combined with high throughput sequencing (ChIP-Seq) to identify DUX4-binding sites in human muscle cells. We used two polyclonal rabbit antisera against DUX4 (Figure S2A and S2B) to immunoprecipitate DUX4-fl from human primary myoblasts 24 hours after transduction with lentiviral expressed DUX4-fl or control non-transduced primary myoblasts. Non-redundant reads unambiguously mapped to the human genome were computationally extended to a total length of 200 nucleotides and “peaks” were defined as regions where the number of reads was higher than a statistical threshold compared to the background (see Methods). Reads mapping to the X and Y chromosomes were excluded from our analysis.
A total of 62,028 and 39,737 peaks were identified at P-value thresholds of 10−10 and 10−15, respectively, after subtracting background peaks in the control samples. DUX4-fl peaks were widely distributed both upstream and downstream of gene transcription start sites (TSSs) with higher numbers in introns and intergenic regions, but showing a relatively constant peak density in all genomic regions when normalized for the size of the genomic compartment (Figure 1B, left). This pattern differs from that reported for many other transcription factors, such as MYOD (Cao et al., 2010), shown for comparison (Figure 1B, right), that show higher average peak density in regions near TSSs.
A de novo motif analysis identified the sequence TAAYBBAATCA (IUPAC nomenclature) (Figure 1C, top) near the center of greater than 90% of peaks. To our knowledge, this motif has not been described for any other transcription factor, but does contain two canonical homeodomain binding motifs (TAAT) arranged in tandem and separated by one nucleotide. Approximately 30% of sequences under the DUX4-fl peaks also contained a second larger motif that encompasses the primary DUX4-fl binding motif. This longer motif matches the long terminal repeat (LTR) of retrotransposons (Figure 1C, bottom). Assessment of the representation of DUX4-fl binding at different annotated repetitive elements in the genome shows a nearly 10-fold enrichment of DUX4-fl binding in the Mammalian apparent LTR-Retrotransposon (MaLR) family of retrotransposons and some enrichment in the related ERV family (Table S3). Note that the quantitative estimate of repeat-associated binding sites is conservative since reads mapping to more than one locus are excluded from our analysis.
MaLR family members expanded in the primate lineages (Smit, 1993). Thus, if DUX4-fl binding sites were carried throughout the genome during this expansion, these newer sites might have a different sequence motif compared to DUX4-fl binding sites located outside of MaLR repeats. To determine if the expansion of MaLR-associated binding sites might obscure the identification of a different DUX4 binding motif in non-repetitive elements, we performed separate motif analysis of MaLR-associated sites and sites not associated with repeats; both yielded nearly identical core motifs, TAAYYBAATCA and TAAYBYAATCA, respectively, but the repeat-associated motifs had slightly more flanking nucleotides preferences reflecting the LTR sequence (Figure S2C). Electrophoretic mobility shift assay (EMSA) confirmed that DUX4-fl binds the core motif present in both MaLR-associated and non-repeat associated sites and that mutation of the core nucleotides abolishes binding (Figure S2D-F), including sites from both repeat and non-repeat regions. Because the DUX4-s alternative splice form retains the N-terminal DNA-binding homeodomains, we hypothesized that it would bind to the same sites as DUX4-fl. EMSA confirmed that DUX4-s specifically binds the same core binding site as DUX4-fl in vitro (Figure S2G). Thus, DUX4-s can bind the same sequences as DUX4-fl but does not activate transcription of the same genes, which supports the prior determination that the C-terminus contains a transactivation domain (Kawamura-Saito et al., 2006).
DUX4-fl is a transcriptional activator
The number of DUX4-fl binding locations exceeds the number of genes that robustly increase expression in muscle cells following transduction with DUX4-fl. A genome-wide analysis of peak height and regional gene expression shows only a weak association of binding and gene expression for DUX4-fl (Figure 2A). To determine whether DUX4-fl binding might function as a transcriptional activator at some of the identified binding sites, DUX4 binding sites from selected genes were cloned upstream of the SV40 promoter in the pGL3-promoter luciferase construct. Co-transfection with DUX4-fl in human rhabdomyosarcoma cell line RD significantly induced luciferase expression independent of orientation or position, and mutation of the DUX4 binding motif eliminated the induction (Figure 2B and 2C). In contrast to DUX4-fl, DUX4-s did not activate expression despite demonstrating in vitro binding to this site (Figure 2B and Figure S2G).
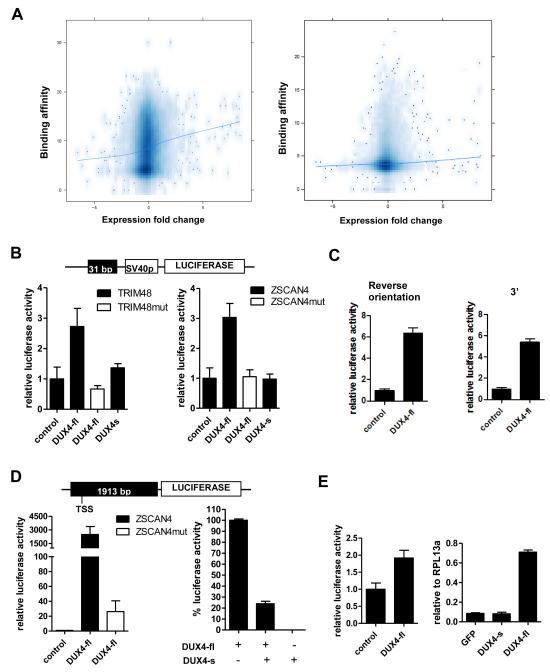
Figure 2. DUX4-fl activates transcription in vivo and DUX4-s can interfere with its activity.
(A) Comparison of DUX4 binding and regional gene transcription. The DUX4 peak height (Y-axis, square root transformation) of binding sites located within the CTCF-flanked domain of the TSS (left panel) or within +/− 2 KB of the TSS (right panel) plotted against the log-2 fold change of mRNA expression for the gene associated with the TSS (X-axis). The trend line shows a weak association between DUX4 binding and regional gene transcription. (B) Genomic fragments near theTRIM48 and ZSCAN4 genes containing DUX4 binding sites were cloned into pGL3-promoter reporter vector (schematic, top) and transfected into human rhabdomyoscaroma cell line RD. Cells were co-transfected with DUX4-fl or DUX4-s. pCS2-β galactosidase (beta-gal) was used to balance DNA amount in control condition. TRIM48mut and ZSCAN4mut, mutated binding sites. (C) DUX4-fl can act as an enhancer. Left: ZSCAN4 DUX4 binding site in reverse orientation relative to panel B upstream of the SV40 promoter. Right: ZSCAN4 DUX4 binding site in original orientation but moved 3′ downstream of the reporter gene. Luciferase activity set relative to control and error bars represent standard deviation of triplicates. (D) Left panel: Genomic fragment upstream of the ZSCAN4 translation start site containing four DUX4 binding sites was cloned into pGL3-basic luciferase vector (schematic, top). DUX4-fl highly activates the luciferase expression, whereas mutation of the binding sites (ZSCAN4mut) drastically reduces this induction. Right panel: Luciferase activity from DUX4-fl is set at 100% (pCS2-beta-gal is used to normalize the amount of plasmid). Co-transfection of equal amounts of DUX4-fl and DUX4-s diminishes luciferase activity. (E) Left: Genomic fragment from the LTR of THE1D MaLR element containing the DUX4 binding site were cloned into pGL3-promoter vector and tested for response to DUX4-fl as in (a). Right: Transcripts from endogenous retroelement MaLRs are upregulated by lentiviral transduction of DUX4-fl into primary human myoblasts. No upregulation is seen with lentiviral transduction of GFP or DUX4-s. Real-time RT-PCR quantitation is reported relative to internal standard RPL13a. All data represent mean +/− SD from at least triplicates.
To determine whether DUX4 binding might directly regulate transcription of select genes, we cloned the 1.9 kb enhancer and promoter region of the ZSCAN4 gene that includes four DUX4 binding sites into pGL3-basic reporter vector. Co-transfection with DUX4-fl significantly induced expression of this reporter and mutation of three of the four DUX4 binding sites nearly abolished the induction (Figure 2D, left). DUX4-s interfered with the activity of DUX4-fl when the two were co-expressed (Figure 2D, right), suggesting that DUX4-s acts as a dominant negative for DUX4-fl activity.
DUX4-fl also activated transcription through DUX4 sites in repetitive elements: DUX4-fl activated transcription of a luciferase reporter containing DUX4 binding sites cloned from LTRs at a MaLR THE1D element (Figure 2E, left) and RT-PCR showed induction of endogenous MaLR transcripts in muscle cells transduced with DUX4-fl (Figure 2E, right).
DUX4-fl directly regulates genes involved in germline development
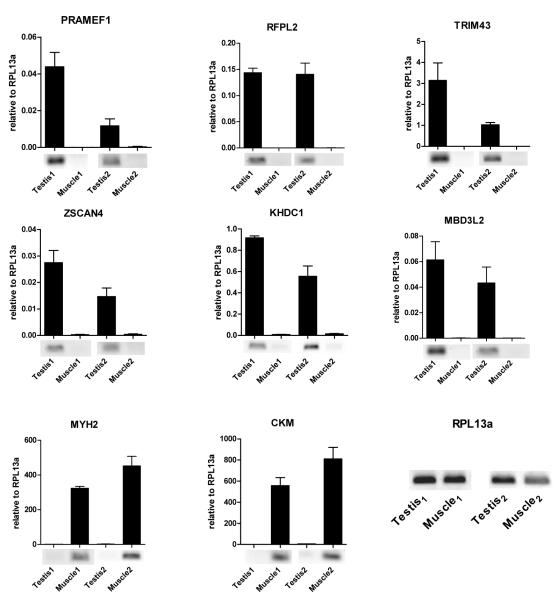
To identify the set of genes that might reflect the function of DUX4-fl prior to the expansion of MaLRs in primates, we identified the subset of genes activated at least 3-fold by DUX4-fl that also contain a non-repeat associated binding site within six kilobases of the TSS and not separated from the TSS by a binding site for the insulator factor CTCF (Table S4). The 74 genes meeting these criteria are highly enriched for genes involved in stem and germ cell functions, RNA processing, and regulated components of the PolII complex, similar to the major GO categories identified for all of the genes regulated by DUX4-fl. Quantitative RT-PCR of six DUX4-regulated genes on paired samples of testis mRNA and skeletal muscle mRNA from two control individuals found high expression of these targets in the testes and absent, or nearly absent, expression in skeletal muscle (Figure 3). We also detected the expression of the related DUXA and DUX1 genes in healthy human testis (data not shown), further supporting the notion that this family of double homeodomain proteins has a role in germ cell biology.
Figure 3. DUX4 targets are normally expressed in human testis but not in healthy skeletal muscle.
DUX4-fl targets expression in human testis versus matched skeletal muscle tissue from two healthy donors. Real-time RT-qPCR analysis of gene expression is presented relative to internal standard RPL13a and error bars represent standard deviation of PCR triplicates. MYH2 and CKM are markers of skeletal muscle. Qualitative RT-PCR gel panels are shown below qPCR graphs.
DUX4-fl-regulated genes are expressed in FSHD muscle
To determine whether the low levels of endogenous DUX4-fl mRNA detected in FSHD skeletal muscle is sufficient to activate DUX4 target genes, we assessed the expression of these genes in a set of control and FSHD muscle. Cultured muscle cells from control biopsies showed low or absent expression of the six DUX4-fl regulated genes, whereas these genes were expressed at significantly higher levels in the FSHD muscle cultures (Figure 4A), including those from both FSHD1 and FSHD2 individuals.
Figure 4. DUX4 regulated genes normally expressed in the testis are aberrantly expressed in FSHD muscle.
Real-time RT-PCR quantitation of six DUX4-fl target genes, PRAMEF1, RFPL2, TRIM43, ZSCAN4, KHDC1 and MBD3L2. Values are expressed as relative to internal standard RPL13a and represent mean +/− SD from triplicates. See Table S5 for all sample names and endogenous DUX4-fl expression status. (A) Cultured control and FSHD muscle cells. Note, sample 11 is from FSHD2 individual. (B) Control and FSHD muscle biopsies. (C) Top: RT-PCR gel showing siRNA knockdown of endogenous DUX4-fl in cultured FSHD muscle cells, done in triplicate with Timm17b as an internal standard. Negative control siRNA is against unrelated luciferase gene. Bottom: Levels of DUX4-fl target genes relative to the control treated samples were also reduced, as measured by qPCR. Error bars represent SD of triplicates; *P<0.05, **P<0.01 between DUX4 siRNA and control siRNA treated cells.
Similar to the expression of DUX4-fl regulated targets in cultured FSHD muscle, muscle biopsies from FSHD individuals had readily detectable mRNA of DUX4-fl regulated genes, although at varying levels in different biopsies (Figure 4B). The DUX4-fl mRNA is at extremely low abundance in FSHD muscle and it is notable that some biopsy samples in which the DUX4-fl mRNA was not detected showed elevation of DUX4 regulated targets (Table S5 and Figure S3A), indicating that the target mRNA is of significantly higher abundance and perhaps more stable than the DUX4 mRNA.
To determine whether the expression of these genes in FSHD muscle cells was directly due to DUX4, we transfected FSHD muscle cells with siRNA to the endogenous DUX4-fl mRNA. The siRNA sequences that decreased the DUX4-fl mRNA also resulted in decreased expression of the DUX4 target genes (Figure 4C, siRNA 1 and siRNA 4), confirming that endogenous DUX4 drives the expression of these genes in FSHD muscle cells. In addition, expression of the dominant negative DUX4-s (see Figure 2D) also inhibited the endogenous expression of the target genes (Figure S3B).
DUX4-induced DEFB103 inhibits the innate immune response and muscle differentiation
As noted above, genes enriched in the innate immunity pathways were expressed at lower levels in myoblasts transduced with lenti-DUX4-fl compared to the lenti-GFP or lenti-DUX4-s (see Table S2B). When compared to non-transduced cells, it was evident that about 350 genes, most of which were in the innate immunity pathway, were unchanged in the lenti-DUX4-fl transduced myoblasts but increased in cells transduced with either control lenti-GFP or lenti-DUX4-s (Table S6). Therefore, lentiviral induction of the innate immune response in human muscle cells appeared to be inhibited by DUX4-fl. RT-qPCR validated that lenti-GFP, lenti-DUX4-s, and multiple other lentivirus constructs induced the innate immune response in myoblasts, whereas similar titers of lenti-DUX4-fl did not (Figure 5A, left, and data not shown). Additionally, supernatant from DUX4-fl infected cells reduced the induction of these genes by lenti-GFP (Figure 5B), indicating that a secreted factor induced by DUX4-fl could mediate this suppressive effect.
Figure 5. DEFB103 inhibits innate immune response to viral infection and inhibits muscle differentiation.
Real time RT-PCR quantitation of innate immune responsive genes and genes involved in muscle differentiation. Values represent mean +/− SD from triplicates and are either expressed as relative to internal standard RPL13a or as percentage relative to control condition after being normalized to RPL13a. (A) Expression levels of innate immune responder IFIH1 and secreted factor DEFB103 after infection of muscle cells with lenti-GFP or lenti-DUX4-fl. (B) Expression levels of innate immune responders IFIH1 and ISG20 after infection with lenti-GFP in the presence of untreated media (control) or media supplemented with human β-defensin 3 peptide (5.0 ug/ml) (DEFB103) or conditioned media from lenti-DUX4-fl (C.M.). (C) Endogenous expression of DEFB103 in control testis and skeletal muscle tissues (Testis 1/2, Muscle 1/2), FSHD muscle biopsies (F-Muscle 3/4) and cultured FSHD and control muscle cells (F-MB and MB). (D) Upregulation of myostatin (MSTN) in myoblasts cultured in growth media supplemented with 2.5 ug/ml DEFB103. (E) Expression levels of various muscle marker genes in response to 2.5 ug/ml uM DEFB103 when added to muscle cells cultured in differentiation media. Myf6 and Desmin were included as genes that were unchanged on the arrays. (F) Cultured human muscle cells in differentiation media for 72 hr either without (a and b) or with (c and d) 2.5 μg/ml DEFB103. a and c, phase contrast showing inhibition of fusion by DEFB103. b and d, myosin heavy chain immunofluorescence (red) and DAPI nuclear stain (blue).
DUX4-fl robustly induced expression of DEFB103A and DEFB103B (β-defensin 3) (Figure 5A, right, Table 1 and S1), which has been shown to inhibit the transcription of pro-inflammatory genes in TLR4-stimulated macrophages (Semple et al., 2011). Indeed, addition of DEFB103 peptide also inhibited the induction of the innate immune response to lenti-GFP when added to the muscle cells at the time of infection (Figure 5B) but did not prevent viral entry and transduction as measured by copies of viral integrants in the genome and levels of GFP mRNA expressed (data not shown). Thus, DUX4 can prevent the innate immune response to viral infection in skeletal muscle cells, at least in part, through the transcriptional induction of DEFB103.
Like other DUX4-regulated genes, endogenous expression of DEFB103 was detected in FSHD cultured muscle cells, FSHD muscle biopsies, and in healthy testes, but little to none was seen in control skeletal muscle (Figure 5C). DEFB103 has been previously shown to bind the CCR6, CCR2, and melanocortin receptors and to be an antagonist ligand for the CXCR4 receptor, which is important for muscle cell migration and differentiation (Candille et al., 2007; Feng et al., 2006; Jin et al., 2010; Yang et al., 1999). To determine whether DEFB103 could affect myoblasts or muscle differentiation, we treated cultured control human muscle cells with DEFB103 peptide at concentrations used in anti-microbial and immunological assays (2.5-5.0 ug/ml) (Funderburg et al., 2007; Midorikawa et al., 2003; Semple et al., 2011) and assessed changes with gene expression arrays. Based on a 2-fold change threshold, DEFB103 did not alter the expression of any genes in myoblasts, although it is of interest that myostatin was upregulated approximately 50% and RT-qPCR confirmed that DEFB103 increased the mRNA for myostatin in myoblasts (Figure 5D). In contrast, exposing differentiating muscle cells to DEFB103 reduced the expression of 44 genes relative to the untreated control, the majority of which were genes associated with muscle differentiation (Table S7), and RT-qPCR on select genes (ACTA1, CKM, MYH2 and TNNT3) validated the array results (Figure 5E). Furthermore, muscle cultures exposed to DEFB103 during 72 hr of differentiation media showed decreased fusion to myotubes and decreased expression of myosin heavy chain, a marker of terminal muscle differentiation (Figure 5F). Therefore, DEFB103 activates the expression of myostatin in myoblasts and inhibits the expression of genes necessary for normal muscle differentiation, although it remains to be determined whether this activity is mediated by the CXCR4 receptor. Therefore, DUX4-mediated expression of DEFB103 in FSHD muscle can modulate the innate immune response and can inhibit myogenic differentiation.
DISCUSSION
Recent genetic and molecular studies indicated DUX4 as the likely candidate gene for FSHD (Dixit et al., 2007; Lemmers et al., 2010; Snider et al., 2010). Although the abundance of DUX4-fl mRNA was extremely low in FSHD muscle, we previously showed that this represented relatively high expression of both DUX4-fl mRNA and protein in a small percentage of muscle nuclei at any time point, either because the gene was on transiently or the expressing nuclei were eliminated (Snider et al., 2010). Yet, it remained unclear whether DUX4-fl expression had a biological consequence in FSHD. In our current study, we identify genes regulated by DUX4-fl and show that they are expressed at readily detectable levels in FSHD skeletal muscle, both cell lines and muscle biopsies, but not in control tissues, providing direct support for the model that misexpression of DUX4-fl is a causal factor for FSHD. The genes regulated by DUX4-fl suggest several specific mechanisms for FSHD pathophysiology.
Many of the genes highly upregulated by DUX4-fl normally function in the germline and/or early stem cells and are not present in healthy adult skeletal muscle. This supports a biological role for DUX4-fl in germ cell development and suggests potential disease mechanisms for FSHD. Activation of the gametogenic program might be incompatible with post-mitotic skeletal muscle, leading to apoptosis or cellular dysfunction. Also, the testis is an immune-privileged site and testis proteins misexpressed in cancers can induce an adaptive immune response (Simpson et al., 2005). In fact, some of the genes regulated by DUX4-fl, such as the PRAME family (Chang et al., 2011), are known cancer testis antigens, so it is reasonable to suggest that expression of these genes in skeletal muscle might also induce an adaptive immune response. An immune-mediated mechanism for FSHD is consistent with the focal inflammation and CD8+ T-cell infiltrates that characterize FSHD muscle biopsies (Frisullo et al., 2011; Molnar et al., 1991).
The induction of DEFB103 by DUX4 might influence both the adaptive and the innate immune response. DEFB103 can have a pro-inflammatory role in the adaptive immune response (Funderburg et al., 2007) and can act as a chemo-attractant for monocytes, lymphocytes and dendritic cells (Lai and Gallo, 2009). In this regard, it might enhance an adaptive immune response to germline antigens expressed in FSHD muscle. Though traditionally known for its role in antimicrobial defense (Sass et al., 2010), DEFB103 has been shown to suppress the innate immune response to LPS and TLR4 stimulation in macrophages (Semple et al., 2011; Semple et al., 2010), and has also been shown to be an antagonistic ligand of the CXCR4 receptor (Feng et al., 2006), which is important for muscle migration, regeneration, and differentiation (Griffin et al., 2010; Melchionna et al., 2010). In this study we show that DEFB103 inhibited the innate immune response to lentiviral infection in skeletal muscle cells, modestly induced myostatin in myoblasts, and impaired muscle cell differentiation. Therefore, DEFB103 might contribute to FSHD pathology by modulating the adaptive and innate immune response, as well as through inhibiting muscle differentiation. In this regard it is interesting to note that expression of murine Defb6 in the skeletal muscle of transgenic mice induced progressive muscle degeneration (Yamaguchi et al., 2007), although the mechanism was not determined.
Reactivation of retroelements can result in genomic instability (Belancio et al., 2010) and transcriptional deregulation (Schulz, 2006). Therefore, DUX4 activation of MaLR transcripts might directly contribute to FSHD pathophysiology. It is interesting that DUX4 both activates retroelement transcription and suppresses the virally induced innate immune response. Although we have shown that DEFB103 can substitute for DUX4 to suppress the innate immune response, products of retroelements and endogenous retroviruses may do the same and, thus, the DUX4-mediated suppression of the innate immune response might be multi-factorial. Since DEFB103 is also expressed in the testis, it is interesting to consider whether the role of DUX4 in the germline might include a simultaneous activation of retroelement transcription and suppression of the innate immune response to those transcripts.
DUX4 regulated targets also include genes involved in RNA splicing, developmentally regulated components of the Pol II transcription complex, and ubiquitin-mediated protein degradation pathways, all of which may have pathophysiological consequences. A recent study indicated that induction of E3 ubiquitin ligases by DUX4 might cause muscle atrophy in FSHD (Vanderplanck et al., 2011), consistent with our findings that multiple ubiquitin ligase family members are induced by DUX4. In addition, myostatin induces some of these ubiquitin ligases in skeletal muscle (Lokireddy et al., 2011) and it is therefore possible that both DUX4 induction of ubiquitin ligases and the modest upregulation of myostatin by DEFB103 that we observed in this study can both contribute to muscle atrophy. DUX4 is also known to induce apoptosis in muscle cells and DUX-4 mediated myopathy in mice has been shown to be p53-dependent (Wallace et al., 2011). As noted above, the activation of retrotransposons or reactivation of the gametogenic program, particularly inducers of cell cycle in post-mitotic muscle, might contribute to apoptosis. In addition, the altered expression of many factors involved in RNA transcription and splicing might affect cell differentiation and survival. In many human diseases a single mutation can effect multiple pathological pathways that collectively account for the complex disease phenotype. Our study of DUX4 regulated genes has identified several candidate pathways and future work will be necessary to determine their relative contributions to the disease phenotype.
In this regard, other genes have been identified as candidates for FSHD. For example, FRG1 expression has been reported to be elevated in FSHD muscle (Gabellini et al., 2002) and FRG1 transgenic mice display a muscular dystrophy phenotype (Gabellini et al., 2006). It is interesting that FRG1 is reported to alter RNA splicing in FSHD muscle (Gabellini et al., 2006) and that our study shows that DUX4-fl also alters the expression of many genes that regulate splicing and RNA processing. It will be important to determine the relative contributions of DUX4 and FRG1 to FSHD pathophysiology; however, the human genetics shows a convincing linkage to polymorphisms necessary for the polyadenylation of the DUX4 mRNA (Lemmers et al., 2010), indicating that DUX4 mRNA is a necessary component of the disease. Therefore, one therapeutic avenue to pursue for FSHD is to reduce the activity of DUX4, either by eliminating its expression in the muscle cells as we have done in vitro with an siRNA or by introducing a dominant negative, such as the DUX4-s splice form.
A previous study identified PITX1 as a DUX4 target gene expressed in FSHD skeletal muscle and in mouse cells transfected with DUX4 (Dixit et al., 2007). Others have expressed DUX4 in mouse muscle cells and identified repression of the glutathione redox pathway (Bosnakovski et al., 2008). Both of these findings are consistent with our expression array data. However, since many of the DUX4 binding sites reside in primate-specific MaLRs and some of the DUX4 targets are not conserved in mice, further studies are necessary to determine the conserved and primate-specific functions of DUX4, an important consideration for evaluating mouse models of FSHD.
In conclusion, our data support the model that inappropriate expression of DUX4 plays a causal role in FSHD skeletal muscle pathophysiology by activating germline gene expression, endogenous retrotransposons, and suppressors of differentiation in skeletal muscle. The set of genes robustly upregulated by DUX4 in FSHD skeletal muscle are candidate biomarkers because they are absent in control muscle and easily detected in FSHD1 and FSHD2 muscle. Furthermore, some target genes encode secreted proteins, which offer the potential for developing blood tests to diagnose FSHD or monitor response to interventions. Beyond their utilities as candidate biomarkers, the DUX4 targets identified in this study point to specific mechanisms of disease and may help guide the development of therapies for FSHD.
EXPERIMENTAL PROCEDURES
Cell culture
Primary human myoblasts were collected and cultured as previously described (Snider et al., 2010). Human RD cells were grown in DMEM in 10% bovine calf serum (Hyclone) and pencillin/streptomycin.
Microarray and GO Analysis
Quadruplicate total RNA samples were collected from control human primary myoblasts transduced with lentivirus carrying DUX4-fl, DUX4-s or GFP (MOI = 15) for 24 h. Samples were analyzed by Illumina HumanHT-12 v4 Expression BeadChip Whole Genome arrays. Probe intensities were corrected, normalized, and summarized by the Lumi package of Bioconductor (Du et al., 2008). Differentially expressed genes were identified by the LIMMA package of Bioconductor (Wettenhall and Smyth, 2004). Gene set enrichment analysis (GSEA) was performed using the Bioconductor GOstats package (Falcon and Gentleman, 2007).
Chromatin Immunoprecipitation and Ultra-High-Throughput Sequencing
ChIP was performed and ChIP DNA samples were prepared as previously described (Cao et al., 2010). Anti-DUX4 C-terminus rabbit polyclonal antibodies MO488 and MO489 were combined to immunoprecipitate DUX4-fl. The samples were sequenced with Illumina Genome Analyzer II.
Defining peaks
Sequences were extracted by Illumina package GApipeline and reads were aligned using BWA to the human genome (hg18). We only kept one of the duplicated sequences to minimize the artifacts of PCR amplification. Reads mapping to multiple locations in the genome were excluded from our analysis. Each read was extended in the sequencing orientation to a total of 200 bases to infer the coverage at each genomic position. We performed Peak calling by an in-house developed R package “peakSig” (pending submission to Bioconductor), which models background reads by a negative binomial distribution. The negative binomial distribution can be viewed as a continuous mixture of Poisson distribution where the mixing distribution of the Poisson rate is modeled as a Gamma prior. This prior distribution is used to capture the variation of background reads density across the genome. Model parameters were estimated by fitting the truncated distribution on the number of bases with low coverage (one to three), to avoid the problem of inferring effective genome size excluding the non-mappable regions, and to eliminate contamination of any foreground signals in the high coverage regions. We also fit a GC dependent mixture model so that the significance of the peaks is determined not only by peak height, but also by the GC content of the neighboring genomic regions.
Motif analysis
We used an in-house developed R package “motifRG” (pending submission to Bioconductor) described previously for discriminative de-novo motif discovery (Palii et al., 2011). Briefly, it finds motifs that distinguish positive and negative sequence datasets, which in this study correspond to DUX4 binding sites and randomly sampled flanking regions of DUX4 binding sites. To generate a more accurate presentation of the DUX4 binding sites from the consensus pattern returned by the motifRG package, we further used a positional weight matrix (PWM) model, using the matches of the consensus pattern as the seed to initialize the iterative expectation-maximization (EM) refinement process similar to MEME. The motifs were extended iteratively as long as there was sequence preference in the flanking region, and refined in the same EM process.
Luciferase Assay
Transient DNA transfections of RD cells were performed using SuperFect (Qiagen) according to manufacturer specifications. Briefly, 3 × 105 cells were seeded per 35 mm plate the day prior to transfection. Cells were co-transfected with pCS2 expression vectors (2 ug/plate) carrying either β-galactosidase, DUX4-fl or DUX4-s and with pGL3-promoter luciferase reporter vectors (1 ug/plate) carrying various putative DUX4 binding sites or mutant sites upstream of the SV40 promoter or pGL3-basic reporter vector (1 μg/plate) carrying test promoter fragment upstream of the firefly luciferase gene. Cells were lysed 24 h post-transfection in Passive Lysis Buffer (Promega). Luciferase activities were quantified using reagents from the Dual-Luciferase Reporter Assay System (Promega) following manufacturer’s instructions. Light emission was measured using BioTek Synergy2 luminometer. Luciferase data are given as the averages ± SD of at least triplicates.
Real-time PCR
One microgram of total RNA was reverse transcribed into first strand cDNA in a 20 uL reaction using SuperScript III (Invitrogen) and digested with 1U of RNase H (Invitrogen) for 20 min at 37°C. cDNA was diluted and used for quantitative PCR with iTaq SYBR Green supermix with ROX (Bio-Rad). The relative expression levels of target genes were normalized to those of ribosomal protein L13A (RPL13A) by 2ΔCt. Undetermined values were equated to zero. Standard deviations from the mean of the ΔCt values were calculated from triplicates. PCR primers used for detecting the transcripts of the selected genes are listed in Supplementary methods. All primers amplify with similar and high (>90%) efficiencies.
Muscle biopsies and human RNA
Muscle biopsy samples were collected at the University of Rochester Fields Center from the vastus lateralis muscle of clinically affected and control individuals as previously described (Snider et al., 2010). RNA from matched tissues from healthy donors were purchased from BioChain (Hayward, CA).
Statistical analyses
Statistical significance between two means was determined by unpaired one-tailed t tests with P-value <0.05. Statistics for the microarray and ChIP-Seq experiments are described separately.
Supplementary Material
Acknowledgments
Supported by NIAMS R01AR045203, NINDS P01NS069539, and Friends of FSH Research; University of Washington Genome Sciences Training Grant (L.N.G.), and University of Washington Child Health Research Center, NIH U5K12HD043376-08 (A.P.F.). We thank M. Conerly, S. Diede, K. MacQuarrie, L. Maves and K. Siebenthall for review of the manuscript; A. Tyler for technical assistance; M. Gale and J. Smith for advice on innate immunity and defensins; and F. Rigo and F. Bennett at ISIS Pharmaceuticals for assistance with siRNA design.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession numbers Microarray and ChIP-seq data have been deposited in Gene Expression Omnibus (GEO) under accession numbers GSE33799 and GSE33838, respectively.
The authors declare no conflict of interest.
REFERENCES
- Belancio VP, Roy-Engel AM, Deininger PL. All y’all need to know ‘bout retroelements in cancer. Semin Cancer Biol. 2010;20:200–210. doi: 10.1016/j.semcancer.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnefont J, Nikolaev SI, Perrier AL, Guo S, Cartier L, Sorce S, Laforge T, Aubry L, Khaitovich P, Peschanski M, et al. Evolutionary forces shape the human RFPL1,2,3 genes toward a role in neocortex development. Am J Hum Genet. 2008;83:208–218. doi: 10.1016/j.ajhg.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth HA, Holland PW. Annotation, nomenclature and evolution of four novel homeobox genes expressed in the human germ line. Gene. 2007;387:7–14. doi: 10.1016/j.gene.2006.07.034. [DOI] [PubMed] [Google Scholar]
- Bosch N, Caceres M, Cardone MF, Carreras A, Ballana E, Rocchi M, Armengol L, Estivill X. Characterization and evolution of the novel gene family FAM90A in primates originated by multiple duplication and rearrangement events. Hum Mol Genet. 2007;16:2572–2582. doi: 10.1093/hmg/ddm209. [DOI] [PubMed] [Google Scholar]
- Bosnakovski D, Xu Z, Gang EJ, Galindo CL, Liu M, Simsek T, Garner HR, Agha-Mohammadi S, Tassin A, Coppee F, et al. An isogenetic myoblast expression screen identifies DUX4-mediated FSHD-associated molecular pathologies. Embo J. 2008;27:2766–2779. doi: 10.1038/emboj.2008.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candille SI, Kaelin CB, Cattanach BM, Yu B, Thompson DA, Nix MA, Kerns JA, Schmutz SM, Millhauser GL, Barsh GS. A -defensin mutation causes black coat color in domestic dogs. Science. 2007;318:1418–1423. doi: 10.1126/science.1147880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Yao Z, Sarkar D, Lawrence M, Sanchez GJ, Parker MH, MacQuarrie KL, Davison J, Morgan MT, Ruzzo WL, et al. Genome-wide MyoD binding in skeletal muscle cells: a potential for broad cellular reprogramming. Dev Cell. 2010;18:662–674. doi: 10.1016/j.devcel.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TC, Yang Y, Yasue H, Bharti AK, Retzel EF, Liu WS. The expansion of the PRAME gene family in Eutheria. PLoS One. 2011;6:e16867. doi: 10.1371/journal.pone.0016867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapp J, Mitchell LM, Bolland DJ, Fantes J, Corcoran AE, Scotting PJ, Armour JA, Hewitt JE. Evolutionary conservation of a coding function for D4Z4, the tandem DNA repeat mutated in facioscapulohumeral muscular dystrophy. Am J Hum Genet. 2007;81:264–279. doi: 10.1086/519311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Greef JC, Lemmers RJ, van Engelen BG, Sacconi S, Venance SL, Frants RR, Tawil R, van der Maarel SM. Common epigenetic changes of D4Z4 in contraction-dependent and contraction-independent FSHD. Hum Mutat. 2009;30:1449–1459. doi: 10.1002/humu.21091. [DOI] [PubMed] [Google Scholar]
- Dixit M, Ansseau E, Tassin A, Winokur S, Shi R, Qian H, Sauvage S, Matteotti C, van Acker AM, Leo O, et al. DUX4, a candidate gene of facioscapulohumeral muscular dystrophy, encodes a transcriptional activator of PITX1. Proc Natl Acad Sci U S A. 2007;104:18157–18162. doi: 10.1073/pnas.0708659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- Falcon S, Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007;23:257–258. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
- Feng Z, Dubyak GR, Lederman MM, Weinberg A. Cutting edge: human beta defensin 3--a novel antagonist of the HIV-1 coreceptor CXCR4. J Immunol. 2006;177:782–786. doi: 10.4049/jimmunol.177.2.782. [DOI] [PubMed] [Google Scholar]
- Frisullo G, Frusciante R, Nociti V, Tasca G, Renna R, Iorio R, Patanella AK, Iannaccone E, Marti A, Rossi M, et al. CD8(+) T cells in facioscapulohumeral muscular dystrophy patients with inflammatory features at muscle MRI. J Clin Immunol. 2011;31:155–166. doi: 10.1007/s10875-010-9474-6. [DOI] [PubMed] [Google Scholar]
- Funderburg N, Lederman MM, Feng Z, Drage MG, Jadlowsky J, Harding CV, Weinberg A, Sieg SF. Human -defensin-3 activates professional antigen-presenting cells via Toll-like receptors 1 and 2. Proc Natl Acad Sci U S A. 2007;104:18631–18635. doi: 10.1073/PNAS.0702130104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabellini D, D’Antona G, Moggio M, Prelle A, Zecca C, Adami R, Angeletti B, Ciscato P, Pellegrino MA, Bottinelli R, et al. Facioscapulohumeral muscular dystrophy in mice overexpressing FRG1. Nature. 2006;439:973–977. doi: 10.1038/nature04422. [DOI] [PubMed] [Google Scholar]
- Gabellini D, Green MR, Tupler R. Inappropriate gene activation in FSHD: a repressor complex binds a chromosomal repeat deleted in dystrophic muscle. Cell. 2002;110:339–348. doi: 10.1016/s0092-8674(02)00826-7. [DOI] [PubMed] [Google Scholar]
- Gabriels J, Beckers MC, Ding H, De Vriese A, Plaisance S, van der Maarel SM, Padberg GW, Frants RR, Hewitt JE, Collen D, et al. Nucleotide sequence of the partially deleted D4Z4 locus in a patient with FSHD identifies a putative gene within each 3.3 kb element. Gene. 1999;236:25–32. doi: 10.1016/s0378-1119(99)00267-x. [DOI] [PubMed] [Google Scholar]
- Geng LN, Tyler AE, Tapscott SJ. Immunodetection of human double homeobox 4. Hybridoma (Larchmt) 2011;30:125–130. doi: 10.1089/hyb.2010.0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin CA, Apponi LH, Long KK, Pavlath GK. Chemokine expression and control of muscle cell migration during myogenesis. J Cell Sci. 2010;123:3052–3060. doi: 10.1242/jcs.066241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland PW, Booth HA, Bruford EA. Classification and nomenclature of all human homeobox genes. BMC Biol. 2007;5:47. doi: 10.1186/1741-7007-5-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang CL, Jin SG, Lee DH, Lan ZJ, Xu X, O’Connor TR, Szabo PE, Mann JR, Cooney AJ, Pfeifer GP. MBD3L1 and MBD3L2, two new proteins homologous to the methyl-CpG-binding proteins MBD2 and MBD3: characterization of MBD3L1 as a testis-specific transcriptional repressor. Genomics. 2002;80:621–629. doi: 10.1006/geno.2002.7001. [DOI] [PubMed] [Google Scholar]
- Jin G, Kawsar HI, Hirsch SA, Zeng C, Jia X, Feng Z, Ghosh SK, Zheng QY, Zhou A, McIntyre TM, et al. An antimicrobial peptide regulates tumor-associated macrophage trafficking via the chemokine receptor CCR2, a model for tumorigenesis. PLoS One. 2010;5:e10993. doi: 10.1371/journal.pone.0010993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SG, Tsark W, Szabo PE, Pfeifer GP. Haploid male germ cell- and oocyte-specific Mbd3l1 and Mbd3l2 genes are dispensable for early development, fertility, and zygotic DNA demethylation in the mouse. Dev Dyn. 2008;237:3435–3443. doi: 10.1002/dvdy.21767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura-Saito M, Yamazaki Y, Kaneko K, Kawaguchi N, Kanda H, Mukai H, Gotoh T, Motoi T, Fukayama M, Aburatani H, et al. Fusion between CIC and DUX4 up-regulates PEA3 family genes in Ewing-like sarcomas with t(4;19)(q35;q13) translocation. Hum Mol Genet. 2006;15:2125–2137. doi: 10.1093/hmg/ddl136. [DOI] [PubMed] [Google Scholar]
- Klooster R, Straasheijm K, Shah B, Sowden J, Frants R, Thornton C, Tawil R, van der Maarel S. Comprehensive expression analysis of FSHD candidate genes at the mRNA and protein level. Eur J Hum Genet. 2009;17:1615–1624. doi: 10.1038/ejhg.2009.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowaljow V, Marcowycz A, Ansseau E, Conde CB, Sauvage S, Matteotti C, Arias C, Corona ED, Nunez NG, Leo O, et al. The DUX4 gene at the FSHD1A locus encodes a pro-apoptotic protein. Neuromuscul Disord. 2007;17:611–623. doi: 10.1016/j.nmd.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Lai Y, Gallo RL. AMPed up immunity: how antimicrobial peptides have multiple roles in immune defense. Trends Immunol. 2009;30:131–141. doi: 10.1016/j.it.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laoudj-Chenivesse D, Carnac G, Bisbal C, Hugon G, Bouillot S, Desnuelle C, Vassetzky Y, Fernandez A. Increased levels of adenine nucleotide translocator 1 protein and response to oxidative stress are early events in facioscapulohumeral muscular dystrophy muscle. J Mol Med. 2005;83:216–224. doi: 10.1007/s00109-004-0583-7. [DOI] [PubMed] [Google Scholar]
- Lemmers RJ, van der Vliet PJ, Klooster R, Sacconi S, Camano P, Dauwerse JG, Snider L, Straasheijm KR, van Ommen GJ, Padberg GW, et al. A unifying genetic model for facioscapulohumeral muscular dystrophy. Science. 2010;329:1650–1653. doi: 10.1126/science.1189044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokireddy S, McFarlane C, Ge X, Zhang H, Sze SK, Sharma M, Kambadur R. Myostatin Induces Degradation of Sarcomeric Proteins through a Smad3 Signaling Mechanism During Skeletal Muscle Wasting. Mol Endocrinol. 2011;25:1936–1949. doi: 10.1210/me.2011-1124. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lyle R, Wright TJ, Clark LN, Hewitt JE. The FSHD-associated repeat, D4Z4, is a member of a dispersed family of homeobox-containing repeats, subsets of which are clustered on the short arms of the acrocentric chromosomes. Genomics. 1995;28:389–397. doi: 10.1006/geno.1995.1166. [DOI] [PubMed] [Google Scholar]
- Melchionna R, Di Carlo A, De Mori R, Cappuzzello C, Barberi L, Musaro A, Cencioni C, Fujii N, Tamamura H, Crescenzi M, et al. Induction of myogenic differentiation by SDF-1 via CXCR4 and CXCR7 receptors. Muscle Nerve. 2010;41:828–835. doi: 10.1002/mus.21611. [DOI] [PubMed] [Google Scholar]
- Midorikawa K, Ouhara K, Komatsuzawa H, Kawai T, Yamada S, Fujiwara T, Yamazaki K, Sayama K, Taubman MA, Kurihara H, et al. Staphylococcus aureus susceptibility to innate antimicrobial peptides, beta-defensins and CAP18, expressed by human keratinocytes. Infect Immun. 2003;71:3730–3739. doi: 10.1128/IAI.71.7.3730-3739.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar M, Dioszeghy P, Mechler F. Inflammatory changes in facioscapulohumeral muscular dystrophy. Eur Arch Psychiatry Clin Neurosci. 1991;241:105–108. doi: 10.1007/BF02191151. [DOI] [PubMed] [Google Scholar]
- Palii CG, Perez-Iratxeta C, Yao Z, Cao Y, Dai F, Davison J, Atkins H, Allan D, Dilworth FJ, Gentleman R, et al. Differential genomic targeting of the transcription factor TAL1 in alternate haematopoietic lineages. Embo J. 2011;30:494–509. doi: 10.1038/emboj.2010.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre A, Gautier M, Callebaut I, Bontoux M, Jeanpierre E, Pontarotti P, Monget P. Atypical structure and phylogenomic evolution of the new eutherian oocyte- and embryo-expressed KHDC1/DPPA5/ECAT1/OOEP gene family. Genomics. 2007;90:583–594. doi: 10.1016/j.ygeno.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Reed PW, Corse AM, Porter NC, Flanigan KM, Bloch RJ. Abnormal expression of mu-crystallin in facioscapulohumeral muscular dystrophy. Exp Neurol. 2007;205:583–586. doi: 10.1016/j.expneurol.2007.03.009. [DOI] [PubMed] [Google Scholar]
- Sass V, Schneider T, Wilmes M, Korner C, Tossi A, Novikova N, Shamova O, Sahl HG. Human beta-defensin 3 inhibits cell wall biosynthesis in Staphylococci. Infect Immun. 2010;78:2793–2800. doi: 10.1128/IAI.00688-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz WA. L1 retrotransposons in human cancers. J Biomed Biotechnol. 2006;2006:83672. doi: 10.1155/JBB/2006/83672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semple F, Macpherson H, Webb S, Cox SL, Mallin LJ, Tyrrell C, Grimes GR, Semple CA, Nix MA, Millhauser GL, et al. Human beta-defensin 3 affects the activity of pro-inflammatory pathways associated with MyD88 and TRIF. Eur J Immunol. 2011 doi: 10.1002/eji.201141648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semple F, Webb S, Li HN, Patel HB, Perretti M, Jackson IJ, Gray M, Davidson DJ, Dorin JR. Human beta-defensin 3 has immunosuppressive activity in vitro and in vivo. Eur J Immunol. 2010;40:1073–1078. doi: 10.1002/eji.200940041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson AJ, Caballero OL, Jungbluth A, Chen YT, Old LJ. Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer. 2005;5:615–625. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- Smit AF. Identification of a new, abundant superfamily of mammalian LTR-transposons. Nucleic Acids Res. 1993;21:1863–1872. doi: 10.1093/nar/21.8.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider L, Geng LN, Lemmers RJ, Kyba M, Ware CB, Nelson AM, Tawil R, Filippova GN, van der Maarel SM, Tapscott SJ, et al. Facioscapulohumeral dystrophy: incomplete suppression of a retrotransposed gene. PLoS Genet. 2010;6:e1001181. doi: 10.1371/journal.pgen.1001181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanghellini I, Falco G, Lee SL, Monti M, Ko MS. Trim43a, Trim43b, and Trim43c: Novel mouse genes expressed specifically in mouse preimplantation embryos. Gene Expr Patterns. 2009;9:595–602. doi: 10.1016/j.gep.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tupler R, Berardinelli A, Barbierato L, Frants R, Hewitt JE, Lanzi G, Maraschio P, Tiepolo L. Monosomy of distal 4q does not cause facioscapulohumeral muscular dystrophy. J Med Genet. 1996;33:366–370. doi: 10.1136/jmg.33.5.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Overveld PG, Lemmers RJ, Sandkuijl LA, Enthoven L, Winokur ST, Bakels F, Padberg GW, van Ommen GJ, Frants RR, van der Maarel SM. Hypomethylation of D4Z4 in 4q-linked and non-4q-linked facioscapulohumeral muscular dystrophy. Nat Genet. 2003;35:315–317. doi: 10.1038/ng1262. [DOI] [PubMed] [Google Scholar]
- Vanderplanck C, Ansseau E, Charron S, Stricwant N, Tassin A, Laoudj-Chenivesse D, Wilton SD, Coppee F, Belayew A. The FSHD Atrophic Myotube Phenotype Is Caused by DUX4 Expression. PLoS One. 2011;6:e26820. doi: 10.1371/journal.pone.0026820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace LM, Garwick SE, Mei W, Belayew A, Coppee F, Ladner KJ, Guttridge D, Yang J, Harper SQ. DUX4, a candidate gene for facioscapulohumeral muscular dystrophy, causes p53-dependent myopathy in vivo. Ann Neurol. 2011;69:540–552. doi: 10.1002/ana.22275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wettenhall JM, Smyth GK. limmaGUI: a graphical user interface for linear modeling of microarray data. Bioinformatics. 2004;20:3705–3706. doi: 10.1093/bioinformatics/bth449. [DOI] [PubMed] [Google Scholar]
- Wijmenga C, Hewitt JE, Sandkuijl LA, Clark LN, Wright TJ, Dauwerse HG, Gruter AM, Hofker MH, Moerer P, Williamson R, et al. Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat Genet. 1992;2:26–30. doi: 10.1038/ng0992-26. [DOI] [PubMed] [Google Scholar]
- Wu SL, Tsai MS, Wong SH, Hsieh-Li HM, Tsai TS, Chang WT, Huang SL, Chiu CC, Wang SH. Characterization of genomic structures and expression profiles of three tandem repeats of a mouse double homeobox gene: Duxbl. Dev Dyn. 2010;239:927–940. doi: 10.1002/dvdy.22210. [DOI] [PubMed] [Google Scholar]
- Wuebbles RD, Long SW, Hanel ML, Jones PL. Testing the effects of FSHD candidate gene expression in vertebrate muscle development. Int J Clin Exp Pathol. 2010;3:386–400. [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Nagase T, Tomita T, Nakamura K, Fukuhara S, Amano T, Yamamoto H, Ide Y, Suzuki M, Teramoto S, et al. Beta-defensin overexpression induces progressive muscle degeneration in mice. Am J Physiol Cell Physiol. 2007;292:C2141–2149. doi: 10.1152/ajpcell.00295.2006. [DOI] [PubMed] [Google Scholar]
- Yang D, Chertov O, Bykovskaia SN, Chen Q, Buffo MJ, Shogan J, Anderson M, Schroder JM, Wang JM, Howard OM, et al. Beta-defensins: linking innate and adaptive immunity through dendritic and T cell CCR6. Science. 1999;286:525–528. doi: 10.1126/science.286.5439.525. [DOI] [PubMed] [Google Scholar]
- Zalzman M, Falco G, Sharova LV, Nishiyama A, Thomas M, Lee SL, Stagg CA, Hoang HG, Yang HT, Indig FE, et al. Zscan4 regulates telomere elongation and genomic stability in ES cells. Nature. 2010;464:858–863. doi: 10.1038/nature08882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, de Greef JC, Chen YY, Chien R, Kong X, Gregson HC, Winokur ST, Pyle A, Robertson KD, Schmiesing JA, et al. Specific loss of histone H3 lysine 9 trimethylation and HP1gamma/cohesin binding at D4Z4 repeats is associated with facioscapulohumeral dystrophy (FSHD) PLoS Genet. 2009;5:e1000559. doi: 10.1371/journal.pgen.1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.