Abstract
Functional stacking of broad spectrum resistance (R) genes could potentially be an effective strategy for more durable disease resistance, for example, to potato late blight caused by Phytophthora infestans (Pi). For this reason, three broad spectrum potato R genes (Rpi), Rpi-sto1 (Solanum stoloniferum), Rpi-vnt1.1 (S. venturii) and Rpi-blb3 (S. bulbocastanum) were selected, combined into a single binary vector pBINPLUS and transformed into the susceptible cultivar Desiree. Among the 550 kanamycin resistant regenerants, 28 were further investigated by gene specific PCRs. All regenerants were positive for the nptII gene and 23 of them contained the three Rpi genes, referred to as triple Rpi gene transformants. Detached leaf assay and agro-infiltration of avirulence (Avr) genes showed that the 23 triple Rpi gene transformants were resistant to the selected isolates and showed HR with the three Avr effectors indicating functional stacking of all the three Rpi genes. It is concluded that Avr genes, corresponding to the R genes to be stacked, must be available in order to assay for functionality of each stack component. No indications were found for silencing or any other negative effects affecting the function of the inserted Rpi genes. The resistance spectrum of these 23 triple Rpi gene transformants was, as expected, a sum of the spectra from the three individual Rpi genes. This is the first example of a one-step approach for the simultaneous domestication of three natural R genes against a single disease by genetic transformation.
Electronic supplementary material
The online version of this article (doi:10.1007/s11248-011-9510-1) contains supplementary material, which is available to authorized users.
Keywords: Potato, Gene stacking, Durable resistance, Functionality
Introduction
Many devastating diseases are threatening plants, such as potato late blight, apple scab and wheat yellow rust. In contrast to higher animals, which have an advanced adaptive immune system, plants mainly rely on innate immunity to resist invading pathogens (Staskawicz et al. 1995; Jones and Dangl 2006). As a consequence of competition, plant resistance and pathogen virulence have been co-evolving up till now. Co-evolution, on the one hand, is diversifying the species; on the other hand, it is also making the competitive species more adapted to the required environment (Rosenthal et al. 1976; Rausher 2001). A good example, is the late blight pathogen Phytophthora infestans (Pi), whose genome has been recently sequenced (Haas et al. 2009). This pathogen evolved by mutation of effectors frequently and rapidly. Consequently, individual resistance genes (Rpi) were overcome relatively fast after their introduction into the potato crop (Black et al. 1953).
A promising strategy for breeding more durable resistance is to stack multiple, broad spectrum, resistance genes in one genotype (Halpin 2005; Douglas and Halpin 2010), especially with resistance genes originating from different gene clusters, representing different HR interactions between R genes and their cognate Avr effectors. In comparison to traditional breeding, with the increasing problem of linkage drag during stacking, genetic transformation is a fast and efficient way to introduce and domesticate multiple R genes into an existing cultivar. For potato, three different transformation methods can be used for R gene stacking: (1) transformation of one or two R genes followed by re-transformation of a selected, well performing, resistant transformant with additional R genes; (2) in one step via co-transformation, by using mixed Agrobacterium strains containing unlinked R genes in two or more vectors; (3) with multiple R genes in one vector (Douglas and Halpin 2010).
Multiple transgene introduction has been practiced to combine different traits using various methods, such as particle bombardment or Agrobacterium-mediated transformation (Altpeter et al. 2005; Sharma et al. 2005; Halpin 2005; Lacorte 2006). Until now, they were transgenes or pathway genes, corresponding to different agronomic traits, and the individual functionality of which could be easily detected in the transformed plants (Campbell et al. 2000; Beyer et al. 2002; Anand et al. 2003; Agrawal et al. 2005; Cao et al. 2005; Chan et al. 2005; Schmidt et al. 2008). The application of gene stacking using GM technology is increasing. In 2010, worldwide 148.1 million hectare of GMO crops were grown, of which 32.3 million hectares with two or three stacked traits. Double traits stacking was mainly restricted to insect and herbicide resistance. Triple traits stacking was restricted to herbicide resistance and two different insect pests (ISAAA 2010). Our approach differs from these examples as we are introducing three natural R genes to render immunity to one disease, potato late blight.
To date, several R genes against Pi, originating from different Rpi gene clusters and various wild species, have been cloned. For example, R1, R2, and R3a from Solanum demissum (Ballvora et al. 2002; Lokossou et al. 2009; Huang et al. 2005), Rpi-blb1, Rpi-blb2 and Rpi-blb3 from S. bulbocastanum (Song et al. 2003; van der Vossen et al. 2003; van der Vossen et al. 2005; Lokossou et al. 2009); and Rpi-vnt1.1 from S. venturii (Foster et al. 2009; Pel et al. 2009). Some of them showed a broader resistance spectrum than others with a selected set of late blight isolates. It is expected that stacking of at least three broad spectrum Rpi genes is needed to obtain durable resistance in the field. Important reasons are the observation that multiple Rpi genes are frequently found in individual resistant plants in nature (Huang et al. 2005; Lokossou et al. 2010; Verzaux 2010). The Rpi gene differentials R8 and R9, which have been reported to exhibit durable resistance (Zhang and Kim 2007; Lehtinen et al. 2008; Dr. G. Kessel, Plant Research International, Wageningen, The Netherlands, unpublished; YL, unpublished), are known to contain multiple R genes (Trognitz and Trognitz 2004; JV, unpublished). These observations show that stacking of broad spectrum Rpi genes in plants could contribute to the durability of resistance for potato in the field.
Therefore, it is important to know how to stack three or more Rpi genes into a potato variety efficiently and how to test the biological functionality of each gene individually. One way to answer this question is to investigate the relationship between the resistance reaction of individual Rpi genes and the HR reactions in response to cognate Avr effectors (Vleeshouwers et al. 2008). The consensus RXLR pattern of Avr genes and the recent genomic sequence of Phytophthora infestans promoted the research on Avr gene(s) considerably. Avr1 (Dr. F. Govers, Phytopathology, Wageningen University and Research Center, Wageningen, The Netherlands), Avr2 (Dr. P. Birch, Plant Pathology, SCRI, United Kingdom), Avr3a (Armstrong et al. 2005), Avr4 (van Poppel et al. 2008), Avrblb2 (Oh et al. 2009), Avrvnt1 (Pel 2010), and IpiO1 (Vleeshouwers et al. 2008) were cloned, and they are recognized by Rpi genes R1, Rpi-blb3, R3a, R4, Rpi-blb2, Rpi-vnt1.1, and Rpi-blb1, respectively.
In this paper, three broad spectrum resistance genes, Rpi-sto1 (Vleeshouwers et al. 2008), Rpi-vnt1.1 (Pel et al. 2009) and Rpi-blb3 (Lokossou et al. 2009), were selected and cloned into a binary vector pBINPLUS, which was used to transfer these three Rpi genes simultaneously into the susceptible potato cultivar Desiree by Agrobacterium tumefaciens-mediated transformation. Co-integration and co-functioning were confirmed by molecular analyses, disease tests and agro-infiltration using corresponding avirulence (Avr) effectors. This approach provides a new foundation to distinguish the functionality of each stack component. Finally, it is concluded that the expected broadened resistance spectrum based on the functional combination of the three individual Rpi genes, was indeed achieved in the triple Rpi gene transformants.
Materials and methods
Materials
Susceptible potato cultivar Desiree was used for Rpi gene transformation. Single Rpi genes were transformed earlier in house and resulted in Desiree transformants harboring R3a (A04-22), Rpi-blb3 (A03-142) and Rpi-vnt1.1 (SF3) and Rpi-sto1 (A09-6). These single Rpi gene transformants were tested for their resistance spectrum. Three plasmids, pBINPLUS:Rpi-blb3, pBIN19:Rpi-vnt1.1 and pBINPLUS:Rpi-sto1 were used as a basis for designing the triple Rpi gene construct. Supplementary Table 1 lists the effectors or avirulence genes used in the agro-infiltration. Sixteen late blight isolates, derived from multiple locations in Europe and America, used for Rpi gene spectrum analysis were maintained at Wageningen UR Plant Breeding, Wageningen. This in house set of isolates was partly selected based on the compatibility to particular Rpi genes such as isolate EC1 to Rpi-vnt1.1; IPO-0, Pic99177, Pic99189 and Pic99183 to Rpi-sto1; and USA618 to Rpi-blb3. Twenty-eight additional isolates (Table 1), from four different provinces in China, were provided by CAAS in Beijing, China.
Table 1.
DLA on transgenic Desiree plants harboring single Rpi genes using 44 isolates
| Isolate Plant | IPO-0 | Pic99177 | Pic99189 | Pic99183 | EC1 | USA618 | NL00228 | VK98014 | F95573 | 89148-09 | NO01096 | IPO-C | Set 1 | Set 2 | Set 3 | Set 4 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Desiree:Rpi-sto1 a | S | S | SQ | SQ | R | R | R | R | R | R | R | Q | 4 R | 10 R | 12 R | 6 R |
| Desiree:Rpi-blb3 a | R | Q | S | R | R | S | Q | RQ | R | R | R | S | 4 R | 9 R;1S | 12 R | 3 R; 3 S |
| Desiree:R3a a | R | S | R | S | S | S | R | RQ | S | R | Q | S | 4 S | 10 S | 12 S | 6 S |
| Desiree:Rpi-vnt1.1 ab | R | R | R | R | S | R | R | R | R | R | R | R | 4 R | 10 R | 12 R | 6 R |
| Desiree | S | S | S | S | S | S | S | S | SQ | S | S | S | S | S | S | S |
Set 1 In house isolates 88069, H3PO4, 90128 and 428-2 gave the same resistance patterns for these four, different Rpi gene containing, transformants
Set 2 Ten isolates from province Inner Mongolia of China
Set 3 Eight isolates from province Hei Longjiang and four isolates from province Fu Jian of China
Set 4 Six isolates from province Yunnan of China
R all of the 8 inoculated spots on each leaf showed resistance
RQ 6–7 out of 8 spots on each leaf showed resistance
Q 3–5 out of 8 spots on each leaf showed resistance
SQ 1–2 out of 8 spots on each leaf showed resistance
S all of the 8 spots on each leaf showed susceptibility
aDesiree:Rpi-xxx: cv. Desiree transformant harbouring Rpi-xxx
bPart of the data of Rpi-vnt1.1 was adopted from Pel (2010)
Construction of triple Rpi gene vector
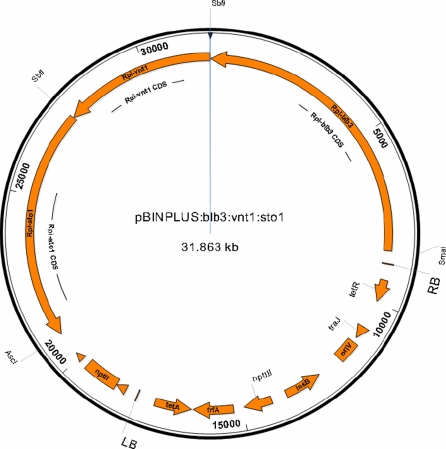
Three Rpi genes Rpi-sto1 (Vleeshouwers et al. 2008), Rpi-vnt1.1 (Pel et al. 2009) and Rpi-blb3 (Lokossou et al. 2009) had been cloned. In order to design the triple Rpi gene vector, first the two Rpi gene construct pBINPLUS:Rpi-blb3:Rpi-sto1 was created by using the unique enzyme sites SmaI and SbfI flanking Rpi-blb3 in the donor vector pBINPLUS:Rpi-blb3 and thus Rpi-blb3 was transferred into the recipient pBINPLUS:Rpi-sto1 vector. Rpi-vnt1.1 was inserted into vector pBINPLUS:Rpi-blb3:Rpi-sto1, using the unique enzyme site SbfI, located between Rpi-blb3 and Rpi-sto1. Since the SbfI site was not available in pBIN19:Rpi-vnt1.1, the Rpi-vnt1.1 fragment flanked by EcoRI was cloned into a modified pGEM-T vector, with SbfI-EcoRI-SbfI enzyme sites in the multiple cloning site (MCS). Subsequently, the Rpi-vnt1.1 fragment flanked by SbfI was subcloned into pBINPLUS:Rpi-blb3:Rpi-sto1. Both anticipated orientations of the Rpi-vnt1.1 gene were found and the clone with the orientation as depicted in Fig. 1 was selected. The resulting pBINPLUS:Rpi-blb3:Rpi-vnt1.1:Rpi-sto1 construct had a total size of 31.8 Kb and harbored a 22 Kb T-DNA insert (Fig. 1).
Fig. 1.
Graphical view of binary vector pBINPLUS:Rpi-blb3:Rpi-vnt1.1:Rpi-sto1. Restriction enzyme cleavage sites, used for cloning of the Rpi genes into pBINPLUS have been indicated
Stability and functionality tests of triple Rpi gene vector
After the vector had been made, we tested if it could replicate stably in Agrobacterium strains. Therefore, the stability of this construct was tested.
DNA was extracted from E. coli and transformed into three Agrobacterium strains (AGL1, AGL1 + VirG and Cor308) using electroporation, and transformants were cultured in three different media: LB (LB Broth: 20 g/l), LB-selected (Pepton-select: Gibco 10 g, yeast extract select Gibco 5 g, NaCl 5 g and agar 8 g per liter) and YEB (5 g beef extract, 5 g bacteriological peptone, 5 g sucrose, 1 g yeast extract and 2 ml 1 M MgSO4 per liter). Two colonies obtained from each of 9 combinations (3 Agrobacterium strains-3 culture media) were cultured for DNA isolation. These 18 DNA samples were transformed back into E. coli. Two colonies from each transformation were selected and DNA from 36 samples (two E. coli colonies were picked up from each of 18 DNA transformations) was extracted and digested with HindIII. The products were loaded on 1% agarose gel to check whether the pattern was the same with the original DNA isolated from E. coli.
Agrobacteria with stable plasmid insertions were used to check the functionality of the genes in the construct. Vector pBINPLUS:Rpi-blb3:Rpi-vnt1.1:Rpi-sto1 was co-infiltrated with Avr effectors Avr2, Avrvnt1 and IpiO1, corresponding to Rpi gene Rpi-blb3, Rpi-vnt1.1 and Rpi-sto1, respectively. Three clones (clone 2–2: vector in COR308 cultured in LB-select media, 5–1: vector in AGL1 cultured in LB-select, and 7–2: vector in AGL1 + virG cultured in LB) showed positive results in both tests, and they could be used for the transformation experiments.
Plant transformation
Stable insertion of the triple Rpi gene cassette into susceptible potato cultivar Desiree was carried out using Agrobacterium-mediated transformation (Filati et al. 1987; Hoekema et al. 1989), as adapted by Pel et al. (2009). Bacterial colony 5–1, selected from the stability and functionality tests, was used for this transformation. The transformation efficiency was calculated as the number of rooted shoots regenerated on kanamycin containing medium divided by the number of explants used within the time course of the experiment.
Molecular analysis of transformants
Presence or absence of specific genes in the regenerated plants was screened by PCR. Primers were listed in Supplementary Table 2. PCR was performed in a 25 μl reaction, including 0.5 μl of forward primer (10 μM), 0.5 μl of reverse primer (10 μM), 0.5 μl of dNTPs (5 mM each), 2.5 μl of 10 × buffer, and 2.5 units of Dream Taq polymerase (Fermentas).
Functional analysis of transformants
Rpi gene transformants were characterized using a detached leaf assay (DLA), in which eight 10 μl droplets of a zoospore suspension (50,000 spores/ml) of a specific Pi isolate (Vleeshouwers et al. 1999) were applied to the abaxial side of the leaf. Criteria for scoring (DLA): R, all of the inoculation spots on a detached leaf showed resistance; RQ, 6–7 out of eight spots on a leaf showed resistance; Q, 3–5 out of eight spots on a leaf showed resistance; SQ, 1–2 out of eight spots on a leaf showed resistance; S, all spots on a leaf showed susceptibility.
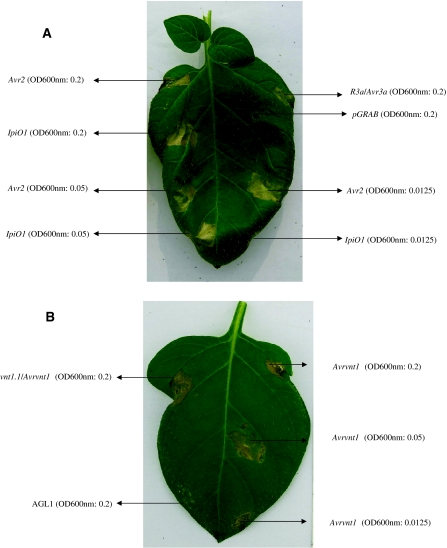
Agro-infiltration, which is referred as Agrobacterium tumefaciens transient assay (ATTA), was used to assay for hypersensitive response (HR) to the cognate Avr effectors (Bendahmane et al. 2000; Vleeshouwers et al. 2008). Nine replicates were performed (three plants per clone, with infiltration of three leaves per plant) using each effector at three different concentrations, with optical density at wavelength 600 nm: 0.2, 0.05 and 0.0125 (Fig. 2). OD600 nm is successively used throughout this paper.
Fig. 2.
Agro-infiltration of Avr effectors in triple Rpi gene regenerant A14 Z-22 using different densities of inoculum. Clear HR reactions were observed at all densities for Avr2, IpiO1 and Avrvnt1. Co-infiltration of R3a/Avr3a, and vnt1.1/Avrvnt-1 served as a positive controls. Empty vector pGRAB, and Agrobacterium strain AGL1 were negative controls. a Showed the infiltration with Avr2 and IpiO1. b Showed the infiltration with Avrvnt1
The functionality of each inserted gene was tested by isolates test and agro-infiltration using the scheme in Supplementary Table 3.
Results
Selection of cloned Rpi genes for broad spectrum resistance
In order to achieve more durable resistance to potato late blight by stacking genes with different resistances, the individual resistance spectra of four cloned Rpi genes (Rpi-sto1, Rpi-blb3, R3a and Rpi-vnt1.1) were compared with transformants of cv. Desiree using a set of 44 isolates, consisting of 16 in house and 28 Chinese isolates.
Table 1 shows the resistance pattern of the four Rpi genes against these 44 Pi isolates. Forty isolates were incompatible to Rpi-sto1, 37 to Rpi-blb3, 5 to R3a and 43 to Rpi-vnt1.1. Although it is known that R3a is overcome frequently, it is remarkable that R3a plants were susceptible to all the 28 Chinese isolates. In contrast to this, the Rpi-vnt1.1 and Rpi-sto1 plants were completely resistant to all Chinese isolates and Rpi-blb3 showed resistance to all isolates from two provinces of China and in a third province to nine out of ten isolates. In Yunnan province, only 3 out of 6 isolates were incompatible on Rpi-blb3 plants. Consequently, Rpi-sto1, Rpi-vnt1.1 and Rpi-blb3 were selected as broad spectrum Rpi genes, and combined into a binary vector pBINPLUS, which included the nptII transgene as a selection marker (Fig. 1).
Transformation efficiency of a triple Rpi gene vector
Vector pBINPLUS:Rpi-blb3:Rpi-vnt1.1:Rpi-sto1, containing native promoters and terminators of each Rpi gene, was used to transform the late blight susceptible cv. Desiree. After co-cultivation with A. tumefaciens using 850 stem explants, 121 and 550 rooting regenerants were produced at the fifth and ninth month after the transformation, which resulted in a transformation efficiency of 14 and 59%, respectively.
Characterization of individual regenerants by PCR, isolates test and agro-infiltration
The first obtained 28 kanamycin resistant regenerants were selected for further characterization. Due to the broad spectrum of Rpi genes used, no isolates were available, which can directly distinguish the functionality of each Rpi gene without the interference from the other Rpi genes. The testing scheme, illustrated in Supplementary Table 3, was developed to determine the presence of individual Rpi genes and their simultaneous biological functionality. This specific way of functional gene testing is needed for plants harboring multiple broad spectrum R genes against the same disease. These regenerants were characterized using Rpi gene specific primers (Supplementary Table 2), selected isolates (EC1 and Pic99189), and the corresponding Avr effectors (Fig. 2). Both isolates were selected due to the fact that (1) isolate EC1 is compatible on Rpi-vnt1.1 plants but incompatible on Rpi-sto1 and Rpi-blb3 plants, and (2) isolate Pic99189 is incompatible to Rpi-vnt1.1, whereas, it is compatible to Rpi-sto1 and Rpi-blb3 plants.
Together with cv. Desiree and the single Rpi gene transformants harboring Rpi-sto1, Rpi-blb3 or Rpi-vnt1.1 in cv. Desiree, the 28 kanamycin resistant regenerants were investigated. PCR analysis showed that all 28 regenerants (Table 2) contained nptII, among which, 23 harbored all three Rpi genes. All five kanamycin resistant plants (18%), without Pi resistance, turned out to lack the three Rpi genes (as evidenced by PCR analysis). DLA and agro-infiltration tests indicated that the 23 triple Rpi gene transformants (82%), also functionally expressed all three Rpi genes. Remarkably, no silencing or other interfering effects were observed for any of the transformed Rpi genes.
Table 2.
Presence and activity of Rpi genes in transgenic Desiree plants
| l (# of) plant | PCR | Pi inoculation | Effector agro-infiltration | ||||||
|---|---|---|---|---|---|---|---|---|---|
| nptII | sto1 | vnt1.1 | blb3 | EC1 | Pic99189 | IpiO1 | Avrvnt1 | Avr2 | |
| Regenerants (28) | |||||||||
| 23 | + | + | + | + | R | R | HR | HR | HR |
| 5 | + | − | − | − | S | S | − | − | − |
| Control | |||||||||
| Desiree:Rpi-sto1 a | + | + | − | − | R | S | HR | − | − |
| Desiree:Rpi-vnt1 | + | − | + | − | S | R | − | HR | − |
| Desiree:Rpi-blb3 | + | − | − | + | R | S | − | − | HR |
| Desiree | − | − | − | − | S | S | − | − | − |
The presence of the Rpi genes was determined by PCR. Biological activity of the Rpi genes was determined by Pi inoculation in detached leaf assays or by agro-infiltration using Avr effectors matching the introduced Rpi genes
HR Hypersensitive response
R Resistant
S Susceptible
“−” in PCR means no PCR product using gene specific primer pairs, and “−” in agro-infiltration means no HR symptom
aDesiree:Rpi-sto1 stands for one cv. Desiree transformant harbouring Rpi-sto1
Discussion
Generally, R gene cloning from donor wild species, followed by one-step transformation into cultivated plants can be considered as an efficient method for domestication of natural R genes into the agricultural environment. Stacking of several R genes is expected to provide more durable and broadened resistance as compared to the introduction of a single R gene, which was proven to be ineffective in the past. In this paper, a study was performed to explore the strategy of functional stacking broad spectrum Rpi genes. Rpi-blb3, Rpi-vnt1.1 and Rpi-sto1 were selected as broad spectrum Rpi genes based on virulence tests in the laboratory and greenhouse studies. It was observed that Rpi-sto1 and Rpi-vnt1.1 were potentially useful in all Chinese provinces and Rpi-blb3 in three provinces and to a lesser extent in Yunnan province of China. Another observation was that not a single Pi isolate from China was avirulent to R3a. This could be confirmed with the molecular data of an overlapping and bigger set of 49 Chinese isolates (Li et al. 2009). Li et al. detected only the presence of the virulence allele for avr3a in these isolates, indicating the general breakage of R3a. In contrast to the results from in house isolates, Rpi-vnt1.1 was still a broad spectrum Rpi gene in Europe, and Rpi-sto1 and Rpi-blb3 were still applicable in the Netherlands, but R3a was widely broken.
Recently, a similar experiment with Pi isolates from Argentina was described. The response of local isolates to ten different cultivars unexpectedly showed that cv. Russet Burbank possessed an Rpi gene, which was useful in this country (Andreu et al. 2010). However, in the United States, this Rpi gene is broken because of the general susceptibility of cv. Russet Burbank to Pi isolates from the USA (Staples 2004). The examples above show that isolates collected from specific geographical areas can predict the value of resistant varieties or help to select broad spectrum R genes for gene stacking. Consequently, this information could contribute to resistance breeding for specific geographical areas, where local field experiments have to be confirmed.
The triple Rpi gene construct showed a transformation efficiency of 14 and 59% at the fifth and ninth month after the transformation. These results showed that the transformation efficiency of this triple Rpi gene construct can be as high as that from transformations with single Rpi gene constructs, according to in house transformation experience (Heeres et al. 2002), but with a certain time delay. It is important to be aware of this phenomenon for future transformation experiments with stacked genes in one vector.
The aim of this study was to achieve durable resistance to Pi by stacking broad-spectrum Rpi genes. This approach inherently causes difficulties to select late blight isolates for determining biological functionality of each individual Rpi gene within the same plant. In this study, biological functionality could be proven for Rpi-vnt1.1 by using isolate Pic99189, but not for the other two Rpi genes simultaneously. Therefore, agro-infiltration with the three Avr effectors IpiO1 (Avrsto1), Avrvnt1 and Avr2 (Avrblb3) was used to determine the biological functionality of each integrated gene. This not only provides a strategy to detect multiple functional genes in the same plant, but also confirmed the necessity of the screening and cloning of cognate Avr effectors of the R genes used. Twenty-eight transformants were investigated in detail in this study, among which, 23 harbored all three Rpi genes. They gave complete resistance in the detached leaf assay, as well as HR after agro-infiltration using corresponding Avr effectors. It was observed that there were no silencing effects among these 23 transformants. All the integrated Rpi genes functioned properly.
On the other hand, there were five out of 28 transformants only harboring the selection marker gene nptII. This phenomenon is not unique. In experiments with rice, transformation was performed with multiple transgenes, VHb (Vitreoscilla hemoglobin gene), tzs (trans-zeatin secretion gene) and EPSP (modified 5-enolpyruvylshikimate-3-phosphate synthase gene) in addition to the hygromycin-gene as a selection marker, located close to the T-DNA left border (Cao et al. 2005). Cao et al. observed that three out of 113 transformants contained only the hyg selection marker gene, but not any target gene (VHb, tzs or EPSP). Our results indicated, at a much higher frequency, that T-DNA transfer might also start in a way only resulting in the functional insertion of the nptII gene close to the left border of T-DNA, but not simultaneously for all three Rpi genes.
Another significant observation is the fact that a 100% match between disease test reaction and agro-infiltration induced hypersensitivity reaction was observed for Rpi-vnt1.1, in 23 triple Rpi gene transformants (Table 2). If a transformant produced resistance in the DLA, it also showed HR response to matching Avr effectors. Similarly, we found that Rpi-blb3 and Rpi-sto1 were biologically active from both disease test and agro-infiltration experiments. This shows that agro-infiltration could be potentially a substitute for DLA on testing multiple Rpi gene containing plants. This predictive potential will be tested in the future for more R-Avr gene combinations and for different varieties. In addition, a 100% match was observed between the presence of Rpi genes indicated by gene specific PCR results and their biological activities after disease test and effector agro-infiltration, respectively. This indicates that, for this construct, PCR on Rpi genes can be directly used as an indication not only for the presence but also for the biological functionality of all three integrated Rpi genes in cv. Desiree transformed with this construct.
This paper adopted the approach to stack broad spectrum Rpi genes by Agrobacterium-mediated transformation using one transformation vector. This one vector and one-step transformation strategy, in comparison with re-transformation or multiple plasmids transformation, has the advantage of creating an artificial cluster of Rpi genes at one locus of the plant genome (Hiei et al. 1997). The plants produced in this paper, harboring these triple Rpi genes at presumably one locus, can be efficiently used as a breeding parent to transfer all three Rpi genes simultaneously into offspring plants.
Testing of durability of Rpi genes is an important phenomenon. The classical differential set R1-R11 has been used for many years as trap plants in potato fields for Pi population studies in order to check the presence or appearance of new (complex) virulent isolates (Dr. G. Kessel, Plant Research International, Wageningen UR, The Netherlands). In this differential set, R2, which is highly homologous to Rpi-blb3, is already a representative of Rpi-blb3. This type of field study can now be extended to differential plants harboring Rpi-sto1 and Rpi-vnt1.1 and compared with triple Rpi gene transformants with Rpi-sto1, Rpi-blb3 and Rpi-vnt1.1, in order to study and compare the potential differences in trapping candidate virulent isolates.
In conclusion, this paper showed an important way to come to more durable resistance by one-step stacking of three resistance genes using one vector in potato, showing an expected broadened resistance spectrum, without the problem of any silencing effects. The use of Avr genes matching the Rpi genes to be stacked in this approach is crucial.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
The authors thank Marjan Bergervoet for assistance in the transformation of cv. Desiree, Maarten Nijenhuis for assistance in making Rpi gene constructs, Hendrik Rietman for the potato agro-infiltration protocol, Ronald Hutten for true to type selection of Desiree transformants, Dirkjan Huigen, Henk Smid and Gerda van Engelenhoven for taking care of plants in the greenhouse, Geert Kessel (Plant Research International, Wageningen UR, The Netherlands), Paul Birch (Plant Pathology, Scottish Crop Research Institute, Dundee, UK), Francine Govers (Phytopathology, Wageningen University and Research Center, Wageningen, The Netherlands) and William Fry (Plant Pathology, Cornell University, Ithaca, USA) for providing Pi isolates and Avr effectors. This project is supported by Wageningen UR Plant Breeding.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Agrawal PK, Kohli A, Twyman RM, Christou P. Transformation of plants with multiple cassettes generates simple transgene integration patterns and high expression levels. Mol Breed. 2005;16:247–260. doi: 10.1007/s11032-005-0239-5. [DOI] [Google Scholar]
- Altpeter F, Baisakh N, Beachy R, Bock R, Capell T, Christou P, Daniell H, Datta K, Datta S, Dix PJ, Fauquet C, Huang N, Kohli A, Mooibroek H, Nicholson L, Nguyen TT, Nugent G, Raemakers K, Romano A, Somers DA, Stoger E, Taylor N, Visser RGF. Particle bombardment and the genetic enhancement of crops: myths and realities. Mol Breed. 2005;15:305–327. doi: 10.1007/s11032-004-8001-y. [DOI] [Google Scholar]
- Anand A, Zhou T, Trick HN, Gill BS, Bockus WW, Muthukrishnan S. Greenhouse and field testing of transgenic wheat plants stably expressing genes for thaumatin-like protein, chitinase and glucanase against Fusarium graminearum. J Exp Bot. 2003;54:1101–1111. doi: 10.1093/jxb/erg110. [DOI] [PubMed] [Google Scholar]
- Andreu AB, Caldiz DO, Forbes GA. Phenotypic expression of resistance to Phytophthora infestans in processing potatoes in Argentina. Am J Pot Res. 2010;87:177–187. doi: 10.1007/s12230-009-9121-z. [DOI] [Google Scholar]
- Armstrong MR, Whisson SC, Pritchard L, Bos JIB, Venter E, Avrova AO, Rehmany AP, Bohme U, Brooks K, Cherevach I, Hamlin N, White B, Fraser A, Lord A, Quail MA, Churcher C, Hall N, Berriman M, Huang SW, Kamoun S, Beynon JL, Birch PRJ. An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognized in the host cytoplasm. PNAS. 2005;102:7766–7771. doi: 10.1073/pnas.0500113102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballvora A, Ercolano MR, Julia W, Meksem K, Bormann CA, Oberhagemann P, Salamini F, Gebhardt C. The R1 gene for potato resistance to late blight (Phytophthora infestans) belongs to the leucine zipper/NBS/LRR class of plant resistance genes. Plant J. 2002;30:361–371. doi: 10.1046/j.1365-313X.2001.01292.x. [DOI] [PubMed] [Google Scholar]
- Bendahmane A, Querci M, Kanyuka K, Baulcombe DC. Agrobacterium transient expression system as a tool for the isolation of disease resistance genes: application to the Rx2 locus in potato. Plant J. 2000;21:73–81. doi: 10.1046/j.1365-313x.2000.00654.x. [DOI] [PubMed] [Google Scholar]
- Beyer P, Al-Babili S, Ye XD, Lucca P, Schaub P, Welsch R, Potrykus I (2002) Golden rice: introducing the beta-carotene biosynthesis pathway into rice endosperm by genetic engineering to defeat vitamin A deficiency. Am Soc Nutr Sci J Nutr 132:506S–510S [DOI] [PubMed]
- Black W, Mastenbroek C, Mills WR, Peterson LC. A proposal for an international nomenclature of races of Phytophthora infestans and of genes controlling immunity in Solanum demissum derivates. Euphytica. 1953;2:173–240. doi: 10.1007/BF00053724. [DOI] [Google Scholar]
- Campbell BT, Baeziger PS, Mitra A, Sato S, Clemente T. Inheritance of multiple transgenes in wheat. Crop Sci. 2000;40:1133–1141. doi: 10.2135/cropsci2000.4041133x. [DOI] [Google Scholar]
- Cao MX, Huang JQ, Wei ZM, Yao QH, Wan CZ, Lu JA. Agrobacterium-mediated multiple gene transformation in rice using a single vector. J Integr Plant Biol. 2005;47:233–242. doi: 10.1111/j.1744-7909.2005.00015.x. [DOI] [Google Scholar]
- Chan YL, Lin KH, Sanjaya, Liao LJ, Chen WH, Chan MT. Gene stacking in Phalaenopsis orchid enhances dual tolerance to pathogen attack. Transgenic Res. 2005;14:279–288. doi: 10.1007/s11248-005-0106-5. [DOI] [PubMed] [Google Scholar]
- Douglas E, Halpin C (2010) Gene stacking. In: Molecular techniques in crop improvement, 2nd edn. Springer, The Netherlands, pp 613–629
- Filati J, Kiser J, Rose R, Comai L. Efficient transfer of a glyphosphate tolerance gene into tomato using a binary Agrobacterium tumefaciens vector. Biotechnology. 1987;5:726–730. doi: 10.1038/nbt0787-726. [DOI] [Google Scholar]
- Foster S, Park TH, Pel M, Brigneti G, Sliwka J, Jagger L, van der Vossen EAG, Jones JDG. Rpi-vnt1.1, a Tm-22 homolog from Solanum venturii, confers resistance to potato late blight. MPMI. 2009;22:589–600. doi: 10.1094/MPMI-22-5-0589. [DOI] [PubMed] [Google Scholar]
- Haas BJ, et al. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature. 2009;461:393–398. doi: 10.1038/nature08358. [DOI] [PubMed] [Google Scholar]
- Halpin C. Gene stacking in transgenic plants-the challenge for 21st century plant biotechnology. Plant Biotechnol J. 2005;3:141–155. doi: 10.1111/j.1467-7652.2004.00113.x. [DOI] [PubMed] [Google Scholar]
- Heeres P, Schippers-Rozenboom M, Jacobsen E, Visser RGF. Transformation of a large number of potato varieties: genotype-dependent variation in efficiency and somaclonal variability. Euphytica. 2002;124:13–22. doi: 10.1023/A:1015689112703. [DOI] [Google Scholar]
- Hiei Y, Komari T, Kubo T. Transformation of rice mediated by Agrobacterium tumefaciens. Plant Mol Biol. 1997;35:205–218. doi: 10.1023/A:1005847615493. [DOI] [PubMed] [Google Scholar]
- Hoekema A, Huisman MJ, Molendijk L, Elzen PJM, Cornelissen BJC. The genetic engineering of two commercial potato cultivars for resistance to potato virus X. Nat Biotechnol. 1989;7:273–278. doi: 10.1038/nbt0389-273. [DOI] [Google Scholar]
- Huang SW, van der Vossen EAG, Kuang HH, Vleeshouwers VGAA, Zhang NW, Borm TJA, van Eck HJ, Baker B, Jacobsen E, Visser RGF. Comparative genomics enabled the isolation of the R3a late blight resistance gene in potato. Plant J. 2005;42:251–261. doi: 10.1111/j.1365-313X.2005.02365.x. [DOI] [PubMed] [Google Scholar]
- ISAAA (2010) Global status of commercialized biotech/GM corps International service for the acquisition of agri-biotech applications. http://www.isaaa.org/resources/publications/briefs/42/executivesummary/default.asp. Accessed 11 March 2011
- Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Lacorte C (2006) Improving and assessing viral vectors for recombinant protein production in plants. Dissertation, Wageningen University
- Lehtinen A, Hannukkala A, Andersson B, Hermansen A, Le VH, Nærstad R, Brurberg MB, Nielsen BJ, Hansen JG, Yuen J. Phenotypic variation in Nordic populations of Phytophthora infestans in 2003. Plant Pathol. 2008;57:227–234. doi: 10.1111/j.1365-3059.2007.01739.x. [DOI] [Google Scholar]
- Li Y, Huang S, Zhang RF, Lan CZ, Yang YL, van der Lee T, Kessel TJT, Jacobsen E, Jin GH, Zhao ZJ, Kamoun S (2009) Allelic diversity of the population of Phytophthora infestans in China. In: Proceedings of the 3rd international late blight conference, Acta Hort 834:123–127
- Lokossou AA, Park T, Arkel G, Arens M, Ruyter-Spira C, Morales J, Whisson SC, Birch PRJ, Visser RGF, Jacobsen E, van der Vossen EAG. Exploiting knowledge of R/Avr genes to rapidly clone a new LZ-NBS-LRR family of late blight resistance genes from potato linkage group IV. MPMI. 2009;22:630–641. doi: 10.1094/MPMI-22-6-0630. [DOI] [PubMed] [Google Scholar]
- Lokossou AA, Rietman H, Wang MQ, Krenek P, Schoot H, Henken B, Hoekstra R, Vleeshouwers VGAA, van der Vossen EAG, Visser RGF, Jacobsen E, Vosman B. Diversity, distribution, and evolution of Solanum bulbocastanum late blight resistance genes. MPMI. 2010;23:1206–1216. doi: 10.1094/MPMI-23-9-1206. [DOI] [PubMed] [Google Scholar]
- Oh SK, Young C, Lee M, Oliva R, Bozkurt TO, Cano LM, Win J, Bos JIB, Liu HY, Damme M, Morgan W, Choi D, van der Vossen EAG, Vleeshouwers VGAA, Kamoun S. In planta expression screens of Phytophthora infestans RXLR effectors reveal diverse phenotypes, including activation of the Solanum bulbocastanum disease resistance protein Rpi-blb2. Plant Cell. 2009;21:2928–2947. doi: 10.1105/tpc.109.068247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pel MA (2010) Mapping, isolation and characterization of genes responsible for late blight resistance in potato. Dissertation, Wageningen UR
- Pel MA, Foster SJ, Park TH, Rietman H, van Arkel G, Jones JDG, Van Eck HJ, Jacobsen E, Visser RGF, Van der Vossen EAG. Mapping and cloning of late blight resistance genes from Solanum venturii using an interspecific candidate gene approach. MPMI. 2009;22:601–615. doi: 10.1094/MPMI-22-5-0601. [DOI] [PubMed] [Google Scholar]
- Rausher MD. Co-evolution and plant resistance to natural enemies. Nature. 2001;411:857–864. doi: 10.1038/35081193. [DOI] [PubMed] [Google Scholar]
- Rosenthal GA, Dahlman DL, Janzen DH. A novel means for dealing with L-canavanine, a toxic metabolite. Science. 1976;192:256–258. doi: 10.1126/science.1257764. [DOI] [PubMed] [Google Scholar]
- Schmidt MA, LaFayette PR, Artelt BA, Parrott WA. A comparison of strategies for transformation with multiple genes via microprojectile-mediated bombardment. In Vitro Cell Dev Biol Plant. 2008;44:162–168. [Google Scholar]
- Sharma KK, Bhatnagar PB, Thorpe TA. Genetic transformation technology: status and problems. In Vitro Cell Dev Biol Plant. 2005;41:102–112. doi: 10.1079/IVP2004618. [DOI] [Google Scholar]
- Song JQ, Bradeen JM, Naess SK, Raasch JA, Wielgus SM, Haberlach GT, Liu J, Kuang HH, Austin-Phillips S, Buell CR, Helgeson JP, Jiang JM. Gene RB cloned from Solanum bulbocastanum confers broad spectrum resistance to potato late blight. PNAS. 2003;100:9128–9133. doi: 10.1073/pnas.1533501100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staples RC. Race nonspecific resistance for potato late blight. Trends Plant Sci. 2004;9:5–6. doi: 10.1016/j.tplants.2003.10.009. [DOI] [PubMed] [Google Scholar]
- Staskawicz BJ, Ausubel FM, Baker BJ, Ellis JG, Jones JDG. Molecular genetics of plant disease resistance. Science. 1995;268:661–667. doi: 10.1126/science.7732374. [DOI] [PubMed] [Google Scholar]
- Trognitz F, Trognitz B (2004) Mapping genes of Solanum caripense involved in resistance to Phytophthora infestans, the causal agent of potato late blight. Genetic variation for plant breeding. In: Proceedings of the 17th Eucarpia general congress, Tulln, Austria, pp. 8–11
- van der Vossen E, Sikkema A, Hekkert BL, Gros J, Stevens P, Muskens M, Wouters D, Pereira A, Stiekema W, Allefs S. An ancient R gene from the wild potato species Solanum bulbocastanum confers broad spectrumresistance to Phytophthora infestans in cultivated potato and tomato. Plant J. 2003;36:867–882. doi: 10.1046/j.1365-313X.2003.01934.x. [DOI] [PubMed] [Google Scholar]
- van der Vossen E, Gros J, Sikkema A, Muskens M, Wouters D, Wolters P, Pereira A, Allefs S. The Rpi-blb2 gene from Solanum bulbocastanum is an Mi-1 gene homolog conferring broad spectrum late blight resistance in potato. Plant J. 2005;44:208–222. doi: 10.1111/j.1365-313X.2005.02527.x. [DOI] [PubMed] [Google Scholar]
- van Poppel PMJA, Guo J, van de Vondervoort PJI, Jung MWM, Birch PRJ, Whisson SC, Govers F. The Phytophthora infestans avirulence gene Avr4 encodes an RXLR-dEER effector. MPMI. 2008;21:1460–1470. doi: 10.1094/MPMI-21-11-1460. [DOI] [PubMed] [Google Scholar]
- Verzaux E (2010) Resistance and susceptibility to late blight in Solanum: gene mapping, cloning and stacking. Dissertation, Wageningen University
- Vleeshouwers VGAA, van Dooijeweert W, Keizer LCP, Sijpkes L, Govers F, Colon LT. A laboratory assay for Phytophthora infestans resistance in various Solanum species reflects the field situation. Eur J Plant Pathol. 1999;105:241–250. doi: 10.1023/A:1008710700363. [DOI] [Google Scholar]
- Vleeshouwers VGGA, Rietman H, Krenek P, Champouret N, Young C, Oh SK, Wang MQ, Bouwmeester K, Ben O, Visser RGF, Jacobsen E, Govers F, Kamoun S, van der Vossen EAG. Effector genomics accelerates discovery and functional profiling of potato disease resistance and Phytophthora infestans avirulence genes. PLOS ONE. 2008;3:e2875. doi: 10.1371/journal.pone.0002875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XZ, Kim B. Physiological races of Phytophthora infestans in Korea. Plant Pathol J. 2007;23:219–222. doi: 10.5423/PPJ.2007.23.3.219. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.