Abstract
The development and testing in the field of genetically modified -so called- orphan crops like cassava in tropical countries is still in its infancy, despite the fact that cassava is not only used for food and feed but is also an important industrial crop. As traditional breeding of cassava is difficult (allodiploid, vegetatively propagated, outbreeding species) it is an ideal crop for improvement through genetic modification. We here report on the results of production and field testing of genetically modified low-amylose transformants of commercial cassava variety Adira4 in Indonesia. Twenty four transformants were produced and selected in the Netherlands based on phenotypic and molecular analyses. Nodal cuttings of these plants were sent to Indonesia where they were grown under biosafety conditions. After two screenhouse tests 15 transformants remained for a field trial. The tuberous root yield of 10 transformants was not significantly different from the control. Starch from transformants in which amylose was very low or absent showed all physical and rheological properties as expected from amylose-free cassava starch. The improved functionality of the starch was shown for an adipate acetate starch which was made into a tomato sauce. This is the first account of a field trial with transgenic cassava which shows that by using genetic modification it is possible to obtain low-amylose cassava plants with commercial potential with good root yield and starch quality.
Electronic supplementary material
The online version of this article (doi:10.1007/s11248-011-9507-9) contains supplementary material, which is available to authorized users.
Keywords: Agrobacterium transformation, Cassava, Field trial, Low-amylose starch, RNAi granule-bound starch synthase construct
Introduction
Cassava (Manihot esculenta Crantz) is considered one of the most important food crops in the tropical areas including Asia, Africa and Latin America. The tuberous roots are rich in carbohydrates, which can constitute up to 35% of the total root weight on a wet basis (Onwueme and Charles 1994). In 2009 242 million tonnes have been harvested (FAO 2009). Cassava is mainly used for human consumption or as a source of animal feed (El-Sharkawy 2004). Increasingly, cassava is used for the production of starch as a source of industrial raw material (Sriroth et al. 2001). With the economic development in many developing countries the demand for starch for industrial purposes—such as in paper and textile—will increase.
As a ‘crisis crop’, cassava plants require few inputs (Cock 1982). The tuberous roots can be left in the ground for well over 1 year and harvested when food shortages arise or when prices of other starch crops become prohibitive. Despite these advantages there are traits that need improvement, such as pest and disease susceptibilities, cyanogenic potential, and post-harvest physiological deterioration (Ceballos et al. 2004; Thro et al. 1998). Due to the high heterozygosity, allopolyploid nature and low natural fertility of the crop traditional breeding of cassava is difficult (Ceballos et al. 2004). Biotechnology has been recognized as an alternative approach for agronomic improvement (Taylor et al. 2004a, b). Genetic transformation of cassava has been described 15 years ago by three independent research groups (Li et al. 1996; Raemakers et al. 1996; Schöpke et al. 1996). Since then, numerous reports have been published with incremental improvements of the transformation protocols. However, none of them have tested the agronomic performance of resulting transformants under field conditions.
Previously, we reported the development of transformants of the Nigerian cassava variety TMS60444 producing amylose-free starch through antisense inhibition of the gene encoding granule bound starch synthase I (GBSSI, Raemakers et al. 2005). Starch consists of two glucan polymers: amylose and amylopectin. Amylopectin is a branched molecule consisting of a linear α-1,4 glucan chain linked by α-1,6 bonds, whereas amylose is essentially a linear α-1,4 glucan chain containing few branches. Because of this difference in structure amylopectin is extremely soluble in water, whereas amylose has a strong tendency to recrystallize after dispersion in water. This so-called retrogradation is undesirable for many applications of starch in which a defined and stable viscosity for hours or days is required. Therefore, for industrial purposes starch is often treated with chemicals in order to make the amylose less sensitive to crystallization (Bruinenberg et al. 1995). As retrogadation is caused mainly by the amylose fraction in the starch amylose-free starches do not have to be treated with chemicals. Indeed, it was shown that amylose-free cassava starch has an increased clarity and stability of the starch gel (Raemakers et al. 2005; Sánchez et al. 2010). These improved functionalities are desired for technical applications in paper and textile, but also in the food industry for the production of dairy products, soups, sauces and noodles.
The full advantage of starch with altered properties from genetically modified plants can only be assessed when the yields of these plants are comparable to those of the untransformed control plants. This paper deals with the agronomic aspects of transgenic cassava plants. To this end an important Indonesian cultivar—Adira4—was transformed with an RNAi GBSSI contruct to obtain transformants producing amylose-free starch. From the obtained transformants 24 low-amylose, backbone DNA-free and low T-DNA copy number events were selected to undergo field testing in Indonesia. All experiments were conducted under permit of the Indonesian Committee for Biosafety and Food Safety. This meant that plants were first tested in green- and screenhouses and subsequently tested in the field. The agronomic performance of the transformants in the field as well as the characteristics of the altered starch are described. This is the first account of the production and agronomic evaluation of low-amylose transformants of a commercially important industrial cassava genotype.
Materials and methods
Construct for transformation
RNAi construct p5IRTCGBa (Fig. 1a) used for transformation contained an inverted repeat of part of the cassava GBSSI gene (Salehuzzaman et al. 1993). The inverted repeat sequence was a 621-bp fragment from the 5′ part of the cassava GBSSI cDNA (from base 175 to base 795 from Genbank sequence X74160). The spacer consisted of a 150 bp fragment from the 5′ part of the cassava GBSSI cDNA (from base 25 to base 174 from Genbank sequence X74160). The GBSSI inverted repeat construct was driven by the potato GBSSI promoter (Visser et al. 1991) and terminated by the nopaline synthase (NOS) terminator. The plasmid contained the bar gene (Strauch et al. 1988) as selectable marker gene and to obtain herbicide (Basta or phosphinothricin) resistance. The bar gene was cloned between the CaMV 35S promoter (Benfey et al. 1990) and the NOS terminator. Construct p5IRTCGBa was made as follows. Binary vector pKGBA50mf-IR1.1 (de Vetten et al. 2003) was digested with BamHI and subsequently self-ligated, removing the potato GBSSI inverted repeat. The T-DNA of the resulting vector pPGBmf only contained the potato GBSSI promoter and the NOS terminator separated by a polylinker containing a BamHI site. The gene cassette CaMV 35S promoter—bar gene—NOS terminator was released from plasmid pDE110 (Van Breusegem et al. 1999) by EcoRI/HindIII digestion and cloned into EcoRI/HindIII-digested plasmid pAAP172 (patent WO03010319). The BamHI site between the bar coding sequence and the NOS terminator was removed by digestion with BamHI, blunting the sticky ends by Klenow treatment, and ligation of the blunt ends. Subsequently, the bar gene cassette was released from this plasmid by NdeI digestion and cloned into VspI-digested pPGBmf, resulting in vector pPGBbar2. Two PCR products were obtained with primers 5CASGBF1 (5′-TAGAATTCACCAGCGGAACCTATTTT-3′) + 5CASGBR1 (5′-TGTCTAGAATGGAAGCAGAGCAGTGT-3′) and 5CASGBF2 (5′-ATGAATTCGGACCCAAACTATCACTC-3′) + 5CASGBR1 using cassava GBSSI cDNA from plasmid pG61 (Salehuzzaman et al. 1993) as template. The forward primers contain an EcoRI site at the 5′ end, whereas the reverse primer contains an XbaI site at the 5′ end. The two PCR fragments were cloned into pGEM-T (Promega), and subsequently released by EcoRI/XbaI digestion. Together these fragments were ligated into XbaI-digested pMTL24 (Chambers et al. 1988), resulting in an inverted repeat of the 5′ part of the cassava GBSSI cDNA. The inverted repeat was released from pMTL24 by BamHI digestion and cloned into BamHI-digested pPGBbar2. The two resulting vectors p5IRTCGBa and p5IRTCGBb differed with respect to the orientation of the spacer sequence. Only vector p5IRTCGBa was used for transformation. It was transferred to Agrobacterium tumefaciens strain AGL0 (Lazo et al. 1991) by electroporation.
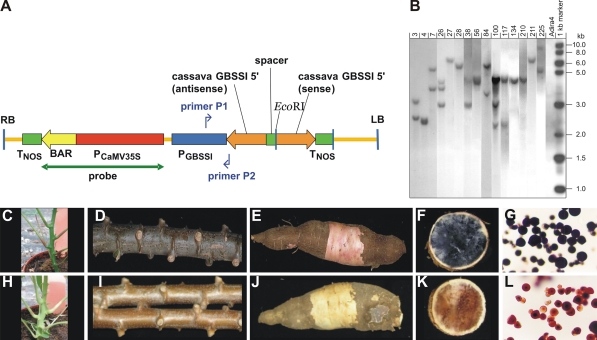
Fig. 1.
a Schematic representation of the T-DNA of vector p5IRTCGBa (5,876 bp) in which primers for PCR analysis and the probe for Southern blot analysis are indicated. b Southern blot of DNA of 15 transformants used in the field trial and of untransformed Adira4, hybridized with the probe indicated in (a) and depicting T-DNA insertion number. c Young plant, d stem, e tuberous root, f tuberous root cross section, and g starch granules of the untransformed Adira4 control. h Young plant, i stem, j tuberous root, k tuberous root cross section, and l starch granules of a low-amylose transformant
Plant material, transformation and regeneration
Three months-old friable embryogenic callus (FEC) cultures initiated on meristems of cassava cultivar Adira4 (Anggraini et al. 2009) were used for transformation. FEC was obtained as described by Raemakers et al. (1997). Proliferation of 10 individual FEC lines took place on ½GD4 medium (medium containing half the concentration of Gresshoff and Doy (1972) salts and vitamins, and half the concentration of Murashige and Skoog (1962) salts and vitamins, supplemented with 40 g/l sucrose), with 10 mg/l Picloram and 10 mg/l micro agar. FEC was inoculated with an overnight culture of Agrobacterium tumefaciens AGL0 containing the plasmid p5IRTCGBa. Agrobacterium inoculation was done on solid and in liquid medium, but FEC inoculated on solid medium was the only method which resulted in transformed calli. A clump of approximately 200 mg of FEC was placed on a membrane filter on MS4 medium (medium with MS salts and vitamins (Murashige and Skoog 1962) and 40 g/l sucrose) with 10 mg/l Picloram. Fifty μl of an overnight Agrobacterium culture was added to the callus and cocultivated at 28°C for 2 days. Per FEC line ten petridishes were inoculated. After cocultivation, FEC was cultured in liquid SH6 medium (medium with Schenk and Hildebrandt (1972) salts and vitamins, and 60 g/l sucrose) plus 10 mg/l picloram and 200 mg/l claforan during 1 week.
After this week FEC was collected and spread on solid (10 mg/l agar) ½GD4 medium with 10 mg/l picloram, 200 mg/l claforan and 1 mg/l phosphinothricin (PPT) for selection. Four to six weeks after Agrobacterium inoculation colonies appeared. Individual colonies were selected and considered independent transformation events. Samples of grown colonies were analyzed by PCR to confirm that they were transgenic. Subsequently, the colonies were propagated on medium containing 5 mg/l PPT. Surviving colonies were propagated on medium containing 10 mg/l PPT. Finally, plant regeneration was induced as described by Raemakers et al. (1997).
Acclimatization of in vitro grown plants in the greenhouse
The transgenic in vitro plants were acclimatized in the greenhouse in small pots covered by plastic to create a 100% humidity situation, which is comparable to the in vitro situation. Perlite was mixed into the soil to prevent dying off of the in vitro plants because of excess water and concomitant fungal diseases in the high humidity conditions. Additionally, after planting the plants were sprayed with Admire (0.03 g/l, Bayer, Germany) against larvae of gnats (Sciara prothalliorum).
DNA analyses
All obtained in vitro plants were analyzed via PCR with forward primer P1 5′- GAGGGAGTTGGTTTAGTTTTTAGA-3′ (annealing to the potato GBSSI promoter sequence) and reverse primer P2 5′-ACACTGCTCTGCTTCCATTCT-3′ (annealing to the cassava GBSSI 5′ antisense sequence) to detect the presence of the inverted repeat construct (Fig. 1a). For the PCR one μg of DNA and 50 ng of primers were added to a standard SuperTaq PCR buffer containing MgCl2 in a final concentration of 1.5 mM. One unit of SuperTaq Polymerase (SphaeroQ) was added. The total PCR reaction volume was 25 μl. The PCR reaction consisted of 4 min at 94°C, 35 cycles of (30 s at 94°C, 30 s at 56°C, 1 min at 72°C), and finally 7 min at 72°C in a Perkin Elmer 9,600 thermocycler. PCR products were fractionated by electrophoresis in a 1.2% (w/v) agarose gel for detection of the 349-bp amplification fragment.
Genomic DNA was isolated from young leaves from in vivo plants using the method described by Rogers and Bendich (1988). PCRs were performed with primers for the nptIII, insB, tetR, and trfA DNA sequence in pBIN19 (Wolters et al. 1998) to analyze the presence of backbone-vector DNA in the transformed plants. The genomic DNA was digested with EcoRI, fragments were separated using gel electrophoresis, and the digested DNA was blotted on a positively charged nylon membrane (Hybond N+, Amersham Biosiences) using standard techniques (Sambrook et al. 1989). The Southern blot was hybridized with a probe (Fig. 1a) consisting of the 2-kb EcoRI/BamHI fragment of CaMV 35S promoter/bar gene cassette of pDE110 (Van Breusegem et al. 1999) labeled with 32P-dCTP using the Rediprime II kit (Amersham) to check the number of T-DNA insertions. After stripping this same blot was hybridized with an 8.0-kb BglII fragment of pBIN19 to analyze the presence or absence of backbone-vector DNA.
Plant tests for agronomical performance
Starch granules from tuberous roots produced in the greenhouse in the Netherlands were stained with LUGOL solution (diluted 1:3 with water). Granules from the untransformed control plants showed blue staining, whereas the granules from transformants showed a range of staining patterns from completely red to completely blue. Transformants showing completely red staining starch granules with a low T-DNA copy number (three or less insertions) and no backbone-vector DNA (24 in total) were selected for propagation by nodal cuttings. Stakes from these plants and from a control untransformed plant were shipped to Bogor in Indonesia where they were planted in biosafety containment for adaptation. After propagation, cuttings were planted in a screenhouse. All transformants grown in this first screenhouse test, except one showing poor growth, were planted again in a second screenhouse test. After this test transformants showing poor growth or instability of the low-amylose phenotype were discarded. The 15 remaining transformants were tested in a field trial. The transformants and untransformed Adira4 control were grown in blocks of 20 plants per transformation event. Planting was performed in ridges with a spacing of 1 m between plants. The distance between ridges was also 1 m. During the growing season, plants were scored for various plant characteristics such as the number of leaves, color of the petiole, color of the stem, number of nodes till appearance of the flower branch. The height of the stems was measured from soil surface to the stem apex. Tuberous roots were harvested after 10 months. The following root characteristics were determined: total weight of roots, total number of roots, underwater weight (measure for starch content, see http://www.starch.dk/isi/methods/13starch.htm), and color of the inner cortex. The field trials were carried out according to the guidelines of the Indonesian Committee for Biosafety and Food Safety and appropriate procedures were followed. One-way ANOVA was used to analyze the statistical differences between agronomic performance data of transformants and untransformed controls. A P-value < 0.05 was considered significant.
Starch isolation and determination of amylose content
Cassava tuberous roots were peeled and homogenized in a coconut grinder directly after harvest. The homogenate was sieved through a 250-μm mesh to remove particulate material. The starch sediment was washed several times with water, and finally air-dried at room temperature for several days. The apparent amylose content was measured with a spectrophotometer according to the method described by Hovenkamp-Hermelink et al. (1998). For amylose-free starch or pure amylopectin an apparent amylose content of 4% is obtained with this method (Kuipers et al. 1992; Raemakers et al. 2005). Therefore, transformants producing starches with an amylose content below 4% were considered amylose-free. Transformants producing starches with an amylose content below 13% were termed low-amylose.
Preparation of tomato sauce
For tomato sauce adipate-acetate starches were prepared. Reactions were carried out with 1.25 mol of starch (202.5 gram dry matter) of untransformed cassava, amylose-free cassava, untransformed potato, amylose-free potato, amylose-free wheat and amylose-free maize. A 39% suspension was prepared and the pH was adjusted to 8.0 with a 4.4% (w/w) NaOH solution at a temperature of 23°C. For steeping 30% H2O2 was added and the suspension was stirred for 10 min at a constant pH. The pH was adjusted to 8.5 with a 4.4% (w/w) NaOH solution and 2 gram 10% (w/w) adipic anhydride was added drop wise in 10 min maintaining the pH at 8.5. Then 9.75 grams of acetic anhydride was added drop wise in 20 min and the pH was kept constant at 8.5 again. Afterwards the suspension was neutralized to pH 5.2 with 10 N H2SO4 and the suspension was dewatered over a Büchner funnel. Then the filter cake was washed with 1 l water. The final product was dried in a stove at 35°C. A mixture was prepared of 2.7% (w/w) starch of the above-mentioned origins (calculated on 17% moisture), 22.5% tomato paste (30 brix), 17% sugar, 8.8% vinegar (8% acetic acid), 2.7% salt, 0.6% potassium sorbate and 45.7% water. Ingredients were mixed in a beaker and stirred for 2 min. Then the mix was poured into the Brabender bowl. The mix was heated according to the following heating profile: 50°C → 95°C → 65°C at a rate of 1.5°C/min with a torque of 700 cm/g and a speed of 75 rpm. Heating from 50 to 95°C took 30 min, then the mix was kept at 95°C during 10 min, and subsequently the mix was cooled down to 65°C during 20 min. At the end of the cycle, the tomato sauces were poured in beakers and cooled down in the refrigerator. Appearance, pulpiness and texture were visually and sensory evaluated by an experienced person.
Results and discussion
Cloning of an RNAi GBSSI construct for improved silencing efficiency
Previously, we reported the production of amylose-free cassava transformants by transformation of Nigerian cultivar TMS60444 with an antisense GBSSI construct (Raemakers et al. 2005). For commercial production of low-amylose starch in Indonesia we set out to transform Indonesian cassava cultivar Adira4. As we expected that transformation efficiency of this cultivar might be lower than for TMS60444—for which the transformation protocol was optimized—we first made an inverted repeat construct that would induce silencing of the endogenous GBSSI gene more efficiently than the previously used antisense construct. However, testing of several constructs in cassava to find the most efficient one is difficult because of the variability among experiments and the long time required before a phenotype can be scored. Therefore, we based our strategy on results obtained in potato by Heilersig et al. (2006). In this study silencing efficiency of several inverted repeat constructs of the potato GBSSI gene were compared. They observed that constructs with inverted repeats of 500–600 bp from the 5′ or middle part of the GBSSI gene, with a spacer sequence of ±150 bp, induced silencing most efficiently. Although results obtained in one species do not necessarily hold true for other species, we assumed that because of the high homology between the coding sequences of potato and cassava GBSSI genes an inverted repeat of ±600 bp of the 5′ part of the cassava GBSSI gene would result in more efficient silencing than the antisense construct described in Raemakers et al. (2005). We made construct p5IRTCGBa, which contained an inverted repeat of cassava GBSSI driven by the potato GBSSI promoter, and a bar gene cassette as selectable marker (growth on PPT-containing medium) and to provide herbicide (Basta) resistance in the field.
Transformation and selection
Ten friable embryogenic callus (FEC) lines derived from Indonesian cassava cultivar Adira4 were inoculated with Agrobacterium tumefaciens strain AGL0 containing the GBSSI cassava inverted repeat construct p5IRTCGBa (Fig. 1a). After PCR analysis of colonies growing on 1 mg/l PPT, PCR-positive calli were obtained from three out of the ten FEC lines. Calli from the other seven lines showed reduced growth, and did not yield PCR-positive colonies. Despite years of experience in transforming cassava (Raemakers et al. 1993a, b, 2001, 2005) it is still very difficult to combine optimum growth conditions of the FEC lines with optimal A. tumefaciens receptiveness.
Transformation of one FEC line (#1) was very successful and yielded more than 200 PCR-positive colonies. In total around 300 colonies from the three FEC lines were transferred to medium with 5 mg/l PPT. After a third selection round on 10 mg/l PPT approximately 200 colonies of the three FEC lines remained, which were transferred to regeneration medium. Each colony was regarded as a derivative from a unique transformation event.
Regeneration
From 157 colonies (150 from FEC line #1, 4 from FEC line #2 and 3 from FEC line #3) plant regeneration was successful, which resulted in a total of 588 plants. It was relatively easy to regenerate the embryogenic callus from FEC line #1. Within 3 months, plants were obtained from 40% of the colonies. Regeneration of plants from FEC lines #2 and #3 proved to be more difficult. The induction of callus of all three lines took place in the same way around the same time and we could not discern visible differences among the lines. Differences in regeneration potential among FEC lines have been described before by Schöpke et al. (1996) and Raemakers et al. (2006). Vanderschuren et al. (2007) reported that from more than 200 hygromycin resistant embryos only three developed into plants, although they used cassava genotype TMS60444, which is known as an easy-to-transform genotype. Similarly, Ihemere et al. (2006) regenerated plants from only 26 out of 872 paromomycin resistant embryos from cassava genotype TMS71173.
Plant development
Several (1–6) plants per transformed colony were regenerated to increase the chance of obtaining normal growing plants. Because not all regenerants grew equally well, they were categorized as N (normal, well-growing plants), S (small [maximum length 2 cm], weakly growing but with normal leaf morphology) and A (aberrant leaves, weakly growing). 65% of the transformants belonged to the N-group, 28% to the S-group and 8% to the A-group. After two rounds of in vitro propagation 71% of the plants produced roots, of which 56% came from the N-group. Rooted plants from all three groups were transferred to the greenhouse in the Netherlands (Fig. 2a, b). Plants derived from 131 of the 157 transformants survived this transfer. As expected, the highest survival rate was obtained for the N-group of in vitro plants. After several weeks to a few months it was evident that plants from 24 transformants of the S- and A-group were lagging behind in growth compared to the rest. These were excluded, which resulted in 107 transformants with at least one well-growing plant per transformant.
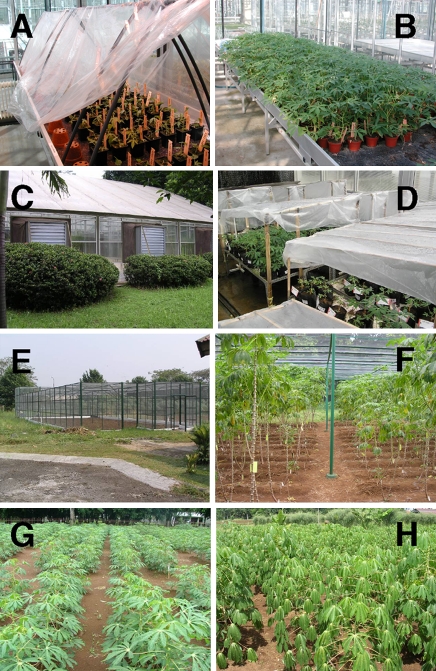
Fig. 2.
Growth of the transformants. a Acclimatization, and b young plants in the greenhouse (the Netherlands). Biosafety containment in Indonesia c outside and d inside the greenhouse. Screenhouse e outside and f inside. Field trial showing g young plants and h older plants
We observed that to a large extent in vitro growth was predictive for and related to growth in the greenhouse. The genotypes that failed to grow in the greenhouse were all from the S- and A-groups of in vitro plants. This means that it is most efficient to focus on the well-growing in vitro plants with normal morphology, i.e. similar to untransformed in vitro plants.
All genetically modified plants in the greenhouse obtained from FEC line #1 showed the same aberration (a difference in the color of young and old stems, and in the color of the inner cortex of the roots) compared to the untransformed Adira4 control obtained from an in vitro plant (Fig. 1c–e, h–j). Also untransformed regenerants from FEC line #1 showed this color variation, whereas transformed and untransformed regenerants from other FEC lines did not show this aberrant phenotype. Therefore, it is likely that the variation was induced during FEC induction, and is not the result of the transformation procedure. Color variation induced by tissue culture was also reported for maize by Rhee et al. (2009). They provided evidence that the variation was induced by transposable element activity. Possible, some genotypes of one species are more prone to somaclonal variation than other genotypes of the same species, as shown by genotype-specific responses after transformation and regeneration in a large study with fourteen potato varieties (Heeres et al. 2002).
Herbicide resistance
Plants were tested for resistance to the herbicide ‘Basta’ because of the presence of the bar-gene. Transformed and untransformed Adira4 plants were sprayed with 1 mg/l Basta (Bayer, Germany). After 1 week the stems of the untransformed Adira4 plants showed necrosis and leaf drop. The transformants displayed no or only slight damage. After 4 weeks it was obvious that plants from the untransformed Adira4 had died whereas the transformants grew well (Fig. S1).
Improved silencing efficiency of the RNAi construct
Starch was isolated from tuberous roots of greenhouse-grown plants and stained with LUGOL iodine solution. The starch granules were microscopically assessed for the presence or absence of amylose. Of the 107 transformants analyzed 41% showed partial or complete inhibition of GBSSI activity, as indicated by the red color of the starch granules instead of the blue color of the untransformed Adira4 genotype (Fig. 1f, g, k, l). This percentage is considerably higher than the 8% low-amylose plants obtained previously after transformation with a construct containing a full-length cassava GBSSI cDNA in antisense orientation (Raemakers et al. 2005), thus showing the superiority of the inverted repeat construct over the antisense construct.
Molecular analyses
Integration of backbone vector DNA into the genome of the host plant is reported to occur in 20–75% of the transformed plants (Kononov et al. 1997; Martineau et al. 1994). Therefore, selected transformants with low amylose content were analyzed for presence or absence of backbone vector DNA by PCR, using primers to four open reading frames of the pBIN19 vector: tetR, trfA, nptIII and insB (Wolters et al. 1998). Of the 44 tested low-amylose transformants 27 were negative for all four DNA fragments (61%). Southern blot hybridization of these 27 vector DNA-free transformants with the full pBIN19 backbone vector DNA confirmed the absence of vector DNA. Subsequently, the backbone vector DNA-free transformants were analyzed by Southern blot hybridization using the 35S-bar-gene as probe (Fig. 1a, RB probe). These analyses showed that 24 out of 27 transformants (90%) contained three or less T-DNA insertions. Moreover, 7 of these contained a single T-DNA insertion (see Fig. 1b). The entire selection process resulting in the transformants to be tested in Indonesia is shown in Fig. S2.
Agronomic evaluation in Indonesia
After adaptation in Biosafety containment in Bogor, twenty-four low-amylose, backbone vector DNA-free and low-copy number transformants were propagated in a screenhouse (1–16 plants per transformant) to produce stakes for subsequent field trials and to verify the amylose content previously determined using greenhouse-grown tuberous roots from the Netherlands (Fig. 2c–h). In the first screenhouse test the transgenic plants were smaller than the control plants and the weight of the roots was around 50% of the control. The control plants were untransformed plants of Adira4 obtained from in vitro culture. One transformant was excluded because of poor growth. After the second screenhouse test (up to 30 plants per transformant) 15 of the 23 transformants remained. Five were excluded because of instability of the low-amylose phenotype and three transformants because of poor growth. During the process of making FEC and later on during regeneration of the FEC, many hormones are added to the medium and somaclonal variation might occur (De Klerk 1990). During the initial propagation cycles in the green- and screenhouse or in the field, the effect of the stress from in vitro culture was still present. Several propagation cycles seemed to be necessary before a good field trial could be performed. After each propagation round, the transformants were more vigorous and more similar to the control plants. Also root weight of the transformants increased in subsequent harvests and approached the weight of the control plants (Fig. 3). All low-amylose transformants developed and matured in the same manner as the untransformed Adira4 variety.
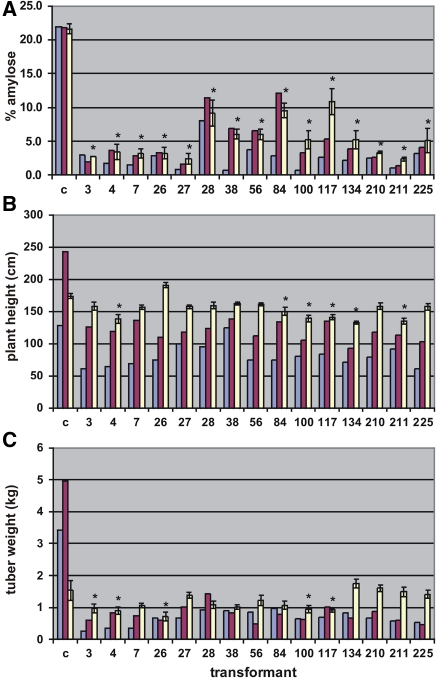
Fig. 3.
a Percentage amylose, b plant height and c tuberous root yield of 15 low-amylose transformants compared to the untransformed Adira4 (c) in respectively first screenhouse test (blue columns), second screenhouse test (red columns) and field trial (white columns, with standard deviation). Significant deviations (P < 0.05) from the control are indicated with an asterisk (*)
The general phenotype of all transformants was comparable to the untransformed Adira4 except for a slight difference in the color of the stem. After harvest of the roots, visual differences were also observed in the color of the inner cortex of the roots between the transformants and the untransformed Adira4, as was seen previously in the greenhouse experiments (Fig. 1c–e, h–j).
For all 15 low-amylose transformants and the untransformed Adira4, amylose content, height and tuberous root yield of each individual plant was determined in the screenhouse as well as in the field trial (Fig. 3). Although the results of the screenhouse and field tests are shown in one figure they are not directly comparable. The plants in the first screenhouse test were harvested after a shorter growing period than in the second screenhouse test. Nevertheless, Fig. 3 shows that the agronomic performance of the transformants improved after every propagation cycle. In the field trial, plots of 20 plants per transformant were used. Therefore, a reliable standard deviation could be given for these measurements and the significance could be calculated by an ANOVA test. Seven transformants showed a stable apparent amylose content of less than 4% after the three propagation cycles, and thus are considered amylose-free (Fig. 3a). In the field trial the average height of the transformants was calculated to range from 76 to 110%, when the control untransformed Adira4 was set at 100%. Six of the 15 transformants were significantly smaller than the untransformed Adira4 (Fig. 3b). Relative yields of the low-amylose transformants were between 47 and 115% of the control. The majority of the transformants (10) was not significantly different from the control with respect to root yield (Fig. 3c). This indicates that no clear yield effect can be attributed to the low-amylose phenotype or to the transformation procedure.
Re-evaluating the entire transformation and selection procedure starting with transformants generated from 157 colonies, 107 grew to normal plants in the greenhouse, of which 44 transformants had a silenced phenotype (41% of the transformants). Of these 44 low-amylose transformants 27 were backbone vector DNA-free (61%). Moreover, of the 27 transformants analyzed 7 contained a single T-DNA insertion. Five of the 7 single-copy, low-amylose, backbone vector DNA-free transformants showed no significant negative effect on yield. Finally, 3 of these transformants were considered amylose-free (apparent amylose content below 4%).
Thus, it is demonstrated that it is feasible to obtain single-copy, backbone vector DNA-free transformants of a commercial cassava cultivar, in which an endogenous gene is completely silenced, and showing a good agronomic performance.
Starch and product evaluations
Cassava starch is used in a wide variety of food and industrial applications. In food applications, starch is used to provide such characteristics as texture, moisture, consistency and shelf stability. To meet the demanding needs of the food industry, starch is often modified to change the behavioural characteristics from that of unmodified starch (Singh et al. 2007). One frequently used modification results in di-starch adipates, prepared by cross-linking native starch with adipic and acetic anhydride reagents. Cross-linking acts to strengthen the starch granules during the process of cooking and prevents changing of the texture into an elastic rubbery state. In general the food industry requires starch pastes with a so-called short, smooth or creamy texture and a shiny appearance for its use in desserts, sauces or soups.
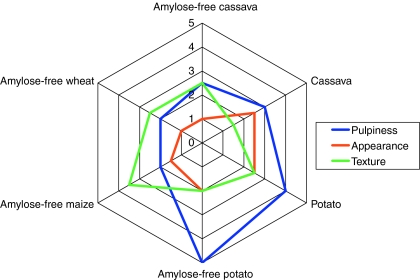
Here, we tested low-amylose cassava starch for its textural behaviour in comparison with normal cassava starch and starches from other botanical origins—regular and amylose-free variants of potato starch, amylose-free wheat and maize starch—in a typical tomato sauce preparation which is representative for soups and sauces (Fig. 4). The amylose-free cassava starch of Adira4 had the same properties as amylose-free cassava starch of genotype TMS60444 (Raemakers et al. 2005), such as a higher T onset and more extreme swelling behaviour than normal cassava starch, and absence of retrogradation. The cross-linked derivative of low-amylose cassava starch gave a smooth, short and shiny texture when it was cooked. A similar derivative of normal cassava starch does not have these features, in that the solution showed a dull appearance. Potato starch, both the amylose-free and amylose-containing, has in general a pulpy mouth feel. In many applications such as dressings, fruit fillings and puddings the pulpy texture is not desired while in other applications such as tomato sauce it is most often preferred. The waxy starches of cereals (amylose-free wheat and maize) are most comparable to amylose-free cassava starch, which is as expected since they have similar small granule sizes. However, cereal starches have the drawback that the pastes are blind (not clear as paste), due to residual fat and protein in the starch.
Fig. 4.
Organoleptic properties of amylose-free cassava starch compared to starches of other botanical origins. Pulpiness: 1 = smooth and 5 = pulpy. Appearance: 1 = shiny and 5 = dull. Texture: 1 = short and 5 = long. For each property 1 is most preferable and 5 is least preferable
Furthermore, for foodstuff, it is in general desirable that a starch is bland or neutral in flavour when used in desserts, sauces, soup-mixes, pie fillings and dressings. The starches generally having the most neutral taste are the non-cereal starches derived from potato or cassava, when compared to starches from cereals such as corn, wheat, rice and waxy variants thereof which, when incorporated into food, give some undesirable flavours to the food.
These results suggest that low-amylose cassava starch has many advantages in its use in food applications having the desired texture and/or organoleptic properties. In view of a worldwide production of roughly 900.000 tonnes of waxy maize starch which is nowadays, despite its shortcomings, the preferred starch for these kinds of products, low-amylose cassava starch could be an important development to benefit the cassava farmers within developing countries.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- bar
Basta-resistance gene
- FEC
Friable Embryogenic Callus
- GBSSI
Granule-bound starch synthase I
- GD
Gresshoff and Doy medium
- LB
Left border
- MS
Murashige and Skoog medium
- NOS
Nopaline synthase
- PPT
Phosphinothricin
- RB
Right border
- SH
Schenk and Hildebrandt medium
Contributor Information
H. J. J. Koehorst-van Putten, Email: herma.koehorst@wur.nl
E. Sudarmonowati, Email: s_enny@hotmail.com
M. Herman, Email: mherman@indo.net.id
I. J. Pereira-Bertram, Email: isolde.pereira@wur.nl
A. M. A. Wolters, Email: anne-marie.wolters@wur.nl
H. Meima, Email: Henk.Meima@AVEBE.COM
N. de Vetten, Email: Nick.de.Vetten@AVEBE.COM
C. J. J. M. Raemakers, Email: c.j.j.m.raemakers@genetwister.nl
R. G. F. Visser, Phone: +31-317-482857, FAX: +31-317-483457, Email: richard.visser@wur.nl
References
- Anggraini V, Sudarmonowati E, Hartati NS, Suurs L, Visser RGF. Characterization of cassava starch attributes of different genotypes. Starch/Stärke. 2009;61:472–481. doi: 10.1002/star.200800121. [DOI] [Google Scholar]
- Benfey PN, Ren L, Chua NH. Tissue-specific expression from CaMV 35S enhancer subdomains in early stages of plant development. EMBO J. 1990;9:677–1684. doi: 10.1002/j.1460-2075.1990.tb08291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruinenberg PM, Jacobsen E, Visser RGF. Starch from genetically engineered crops. ChemInd. 1995;95:881–884. [Google Scholar]
- Ceballos H, Iglesias CA, Pérez JC, Dixon AGO. Cassava breeding: opportunities and challenges. Plant Mol Biol. 2004;56:503–516. doi: 10.1007/s11103-004-5010-5. [DOI] [PubMed] [Google Scholar]
- Chambers SP, Prior SE, Barstow DA, Minton NP. The pMTL nic− cloning vectors. Improved pUC polylinker regions to facilitate the use of sonicated DNA for nucleotide sequencing. Gene. 1988;68:139–149. doi: 10.1016/0378-1119(88)90606-3. [DOI] [PubMed] [Google Scholar]
- Cock JH. Cassava: a basic energy source in the tropics. Science. 1982;218:755–762. doi: 10.1126/science.7134971. [DOI] [PubMed] [Google Scholar]
- De Klerk GJ. How to measure somaclonal variation. Acta Bot Neerl. 1990;39:129–144. [Google Scholar]
- De Vetten N, Wolters AM, Raemakers K, van der Meer I, ter Stege R, Heeres E, Heeres P, Visser R. A transformation method for obtaining marker-free plants of a cross-pollinating and vegetatively propagated crop. Nature Biotechnol. 2003;21:439–442. doi: 10.1038/nbt801. [DOI] [PubMed] [Google Scholar]
- El-Sharkawy MA. Cassava biology and physiology. Plant Mol Biol. 2004;56:481–501. doi: 10.1007/s11103-005-2270-7. [DOI] [PubMed] [Google Scholar]
- FAO (2009) Food Outlook, global market analysis, December 2009. GIEWS, p 26 http://www.fao.org/docrep/012/ak341e/ak341e06.htm
- Gresshoff PM, Doy CH. Development and differentiation of haploid Lycopersicon esculentum (tomato) Planta. 1972;107:161–170. doi: 10.1007/BF00387721. [DOI] [PubMed] [Google Scholar]
- Heeres P, Schippers-Rozenboom M, Jacobsen E, Visser RGF. Transformation of a large number of potato varieties: genotype-dependent variation in efficiency and somaclonal variability. Euphytica. 2002;124:13–22. doi: 10.1023/A:1015689112703. [DOI] [Google Scholar]
- Heilersig HJB, Loonen A, Bergervoet M, Wolters AMA, Visser RGF. Post-transcriptional gene silencing of GBSSI in potato: effects of size and sequence of the inverted repeats. Plant Mol Biol. 2006;60:647–662. doi: 10.1007/s11103-005-5280-6. [DOI] [PubMed] [Google Scholar]
- Hovenkamp-Hermelink JHM, De Vries JN, Adamse P, Jacobsen E, Witholt B, Feenstra WJ. Rapid estimation of the amylose/amylopectin ratio in small amounts of tuber and leaf tissue of the potato. Potato Res. 1998;31:241–246. doi: 10.1007/BF02365532. [DOI] [Google Scholar]
- Ihemere U, Arias-Garzon D, Lawrence S, Sayre R. Genetic modification of cassava for enhanced starch production. Plant Biotechnol. 2006;4:453–465. doi: 10.1111/j.1467-7652.2006.00195.x. [DOI] [PubMed] [Google Scholar]
- Kononov ME, Bassuner B, Gelvin SB. Integration of T-DNA binary vector ‘backbone’ sequences into the tobacco genome: evidence for multiple complex patterns of integration. Plant J. 1997;11:945–957. doi: 10.1046/j.1365-313X.1997.11050945.x. [DOI] [PubMed] [Google Scholar]
- Kuipers GJ, Vreem JTM, Meyer H, Jacobsen E, Feenstra WJ, Visser RGF. Field evaluation of antisense RNA mediated inhibition of GBSS gene expression in potato. Euphytica. 1992;59:83–91. [Google Scholar]
- Lazo GR, Stein PA, Ludwig RA. A DNA transformation-competent Arabidopsis genomic library in Agrobacterium. Bio/Technology. 1991;9:963–967. doi: 10.1038/nbt1091-963. [DOI] [PubMed] [Google Scholar]
- Li HQ, Sautter C, Potrykus I, Puonti-Kaerlas J. Genetic transformation of cassava (Manihot esculenta Crantz) Nature Biotechnol. 1996;14:736–740. doi: 10.1038/nbt0696-736. [DOI] [PubMed] [Google Scholar]
- Martineau B, Voelker TA, Sanders RA. On Defining T-DNA. Plant Cell. 1994;6:1032–1033. doi: 10.1105/tpc.6.8.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
- Onwueme IC, Charles WB. Tropical root and tuber crops: production, perspectives and future prospects. FAO plant production and protection paper 126. Rome: FAO; 1994. [Google Scholar]
- Raemakers CJJM, Amati M, Staritsky G, Jacobsen E, Visser RGF. Cyclic somatic embryogenesis and plant regeneration in cassava. Ann Bot. 1993;71:289–294. doi: 10.1006/anbo.1993.1037. [DOI] [Google Scholar]
- Raemakers CJJM, Schavemaker CM, Jacobsen E, Visser RGF. Improvements of cyclic somatic embryogenesis of cassava (Manihot esculenta Crantz) Plant Cell Rep. 1993;12:226–229. doi: 10.1007/BF00237059. [DOI] [PubMed] [Google Scholar]
- Raemakers CJJM, Sofiari E, Taylor N, Henshaw G, Jacobsen E, Visser RGF. Production of transgenic cassava (Manihot esculenta Crantz) plants by particle bombardment using luciferase activity as selection marker. Mol Breeding. 1996;2:339–349. doi: 10.1007/BF00437912. [DOI] [Google Scholar]
- Raemakers CJJM, Sofiari E, Jacobsen E, Visser RGF. Regeneration and transformation of cassava. Euphytica. 1997;96:153–161. doi: 10.1023/A:1002970925897. [DOI] [Google Scholar]
- Raemakers K, Schreuder M, Pereira I, Munyikwa T, Jacobsen E, Visser R. Progress made in FEC transformation of cassava. Euphytica. 2001;120:15–24. doi: 10.1023/A:1017574713880. [DOI] [Google Scholar]
- Raemakers K, Schreuder M, Suurs L, Furrer-Verhorst H, Vincken JP, de Vetten N, Jacobsen E, Visser RGF. Improved cassava starch by antisense inhibition of granule-bound starch synthase I. Mol Breeding. 2005;16:163–172. doi: 10.1007/s11032-005-7874-8. [DOI] [Google Scholar]
- Raemakers K, Pereira I, Koehorst van Putten H, Visser R. Indirect somatic embryogenesis in cassava for genetic modification purposes. In: Loyola-Vargas VM, Vázquez-Flota F, editors. Methods in molecular biology vol 318: plant cell culture protocols. 2. Totowa: Humana Press; 2006. pp. 101–109. [DOI] [PubMed] [Google Scholar]
- Rhee Y, Lin H, Buell R, Childs K, Kaeppler S. A c2 allele of maize identified in regenerant-derived progeny from tissue culture results from insertion of a novel transposon. Maydica. 2009;54:429–437. [Google Scholar]
- Rogers SO, Bendich AJ. Extraction of DNA from plant tissues. In: Gelvin SB, Schilperoort RA, editors. Plant molecular biology manual. Dordrecht: Kluwer; 1988. pp. A6/1–A6/10. [Google Scholar]
- Salehuzzaman SNIM, Jacobsen E, Visser RGF. Isolation and characterization of a cDNA encoding granule-bound starch synthase in cassava (Manihot esculenta Crantz) and its antisense expression in potato. Plant Mol Biol. 1993;23:947–962. doi: 10.1007/BF00021811. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sánchez T, Dufour D, Moreno IX, Ceballos H. Comparison of pasting and gel stabilities of waxy and normal starches from potato, maize, and rice with those of a novel waxy cassava starch under thermal, chemical, and mechanical stress. J Agric Food Chem. 2010;58:5093–5099. doi: 10.1021/jf1001606. [DOI] [PubMed] [Google Scholar]
- Schenk RU, Hildebrandt AC. Medium and techniques for induction and growth of monocotyledonous and dicotyledonous plant cell cultures. Can J Bot. 1972;50:199–204. doi: 10.1139/b72-026. [DOI] [Google Scholar]
- Schöpke C, Taylor N, Cárcamo R, Konan N, Marmey P, Henshaw GG, Beachy RN, Fauquet C. Regeneration of transgenic cassava plants (Manihot esculenta Crantz) from microbombarded embryogenic suspension cultures. Nature Biotechnol. 1996;14:731–735. doi: 10.1038/nbt0696-731. [DOI] [PubMed] [Google Scholar]
- Singh J, Kaur L, McCarthy OJ. Factors influencing the physico-chemical, morphological, thermal and rheological properties of some chemically modified starches for food applications–a review. Food Hydrocoll. 2007;21:1–22. doi: 10.1016/j.foodhyd.2006.02.006. [DOI] [Google Scholar]
- Sriroth K, Piyachomkwan K, Santisopasri V, Oates CG. Environmental conditions during root development: Drought constraint on cassava starch quality. Euphytica. 2001;120:95–101. doi: 10.1023/A:1017511806128. [DOI] [Google Scholar]
- Strauch E, Wohlleben W, Pühler A. Cloning of a phosphinothricin N-acetyltransferase gene from Streptomyces viridochromogenes Tü494 and its expression in Streptomyces lividans and Escherichia coli. Gene. 1988;63:65–74. doi: 10.1016/0378-1119(88)90546-X. [DOI] [PubMed] [Google Scholar]
- Taylor N, Chavarriaga P, Raemakers K, Siritunga D, Zhang P. Development and application of transgenic technologies in cassava. Plant Mol Biol. 2004;56:671–688. doi: 10.1007/s11103-004-4872-x. [DOI] [PubMed] [Google Scholar]
- Taylor N, Kent L, Fauquet C. Progress and challenges for the deployment of transgenic technologies in cassava. AgBioForum. 2004;7:51–56. [Google Scholar]
- Thro AM, Taylor N, Raemakers K, Puonti-Kaerlas J, Schöpke C, Visser R, Iglesias C, Sampaio MJ, Fauquet C, Roca W, Potrykus I. Maintaining the cassava biotechnology network. Nature Biotechnol. 1998;16:428–430. doi: 10.1038/nbt0598-428. [DOI] [Google Scholar]
- Van Breusegem F, Slooten L, Stassart J, Botterman J, Moens T, Van Montagu M, Inze D. Effects of overproduction of tobacco MnSOD in maize chloroplasts on foliar tolerance to cold and oxidative stress. J Exp Bot. 1999;50:71–78. doi: 10.1093/jexbot/50.330.71. [DOI] [Google Scholar]
- Vanderschuren H, Akbergenov R, Pooggin MM, Hohn T, Gruissem W, Zhang P. Transgenic cassava resistance to African cassava mosaic virus is enhanced by viral DNA-A bidirectional promoter-derived siRNAs. Plant Mol Biol. 2007;64:549–557. doi: 10.1007/s11103-007-9175-6. [DOI] [PubMed] [Google Scholar]
- Visser RGF, Stolte A, Jacobsen E. Expression of a chimaeric granule-bound starch synthase-GUS gene in transgenic potato plants. Plant Mol Biol. 1991;17:691–699. doi: 10.1007/BF00037054. [DOI] [PubMed] [Google Scholar]
- Wolters AMA, Trindade LM, Jacobsen E, Visser RGF. Fluorescence in situ hybridization on extended DNA fibres as a tool to analyse complex T-DNA loci in potato. Plant J. 1998;13:837–847. doi: 10.1046/j.1365-313X.1998.00079.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.