Abstract
A rapid and sensitive UPLC/Q–TOF–MS method has been established for analysis of the constituents in rat serum after oral administration of Fufang Zhenzhu Tiaozhi (FTZ) capsule, an effective compound prescription for treating hyperlipidemia in the clinic. The UPLC/MS information of samples was obtained first in FTZ preparation and FTZ-treated rat serum. Mass spectra were acquired in both negative and positive ion modes. Thirty-six constituents in rat serum after oral administration of FTZ were detected, including the alkaloids, ginsenosides, pentacyclic triterpenes, and their metabolites. These chemicals were identified based on the retention time and mass spectrometry data with those of authentic standards or comparison of the literatures reports. Twenty-seven prototype components originated from FTZ and nine were the metabolites of the FTZ constituents. These results shed light on the potential active constituents of the complex traditional Chinese medicinal formulas.
Keywords: UPLC/Q–TOF–MS, Serum pharmacology, Metabolite, Fufang Zhenzhu Tiaozhi capsule
Introduction
Fufang Zhenzhu Tiaozhi (FTZ) capsule, the patentable Chinese herbal medicine prescription, including Rhizoma Coptidis, Radix Salvia Miltiorrhiza, Radix Notoginseng, Fructus Ligustri Lucidi, Herba Cirsii Jeponici, Cortex Eucommiae, Fructus Citri Sarcodactylis and Radix Atractylodes Macrocephala. FTZ has been prescribed for 12 years by virtue of the potential to regulate abnormal lipid metabolism for treatment of dyslipidemia, atherosclerosis, and related disease [1, 2]. Clinical practice on more than 3,000 dyslipidemic patients demonstrated that FTZ is very safe and less harmful side effects [3, 4]. Giving FTZ not only markedly decrease the levels serum total cholesterol, glycerinate and low-density lipoprotein cholesterol while raising high-density lipoprotein cholesterol, but also improves hepatic tissue pathologic states, and prevents atherosclerosis [5, 6].
At present, hundreds of constituents have been identified, respectively and systematically, from the herbs that compose FTZ [7]. Constituents such as oleanolic acid, salvianolic acid A, salvianolic acid B, notoginsenoside R1, ginsenoside Rb1, ginsenoside Rg1, berberine, palmatine and jateorhizine have been experimentally verified [8, 9]. However, it remains unclear as to which constituents are responsible for the lipid-modulating functions of the drug; furthermore, there has been no integrated study of the constituents of the formula which is not simply a blend of the individual herbs but an integrated prescription.
Serum pharmacochemistry, which is an experimental technique focusing on the analysis of serum samples obtained after dosing, is based on the hypothesis that most effective constituents need to be absorbed into the blood to elicit activities after administration of traditional Chinese medicines (TCMs), and the components absorbed and metabolites formed can be determined simultaneously in order to identify the in vivo active forms from TCM formulas [10–14]. On the other hand, the rapid development of analytical techniques, such as UPLC coupled with HDMS technique in recent years provide a powerful tool for qualitative and quantitative analysis of complicated samples such as TCMs [15, 16].
The present study examined the constituents of rat serum after oral administration of FTZ using combined UPLC/Q−TOF−MS/MS. From a comprehensive analysis of a FTZ preparation, rat serum collected from FTZ-treated group and control group, 27 prototype components, and nine metabolites originating from FTZ were identified. To the best of our knowledge, this is the first systematical study on identifying the possible effective constituents in FTZ. The information will guide us to explore the mechanism under the lipid-modulating effect of FTZ in the following investigation.
Experimental
Chemicals and Materials
Authentic standards such as chloramphenicol, danshensu, protocatechuic acid, protocatechuic aldehyde, salidroside, rosmarinic acid, salvianolic acid B, specnuezhenide, salvianolic acid A, jatrorrhizine, notoginsenoside R1, palmatine, berberine, ginsenoside Rg1, ginsenoside Re, 5,7-dimethoxycoumarin, ginsenoside Rb1, cryptotanshinone, tanshinone IIA, and oleanolic acid were purchased from the National Institute for the Control of Pharmaceutical and Biological Products (Beijing, P. R. China). Acetonitrile was of HPLC grade (Merck, Darmstadt, Germany). HPLC grade methanol was provided by Honeywell International Inc. (Burdick and Jackson, Muskegon, MI, USA). Phosphoric acid and acetic acid glacial were of HPLC grade and purchased from TianJin Chemical Reagents Development Center (TianJin, China). Ultrapure water for the preparation of samples and mobile phase was prepared with PURELAB Ultra GE MK2 water system (ELGA, High Wycombe, UK). Other reagents were of analytical grade. FTZ capsules were prepared by the Institute of Materia Medica, Guangdong Pharmaceutical University (batch number 20090607). Eight comprised crude herbs were purchased from Zhixin Chinese Herbal Medicine Co., Ltd. (Guangzhou, China) and all the herbs were authenticated by Professor Shu-Yuan Li (Guangdong Pharmaceutical University). A voucher specimen was deposited in the Institute of Traditional Chinese Medicine, Guangdong Pharmaceutical University, Guangzhou, P. R. China.
Instrumentation and Analytical Conditions
The Waters AcQuity™ Ultra Performance LC system (Waters Corporation, Milford, USA) was equipped with quaternary pump, vacuum degasser, a cooling autosampler, and a diode-array detector. A UPLC™ BEH C18 column (50 × 2.1 mm, 1.7 μm) was utilized for separation with the column temperature at 30 °C. A binary gradient elution was adopted with mobile phase consisting of (A) 0.25% acetic acid glacial and 10 mM ammonium acetate in water and (B) acetonitrile: 0−1.6 min, B 2−5%; 1.6−7.6 min, B 5−20%; 7.6−9.6 min, B 20%; 9.6−14.6 min, B 20−35%; 14.6−17.6 min, B 35−80%; 17.6−18 min, B 80−100%; 18−18.4 min, B 100%. The flow rate was set at 0.40 mL min−1. The autosampler was conditioned at 4 °C, and the injection volume was 10 μL.
The instrument Waters Micromass Q–TOF–micro™ (Waters Co., UK) was equipped with the Lock Spray and ESI interface operating in both positive ion mode and negative ion mode, and with MassLynx data analysis software. The capillary voltage was set at 3 kV; the cone voltage was set at 30 V for both positive ionization mode and negative ionization mode. The ion source temperature was set at 100 °C and desolvation temperature at 350 °C. Nitrogen and argon were used for cone and collision gases, respectively. The cone and desolvation gas flows were 60 and 600 L h−1, respectively. The mass spectrometric data was collected in full scan mode with the mass range of m/z 100–1,500, using independent reference lock-mass ions via the Lock Spray interface to ensure mass accuracy and reproducibility. The solution of chloramphenicol was used as lock-mass, with an [M+H]+ ion of m/z 345.0021 and an [M−H]− ion of m/z 321.0045. The MS/MS analysis was performed using a variable collision energy (20–50 eV), which was optimized for each individual constituent. The Lock Spray frequency was set at 10 s. Acquity UPLC/Q–TOF micro system was operated using MassLynx 4.1 software (Waters Co., USA). The accurate mass and composition for the precursor and fragment ions were calculated by MassLynx 4.1.
Animals
Ten male Sprague–Dawley rats (body weight 200 ± 20 g) were obtained from the Medical Experimental Animal Center of Guangdong Province (Foshan, China). Animals were housed under standard conditions of temperature, humidity and light with food (laboratory rodent chow) and water provided ad libitum and were acclimated in the laboratory for at least 1 week prior to experiment. Before administration, the animals were fasted overnight with free access of water. All experimental protocols have been approved by Institutional Animal Ethics Committee of Guangdong Pharmaceutical University (GDPUIAEC No.200902), and are also in a compliance with national and international guidelines of animal welfare (NIH Guide for the Care and Use of Laboratory Animals, NIH publication No. 85-23, 1985).
Sample Preparation
Preparation of FTZ Extract
The preparation of FTZ extract from eight constituent herbs was consistent with the protocol described previously [1], and as follows: Radix Salvia Miltiorrhiza (50 g), Radix Atractylodes Macrocephala (60 g), Fructus Citri Sarcodactylis (50 g), Cortex Eucommiae (40 g), and Herba Cirsii Jeponici (30 g) were extracted with boiling water twice (volume over weight values per reflux extraction = 12- and 9-fold, respectively; duration = 1.5 h per reflux extraction); Fructus Ligustri Lucidi (60 g) and Rhizoma Coptidis (20 g) were extracted with 70% ethanol twice (volume over weight values per reflux extraction = 10- and 8-fold, respectively; duration = 2 h per reflux extraction); Radix Notoginseng (20 g) was extracted with 50% ethanol twice (volume over weight values per reflux extraction = 10- and 8-fold, respectively; duration = 2 h per reflux extraction). The above three extracts were combined, filtered by gauzes, and the combined solution was freeze-dried. Five hundred milligrams of the freeze-dried powder was extracted with 50 mL methanol for 20 min under ultrasonics. The methanol extraction was centrifuged at 15,000 rpm for 15 min at 4 °C, and the supernatant was filtered through a 0.20-μm filter, the filtrate was applied for UPLC analysis. All authentic standards were accurately weighed, and dissolved in methanol to obtain stock solutions with indicated concentrations. All the stock solutions were stored in the refrigerator at 4 °C until analysis.
Preparation of Serum Samples
Capsule contents of FTZ, originated from the above extraction, were dispersed with distilled water as stock solution (0.8 g mL−1). The above suspension was orally administered to five rats (2.0 mL/100 g body weight). An equal volume of distilled water was orally administered to the other five rats as control; 30 min after drug administration, the animals were anaesthetized by ether inhalation. The blood was collected from the vena ophthalmica and then centrifuged at 10,000 rpm for 5 min at 4 °C. The supernatant obtained was frozen immediately and stored at −80 °C before use.
Phosphoric acid (240 μL) was added to 6.0 mL of the above supernatant and ultrasonicated for 1 min, and vortexed for 1 min. The mixed solution was applied to three pre-activated OASIS HLB solid phase extraction C18 columns (1 cc, 30 mg, Waters Corporation, USA). The column was washed with 4 mL of water, 2 mL of 100% methanol and 2 mL of 2% acetic acid glacial–methanol (1:9). The 100% methanol elutes and 2% acetic acid glacial–methanol (1:9) elutes were collected and dried under nitrogen gas at 50 °C. The residues were re-dissolved in 300 μL of methanol, centrifuged at 15,000 rpm for 15 min and an aliquot of supernatant was subjected to UPLC analysis.
Results and Discussions
UPLC–MS/MS Analysis and Identification the Constituents of FTZ
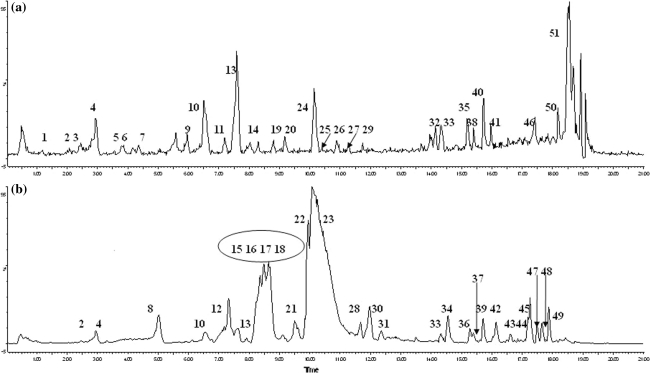
ESI in both negative and positive ion modes was applied to analyze and identify the constituents in the FTZ. The total ion current chromatograms at the two ESI modes are shown in Fig. 1. Fifty-one peaks in FTZ were detected using UPLC–MS/MS, and 44 constituents were identified by comparing their retention behavior, the MS fragments characteristics to those of authentic standards. The names and structures of the identified constituents from Rhizoma Coptidis, Radix Notoginseng, Fructus Ligustri Lucidi, Radix Salvia miltiorrhiza, and other three herbs in both herbal preparation and the serum samples for FTZ-treated rats are listed in Tables 1, 2, 3, 4 and 5. The identified compounds are summarized in Table 6.
Fig. 1.
UPLC–ESI–MS total ion current chromatograms of FTZ at the negative ion (a) and positive (b) mode. The numbers appearing in the circle. These peaks in the circle have similar structures and retention times, each peak separate incompletely and cannot be marked clearly, the number in the circle represents four peaks including
Table 1.
Structures of the Rhizoma Coptidis constituents identified in FTZ preparation and serum samples from FTZ-treated rats
| Type | Substituent group | Name of constituent | MW | Observed |
|---|---|---|---|---|

|
R1=R2=R3=R4=CH3,R5=H, R6=OH | Hydroxyl-palmatine | 368 | In serum and in preparation |
| R1+R2=CH2, R3=H, R4=CH3, R5=R6=H | Thalifendine | 322 | In serum and in preparation | |
| R1=H, R2=R3=R4=CH3, R5=R6=H | Columbamine | 338 | In serum and in preparation | |
| R1=R2=CH3, R3+R4=CH2, R5=R6=H | Epiberberine | 336 | In serum and in preparation | |
| R1+R2=R3+R4=CH2, R5=R6=H | Coptisine | 320 | In serum and in preparation | |
| R1=CH3, R2=H, R3=R4=CH3, R5=R6=H | Jatrorrhizine | 338 | In serum and in preparation | |
| R1+R2=CH2, R3=CH3, R4=H, R5=R6=H | Berberrubine | 322 | In serum and in preparation | |
| R1=R2=R3=R4=CH3, R5=R6=H | Palmatine | 352 | In serum and in preparation | |
| R1+R2=CH2, R3=R4=CH3, R5=R6=H | Berberine | 336 | In serum and in preparation | |
| R1=R2=R3=R4=CH3, R5=CH3, R6=H | Dehydrocorydaline | 366 | In serum and in preparation | |
| R1+R2=CH2, R3=R4=CH3, R5=CH3, R6=H | 13-Methylberberine | 350 | In serum and in preparation | |
| R1=R2=R3=R4=CH3, R5=CH2CH3, R6=H | 13-Ethyl-5,6-dihydro-2,3,9,10-tetramethoxy-dibenzo[a,g]quinolizinium | 380 | In preparation only | |
| R1=CH3, R2=H, R3=R4=CH3, R5=CH3, R6=H | Dehydrocorybulbine | 352 | In preparation only | |
| R1=CH3, R2=GlcUA, R3=R4=CH3, R5=R6=H | Jatrorrhizine 3-O-β-d-glucuronide | 514 | In preparation only | |
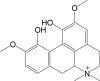
|
Magnoflorine | 342 | In serum only |
In serum and in preparation: the constituent was observed both in FTZ-treated serum and FTZ preparation, in preparation only: the constituent was only observed in FTZ preparation, in serum only: the constituent was only observed in FTZ-treated serum
MW molecular weight
Table 2.
Structures of the Radix Notoginseng constituents identified in FTZ preparation and serum samples from FTZ-treated rats
| Type | Substituent group | Name of constituent | MW | Observed |
|---|---|---|---|---|

|
R1=OH, R2=OGlc(2–1)Xyl, R3=OGlc | Notoginsenoside R1 | 932 | In serum and in preparation |
| R1=OH, R2=OGlc, R3=OGlc | Ginsenoside Rg1 | 800 | In serum and in preparation | |
| R1=OH, R2=OGlc(2–1)Rha, R3=OGlc | Ginsenoside Re | 946 | In preparation only | |
| R1=OH, R2=OGlc, R3=OH | Ginsenoside Rh1 | 638 | In serum and in preparation | |
| R1=OH, R2=OH, R3=OGlc | Ginsenoside F1 | 638 | In serum and in preparation | |
| R1=OGlc(2–1)Glc, R2=H, R3=OGlc(6–1)Glc | Ginsenoside Rb1 | 1108 | In serum and in preparation | |
| R1=OGlc(2–1)Glc, R2=H, R3=OGlc | Ginsenoside Rd | 946 | In serum and in preparation | |
| R1=OH, R2=OH, R3=OH | Protopanaxatriol | 476 | In serum only | |
| R1=OGlc(2–1)Glc, R2=H, R3=OH | Ginsenoside Rg3 | 784 | In serum only | |

|
R1=OGlc, R2=OH | 25-Hydroxy-ginsenoside Rh1 | 656 | In serum only |
| R1=OH, R2=OGlc | 25-Hydroxy-ginsenoside F1 | 656 | In serum only |
In serum and in preparation: the constituent was observed both in FTZ-treated serum and FTZ preparation, in preparation only: the constituent was only observed in FTZ preparation, in serum only: the constituent was only observed in FTZ-treated serum
MW molecular weight
Table 3.
Structures of the Fructus Ligustri Lucidi constituents identified in FTZ preparation and serum samples from FTZ-treated rats
| Type | Substituent group | Name of constituent | MW | Observed |
|---|---|---|---|---|
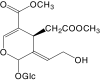
|
10-Hydroxyoleoside dimethyl ester | 434 | In serum and in preparation | |
|
|
Salidroside | 300 | In serum and in preparation | |
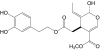
|
Oleuropeine aglycone | 378 | In preparation only | |
|
|
Oleuropeine | 540 | In preparation only | |
|
|
(+)-Pinoresinol-O-β-d-glucopyranoside | 520 | In preparation only | |
|
|
Coniferin | 342 | In preparation only | |

|
R1=OH, R2=OH, R3=R4=CH3, R5=R6=H | Masilinic acid | 472 | In serum and in preparation |
| R1=CH3COO, R2=H, R3=CH3, | ||||
| R4=H, R5=CH3, R6=OH | Pomolic acid acetate | 514 | In serum and in preparation | |
| R1=OH, R2=H, R3=R4=CH3, R5=R6=H | Oleanolic acid | 456 | In serum and in preparation | |

|
R=OH | Specnuezhenide | 686 | In preparation only |
|
|
Oleonuezhenide | 1072 | In preparation only |
In serum and in preparation: the constituent was observed both in FTZ-treated serum and FTZ preparation, in preparation only: the constituent was only observed in FTZ preparation, in serum only: the constituent was only observed in FTZ-treated serum
MW molecular weight
Table 4.
Structures of the Radix Salvia Miltiorrhiza constituents identified in FTZ preparation and serum samples from FTZ-treated rats
| Type | Substituent group | Name of constituent | MW | Observed |
|---|---|---|---|---|

|
R=CH2CH(OH)COOH | Danshensu | 198 | In preparation only |
| R=CHO | Protocatechuic aldehyde | 138 | In preparation only | |
| R=COOH | Protocatechuic acid | 154 | In serum and in preparation | |

|
Salvianolic acid B | 718 | In preparation only | |
|
|
|
Salvianolic acid A | 494 | In preparation only |
|
|
Rosmarinic acid | 360 | In preparation only | |

|
Dihydrotanshinone I | 278 | In preparation only | |

|
Cryptotanshinone | 296 | In preparation only | |

|
Tanshinone IIA | 294 | In preparation only |
In serum and in preparation: the constituent was observed both in FTZ-treated serum and FTZ preparation, in preparation only: the constituent was only observed in FTZ preparation, in serum only: the constituent was only observed in FTZ-treated serum
MW molecular weight
Table 5.
Structures of the remaining herbs constituents identified in FTZ preparation and serum samples from FTZ-treated rats
| Type | Origin | Name of constituent | MW | Observed |
|---|---|---|---|---|

|
Cortex Eucommiae | Eucommiol | 188 | In serum and in preparation |
|
|
Herbal Cirsii Jeponici and Fructus Citri Sarcodactylis | Diosmetin | 300 | In preparation only |

|
Fructus Citri Sarcodactylis | 5,7-Dimethoxycoumarin | 206 | In serum and in preparation |
|
|
Cortex Eucommiae | Pinoresinol | 358 | In serum and in preparation |
In serum and in preparation: the constituent was observed both in FTZ-treated serum and FTZ preparation; in preparation only: the constituent was only observed in FTZ preparation; in serum only: the constituent was only observed in FTZ-treated serum
MW molecular weight
Table 6.
MS data of (+) ESI–MS spectra and (−) ESI–MS spectra, and the identification results of the constituents of FTZ preparation and FTZ-treated rats serum
| No. | Name | R t (min) | MW | MS+ (m/z) | MS− (m/z) |
|---|---|---|---|---|---|
| 1 | Danshensua | 1.14 | 198 | 197[M−H]− | |
| 179[M−H−H2O]− | |||||
| 135[M−H−H2O−CO2]− | |||||
| 2(1)b | 10-Hydroxyoleoside dimethyl ester | 2.45 | 434 | 452[M+NH4]+ | 433[M−H]− |
| 209[M−H−Glu−CH3CO−H2O]− | |||||
| 177[M−H−Glu−CH3CO−CH3O−H2O]− | |||||
| 165[M−H−Glu−2CH3CO−H2O]− | |||||
| 3 | Protocatechuic aldehydea | 2.62 | 138 | 137[M−H]− | |
| 109[M−H–CO]− | |||||
| 4(2) | Salidrosidea | 2.99 | 300 | 318[M+NH4]+ | 299[M−H]− |
| 179[M−H−C8H8O]− | |||||
| 119[M−H−C6H12O6]− | |||||
| 101[M−H−Glu]− | |||||
| 5(3) | Hydroxyl-palmatine | 3.85 | 368 | 367[M−H]− | |
| 352; 191 | |||||
| 6(4) | Protocatechuic acida | 3.89 | 154 | 153[M−H]− | |
| 109[M−H−CO2]− | |||||
| 7 | Oleuropeine aglycone | 4.36 | 378 | 377[M−H]− | |
| 197[M−H−C9H8O4]−; 153 | |||||
| 8(5) | Magnoflorine | 5.04 | 342 | 342[M]+ | |
| 297[M+H−CH3−CH3O]+ | |||||
| 282[M+H−2CH3−CH3O]+ | |||||
| 265[M−CH3−2CH3O]+ | |||||
| 237[M−CH3−2CH3O−CO]+ | |||||
| M1 | Jatrorrhizine3-O-β-d-glucuronide | 5.77 | 514 |
514[M]+ 338[M−GlcUA]+ |
|
| 9 | Rosmarinic acida | 5.96 | 360 |
359[M−H]− 197[M−H−C9H6O3]− 179[M−H−C9H6O3−H2O]− 161[M−H−C9H6O3−2H2O]− |
|
| 10 | Salvianolic acid Ba | 6.52 | 718 | 736[M+NH4]+ |
717[M−H]− 519[M−H−C9H10O5]− 321[M−H−2C9H10O5]− 339[M−H−C9H10O5−C9H8O4]− |
| 11(6) | Eucommiol | 7.19 | 188 | 187[M−H]−; 125; 97 | |
| 12(7) | Thalifendine | 7.33 | 322 |
322[M]+ 307[M−CH3]+ 279[M−CO−CH3]+ 251[M−2CO−CH3]+ |
|
| 13 | Specnuezhenidea | 7.59 | 686 | 704[M+NH4]+ |
685[M−H]− 523[M−H−Glu]− 453[M−H−Glu−C3H2O2]− 423[M−H−Glu−C3H2O2−CH2O]− 299[M−H−Glu−C3H2O2−CH2O−C7H8O2]−; 223; 197 |
| 14 | Salvianolic acid Aa | 8.03 | 494 |
493[M−H]− 313[M−H−C9H8O4]− 295[M−H−C9H10O5]− 185[M−H−C9H10O5−C6H6O2]− |
|
| M2 | 25-Hydroxy-ginsenoside Rh1/F1 | 8.23 | 656 |
715[M−H+CH3COOH]− 655[M−H]−; 493[M−H−Glu]− |
|
| 15(8) | Columbamine | 8.33 | 338 |
338[M]+ 322[M−CH4]+ 308[M−2CH3]+ 294[M−CH4−CO]+ 280[M−2CH3−CO]+ |
|
| 16(9) | Epiberberine | 8.41 | 336 |
336[M]+ 320[M−CH4]+ 294[M−CH4−CO]+ 263[M−CH3−CH2O−CO]+ |
|
| 17(10) | Coptisine | 8.47 | 320 |
320[M]+ 292[M−CO]+ 262[M−CO−CH2O]+ 234[M−2CO−CH2O]+ 204[M−2CO−2CH2O]+ |
|
| 18(11) | Jatrorrhizinea | 8.66 | 338 |
338[M]+ 322[M−CH4]+ 308[M−2CH3]+ 294[M−CH4−CO]+ 280[M−2CH3−CO]+ 265[M−CH3−CO−CH2O]+ 250[M−2CH3−CO−CH2O]+ |
|
| M3 | 25-Hydroxy-ginsenoside Rh1/F1 | 8.74 | 656 |
715[M−H+CH3COOH]− 655[M−H]−; 493[M−H−Glu]− |
|
| 19 | Oleuropein | 8.81 | 540 |
539[M−H]− 377[M−H−Glu]− 307[M−H−Glu−C4H6O]− 275[M−H−Glu−C4H6O−CH3OH]− |
|
| 20(12) | Notoginsenoside Ra1 | 9.17 | 932 |
991[M−H+CH3COOH]− 931[M−H]− 799[M−H−Xyl]− 637[M−H−Xyl−Glu]− 475[M−H−Xyl−2Glu]− |
|
| 21(13) | Berberrubine | 9.49 | 322 |
322[M]+ 307[M−CH3]+ 279[M−CO−CH3]+ 251[M−2CO−CH3]+ |
|
| 22(14) | Palmatinea | 9.93 | 352 |
352[M]+ 337[M−CH3]+ 336[M−CH4]+ 322[M−2CH3]+ 308[M−CH4−CO]+ 294[M−2CH3−CO]+ 279[M−CH3−CO−CH2O]+ |
|
| 23(15) | Berberinea | 10.06 | 336 |
336[M]+ 321[M−CH3]+ 320[M−CH4]+ 306[M−2CH3]+ 292[M−CH4−CO]+ 278[M−2CH3−CO]+ 262[M−CH4−CO−CH2O]+ 234[M−CH4−2CO−CH2O]+ |
|
| 24(16) | Ginsenoside Rga1 | 10.14 | 800 |
859[M−H+CH3COOH]− 637[M−H−Glu]−; 475[Agl]− |
|
| 25 | Ginsenoside Rea | 10.39 | 946 |
1005[M−H+CH3COOH]− 945[M−H]− 799[M−H−Rha]− 637[M−H−Rha−Glu]− 475[M−H−Rha−2Glu]− |
|
| 26 | Diosmetin | 10.88 | 300 |
299[M−H]− 284[M−H−CH3]− 256[M−H−CH3−CO]− |
|
| 27 | (+)-Pinoresinol-O-β-d-glucopyranoside | 11.27 | 520 | 521[M+H]+ |
519[M−H]− 357[M−H−Glu]− 235[M−H−Glu−CH2O−C6H4O]− |
| 28(17) | Dehydrocorydaline | 11.67 | 366 |
366[M]+ 350[M−CH4]+ 336[M−2CH3]+ 322[M−CH4−CO]+ 308[M−2CH3−CO]+ 278[M−2CH3−CO−CH2O]+ |
|
| 29 | Oleonuezhenide | 11.74 | 1072 |
1071[M−H]− 771[M−H−C14H20O7]− 685[M−H−C17H22O10]− 523[M−H−C17H22O10−Glu]− |
|
| 30(18) | 13-Methylberberine | 11.97 | 350 |
350[M]+ 335[M−CH3]+ 334[M−CH4]+ 320[M−2CH3]+ 306[M−CH4−CO]+ 292[M−2CH3−CO]+ 278[M−CH4−2CO]+ |
|
| 31(19) | 5,7-Dimethoxycoumarina | 12.35 | 206 |
207[M+H]+ 192[M+H−CH3]+ 164[M+H−CH3−CO]+ 149[M+H−2CH3−CO]+ |
|
| M4 |
20(S)-Ginsenoside Rh1/ 20(R)-Ginsenoside Rh1/ Ginsenoside F1 |
12.66 | 638 |
697[M−H+CH3COOH]− 441; 423; 405 |
|
| M5 | 20(S)/(R)-Protopanaxatriol | 13.69 | 476 | 477[M+H]+ |
493[M−H+H2O]− 553[M−H+H2O+CH3COOH]− |
| M6 | 20(S)/(R)-Protopanaxatriol | 14.02 | 476 | 477[M+H]+ |
493[M−H+H2O]− 553[M−H+H2O+CH3COOH]− |
| 32(20) |
Ginsenoside F1/ Ginsenoside Rh1 |
14.14 | 638 |
697[M−H+CH3COOH]− 637[M−H]− 475[M−H−Glu]− |
|
| 33(21) | Ginsenoside Rba1 | 14.31 | 1108 |
1109[M+H]+ 1126[M+NH4]+ 1091[M+H−H2O] + |
1107[M−H]− 1167[M−H+CH3COOH]− 945[M−H−Glu]− 783[M−H−2Glu]− 603[M−H−3Glu−H2O]−; 459[Agl]− |
| M7 |
20(S)-Ginsenoside Rh1/ 20(R)-Ginsenoside Rh1/ Ginsenoside F1 |
14.39 | 638 |
697[M−H+CH3COOH]− 441; 423; 405 |
|
| 34 | Coniferin | 14.52 | 342 |
343[M+H]+; 295 181[M+H−Glu]+ 164[M+H−Glu−OH]+; 120 |
|
| 35(22) | Not identified | 15.18 | 532 |
531[M−H]− 1063[2M−H]− 489[M−H−CH2−CO]− 471[M−H−CH2−CO−H2O]− 427[M−H−CH2−CO−H2O−CO2]− |
|
| 36 | Not identified | 15.27 | 357; 328; 293; 249 | ||
| 37 | 13-Ethyl-5,6-dihydro-2,3,9,10-tetramethoxy-dibenzo[a,g]quinolizinium | 15.36 | 380 |
398[M+NH4]+ 366[M−CH2] + 351[M−CH2−CH3]+ 336[M−CH2−2CH3]+ 322[M−CH2−CH4−CO]+ |
|
| 38(23) | Ginsenoside Rd | 15.39 | 946 | 947[M+H]+; 965[M+NH4]+ |
945[M−H]− 1005[M−H+CH3COOH]− 783[M−H−Glu]− 621[M−H−2Glu]−; 459[Agl]− |
| M8 | Salvianolic acid B sulfates | 15.65 | 798 |
797[M−H]− 717[M−H−SO3]− |
|
| 39 | Dehydrocorybulbine | 15.70 | 352 |
352[M]+ 337[M−CH3]+ 322[M−2CH3]+ 308[M−CH4−CO]+ 294[M−2CH3−CO]+ |
|
| 40(24) | Pinoresinol | 15.71 | 358 |
357[M−H]− 342[M−H−CH3]− 311[M−H−CH3−2CH3O]−; 151 |
|
| 41 | Not identified | 15.96 | 259; 244; 201; 189 | ||
| M9 | Ginsenoside Rg3 | 15.97 | 784 |
783[M−H]− 621[M−H−Glu]− 459[M−H−2Glu]− |
|
| 42 | Not identified | 16.14 | 614; 336; 321 | ||
| 43 | Dihydrotanshinone I | 16.60 | 278 |
279[M+H]+ 261[M+H−H2O]+ 233[M+H−H2O−CO]+ |
|
| 44 | Not identified | 17.19 | 519 |
520[M+H]+ 502[M+H−H2O]+ |
578[M−H+CH3COOH]− |
| 45 | Cryptotanshinonea | 17.24 | 296 |
297[M+H]+ 279[M+H−H2O]+ 268[M+H−C3H6]+ 251[M+H−H2O−CO]+ 227[M+H−C2H5]+ |
|
| 46(25) | Masilinic acid | 17.39 | 472 |
471[M−H]− 411[M−2H−CH3−CO2]− 393[M−2H−CH3−CO2−H2O]− |
|
| 47 | Not identified | 17.49 | 496; 478; 184; 104 | ||
| 48 | Not identified | 17.64 | 522; 503; 184; 104 | ||
| 49 | Tanshinone IIAa | 17.87 | 294 |
295[M+H]+ 277[M+H−H2O]+ 249[M+H−H2O−CO]+ 266[M+H−C2H5]+ |
|
| 50(26) | Pomolic acid acetate | 18.19 | 514 |
513[M−H]− 495[M−H−H2O]− 453[M−H−CH2CO−H2O]− |
|
| 51(27) | Oleanolic acida | 18.51 | 456 |
455[M−H]− 407[M−H−HCHO−H2O]− 391[M−H−HCOOH−H2O]− 373[M−H−HCOOH−2H2O]− |
MW molecular weight, Glu glucose, Rha rhamnose, Xyl xylose, Agl aglycone, GlcUA glucuronic acid, M metabolites in serum after oral administration of FTZ
aStructure confirmed by comparison with authentic standards
bNumber in parenthesis represents the no. of peak detected in serum after oral administration of FTZ
In order to obtain MS fragmentation patterns of constituents in FTZ, MS2 spectra of 19 authentic standards were recorded by UPLC–MS/MS. Peaks 1, 3, 4, 6, 9, 10, 13, 14, 18, 20, 22, 23, 24, 25, 31, 33, 45, 49 and 51 were attributed to danshensu, protocatechuic acid, protocatechuic aldehyde, salidroside, rosmarinic acid, salvianolic acid B, specnuezhenide, salvianolic acid A, jatrorrhizine, notoginsenoside R1, palmatine, berberine, ginsenoside Rg1, ginsenoside Re, 5,7-dimethoxycoumarin, ginsenoside Rb1, cryptotanshinone, tanshinone IIA and oleanolic acid, respectively, by comparing the retention time and mass data with those of the authentic standards. Other peaks were identified, utilizing elemental composition analysis of their MS and MS2 data with software MassLynx from data and comparing with the literature data as well.
In the negative ion mode, ginsenosides, iridoid/secoiridoid glycosides, triterpene acids, and phenolic acids were observed in the FTZ, which originated from Radix Notoginseng, Fructus Ligustri Lucidi and Radix Salvia miltiorrhiza, respectively. Among them, six ginsenosides, peaks 20, 24, 25, 32, 33, and 38, were identified as notoginsenoside R1, ginsenoside Rg1, ginsenoside Re, ginsenoside Rh1/F1 and ginsenoside Rb1 and ginsenoside Rd, respectively, by comparison with authentic standards and literature data [17, 18]. The mass spectra of the ginsenosides exhibited the molecular ion peaks at [M−H]− and [M−H+CH3COOH]−. In the MS2 spectra, aglycone ions m/z 475 and 459 were finally formed by loss of several glycosidic units, which were the characteristic ions of panaxatriols and panaxadiols, respectively [19]. Thus, these peaks could be identified as ginsenosides. For example, peak 24 showed a molecular ion at m/z 859 [M−H+CH3COOH]− in MS spectra and exhibited m/z 637 [M–H–Glu]− and m/z 475 [Agl]− ions in the MS2 spectra. The fragmentation ion at m/z 475 was produced by loss of all linked glucosidic bonds, which was a characteristic fragmentation of protopanaxatriol type ginsenosides [19]. Peak 33 showed a molecular ion at m/z 1107 [M−H]− in MS spectra; m/z 945 [M−H−Glu]−, m/z 783 [M−H−2Glu]−, m/z 603 [M−H−3Glu−H2O] and m/z 459 [Agl]− ions could be detected in the MS2 spectra, which exhibited a fragmentation pathway corresponding to the loss of glycosidic units. The fragmentation ion at m/z 459 corresponds to a characteristic ion of the protopanaxadiol moiety [20].
Iridoid glycosides, secoiridoid glycosides and triterpene acids are the essential constituents in the Fructus Ligustri Lucidi extract of FTZ, which include salidroside, oleuropeine aglycone, oleuropein, specnuezhenide, masilinic acid, pomolic acid acetate, oleanolic acid [21, 22]. Peak 13 showed a molecular ion at m/z 685 [M−H]− in MS spectra and exhibited m/z 523, 453, 423, 299, 223 and 197 ions in the MS2 spectra. By comparison with the authentic standard, peak 13 was unambiguously identified as specnuezhenide. The identification of peak 19 as oleuropein was corroborated by detection of the molecular ion at m/z 539 and its aglycone fragment at m/z 377. The MS spectrum showed a quasi-molecular ion at m/z 539 [M−H]− and the fragments were consistent with the following fragmentation pattern: the ion at m/z 377 arose from the loss of glucose, the ion at m/z 307 was characteristic of the loss of a C4H6O fragment and the fragment at m/z 275 might derive from the loss of CH3OH from the elenolic fragment of the molecule [21]. Peak 7 exhibited the pseudo-molecular ion at m/z 377 in MS and characteristic ions at m/z 197 and m/z 153 in its MS2 spectrum, corresponding to the oleuropein aglycone or its isomer. By retrieving of literature data [22], peak 7 was identified as oleuropein aglycone.
Among 51 analytes, there are six phenolic acids and three diterpenoids originated from Radix Salvia Miltiorrhiza. Phenolic acids could be classified into monomer and polymer. Polymers could be composed of one or several different monomers such as danshensu, caffeic acid or others. In the MS2 spectra of three monomer standards, including small molecules such as CO2, CO and H2O were produced in the fragmentation pathways, which indicated the presence of carboxyl, carbonyl or hydroxyl groups [23]. Danshensu showed a [M−H]− ion at m/z 197, and produced m/z 179 [M−H−H2O]− and m/z 135 [M−H−H2O−CO2]−. Similar to danshensu, both of the [M−H]− ions at m/z 137 of protocatechuic aldehyde and m/z 153 of protocatechuic acid produced the same ion at m/z 109 corresponding to the loss of CO and CO2, respectively.
As to three polymers, which contained an ester bond or ester bonds, the predominant fragmentation of their [M−H]− ions was the cleavage of the ester bond to lose danshensu [M−H−198]− and caffeic acid [M−H−180]−. For instance, peak 10 exhibited a quasi-molecular ion [M−H]− of m/z 717. Its MS2 spectra gave rise to prominent ion at m/z 519 corresponding to the loss of a molecule of danshensu. Other two fragment ions, [M−H−198−198]− ion at m/z 321 and [M−H−198−180]− ion at m/z 339 corresponding to the loss of the second danshensu and the first caffeic acid. These data are consistent with those in the literature [23]. Therefore, peak 10 was tentatively identified as salvianolic acid B. Similarly, peaks 9, 14 were identified as rosmarinic acid and salvianolic acid A separately [23, 24].
Rhizoma Coptids alkaloids, which were the most abundant constituents in the alcohol extra of FTZ, exhibited a special fragmentation pathway in the positive ion mode. It is well known that loss the neutral species such as CO, CH3, CH4 and CH2O were observed in the MS2 spectra of Rhizoma Coptids alkaloids [9]. Peak 23 showed a molecular ion at m/z 336 [M]+ in MS spectra, and exhibited some ions at m/z 320 [M−CH4]+, 306 [M−2CH3]+, 292 [M−CH4−CO]+, 278 [M−2CH3−CO]+, 262 [M−CH4−CO−CH2O]+ and 234 [M−CH4−2CO−CH2O]+ in MS2 spectra, showing the neutral loss of CO, CH3, CH4 and CH2O in the fragmentation pathway. These data are typical for the Rhizoma Coptids alkaloids in the present study and consistent with those in the literature [9]. Thus, the compound was identified as berberine. Similarly, peaks 8, 12, 15, 16, 17, 18, 21, 22, 28, 30 and 39 were identified as magnoflorine, thalifendine, columbamine, epiberberine, coptisine, jatrorrhizine, berberrubine, palmatine, dehydrocorydaline, 13-methylberberine and dehydrocorybulbine, respectively [9, 25–27]. Peak 37 showed the molecular ion at m/z 398 [M+NH4]+ and its product typical fragments at m/z 366 [M−CH2]+, 351 [M−CH2−CH3]+, 336 [M−CH2−2CH3]+ and 322 [M−CH2−CH4−CO]+ respectively in the MS2 spectrum. According to the literature data [28], we suggested that peak 37 could be 13-ethyl-5,6-dihydro-2,3,9,10-tetramethoxy-dibenzo[a,g]quinolizinium.
In addition to Rhizoma Coptids alkaloids in positive ion mode, three diterpenoids (dihydrotanshinone I, cryptotanshinone and tanshinone IIA) also exhibited [M+H]+ ions in positive ion mode. It is well known that hydrogen at C-1 and oxygen at C-11 of tanshinones were the source of the dissociated H2O and the neutral species such as CO, H2O, C2H5 and C3H6 were also observed in the MS2 spectra [29]. Peak 45 showed a molecular ion at m/z 297 [M+H]+ in MS spectra, and exhibited an ion at m/z 279 [M+H−H2O]+ in MS2 spectra, which corresponded to three fragment ions at m/z 268 [M+H−C3H6]+, m/z 227 [M+H−C2H5]+ and m/z 251 [M+H–H2O–CO]+, showing the neutral loss of CO, H2O, C2H5 and C3H6 in the fragmentation pathway. According to these data, peak 45 was tentatively identified as cryptotanshinone [30]. Using the same method, peak 43 and peak 49 were identified as dihydrotanshinone I and tanshinone IIA by comparison with literature data and authentic standards [30, 31].
In addition, the molecular ion [M+H]+ of peak 31 was observed in the MS spectra, which dissociated in MS2 to generate several ions at m/z 192, 164, 149 and 121. The ion at m/z 192 can be attributed to the loss of a methyl radical from the parent ion, this ion fragmented further with the loss of CO to give a signal at m/z 164. Subsequent loss of a methyl and a CO group radical to exhibited ions at m/z 149 and 121, were observed. Comparing with the authentic standard and literature data [32], peak 31 was tentatively identified as 5, 7–dimethoxycoumarin. Peak 34 showed a molecular ion at m/z 343 [M+H]+ in MS spectra, and exhibited four ions at m/z 295, m/z 181 [M+H−Glu]+, m/z 164 [M+H−Glu−OH]+ and m/z 120 in MS2 spectra, showing the loss of glucoside and hydroxy group in the fragmentation pathway. By comparison with literature data [33], this component was ascertained as coniferin.
UPLC–MS/MS Analysis and Identification the Constituents of FTZ in Rat Serum
Analysis of FTZ Constituents in Rat Serum
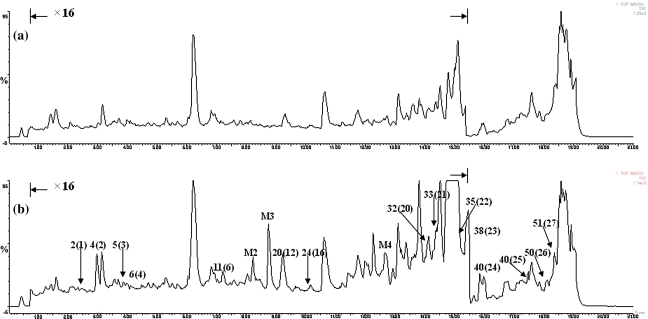
By comparison with the mass chromatography of FTZ and the rat serum samples from control group, the MS spectra for rat serum samples from FTZ-treated group exhibited 27 peaks in common, which demonstrated that the 27 components from FTZ were absorbed into the rat blood after oral administration. In addition, there were another nine peaks, which were only detected in the dosed serum, indicating that those components were metabolites of constituents from FTZ. Ion chromatograms of dosed and controlled rat serum are shown in Figs. 2, 3 and 4. The MS spectra and retention behavior of 36 peaks for prototype components and metabolites are summarized in Table 6.
Fig. 2.
Total ion current chromatograms at the positive ion mode of rat serum samples collected from control (a) and FTZ-treated (b) group
Fig. 3.
Total ion current chromatograms at the negative ion mode of rat serum samples collected from control (a) and FTZ-treated (b) group
Fig. 4.
Total ion current chromatograms of 9 metabolites identified from FTZ-treated rat serum samples
Analysis of Prototype Constituents of FTZ in Rat Serum
The constituents in rat serum after oral administration of FTZ were identified using their retention time and mass spectra. As a result, peaks 1, 2, 22, 26 and 27 were original form compounds existing in Fructus Ligustri Lucidi; peaks 3, 5, 7, 8, 9, 10, 11, 13, 14, 15, 17 and 18 came from Rhizoma Coptidis; peaks 12, 16, 20, 21 and 23 resulted from Radix Notoginseng; peak 19 and 22 originated from Fructus Citri Sarcodactylis; peak 6 and 24 came from Cortex Eucommiae; peak 4 originated from Radix Salvia Miltiorrhiza. It displayed that most of alkaloids, ginsenosides and pentacyclic triterpenes could be unambiguously detected in their original forms from the rat serum after FTZ administration.
Analysis of Metabolites of FTZ in Rat Serum
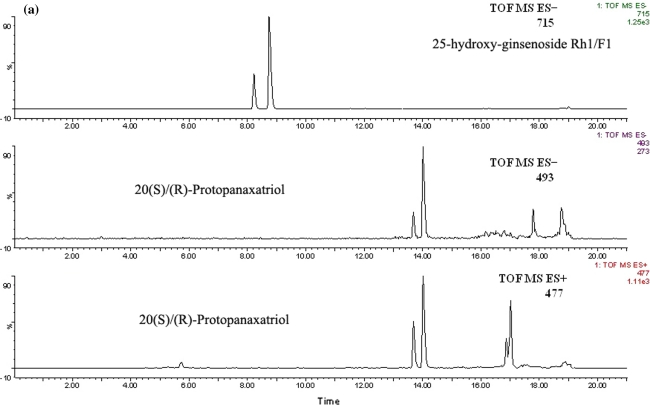
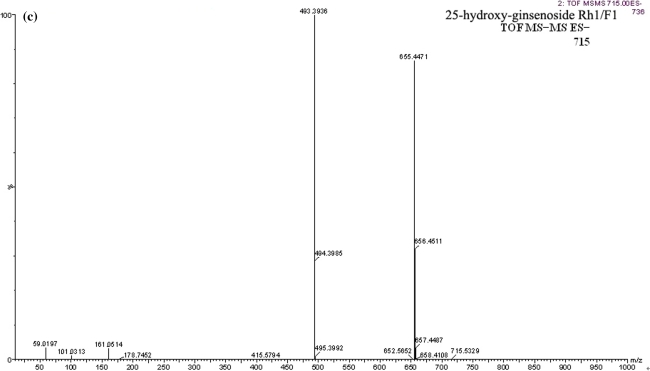
To identify the metabolites accurately, probable structures were first postulated in accordance with the rules and characteristics of drug metabolism in vivo. In this study, the constituents of FTZ extract have been identified. These data may provide guidance for investigating the metabolites of FTZ in rat serum. M1 was identified as the glucuronide conjugate of alkaloids, jatrorrhizine3-O-β-d-glucuronide, since it showed the m/z 514 [M]+ in MS spectra, and exhibited m/z 338 [M−GlcUA]+ in MS2 spectra, which was confirmed by comparison with literature data [34]. M2 and M3 were suspected to be metabolite of ginsenoside Rh1/F1, both of them showed the same molecular ion at m/z 715 [M−H+CH3COOH]− in MS spectra, and exhibited product ions m/z 655 [M−H]− and m/z 493 [M−H−Glu]− in MS2 spectra. By comparison with the literature data [35, 36], this showed the same fragmentation pathway as the metabolite of ginsenoside Rh1/F1, so the two constituents were identified as the 25-hydroxyl-ginsenoside Rh1/F1. Using the same method, M5 and M6 were identified as 20(S)/(R)-protopanaxatriol because they showed the m/z 477 [M+H]+ ion in positive ion mode and m/z 493 [M−H+H2O]− and m/z 553 [M−H+H2O+CH3COOH]− ions in negative ion mode. By comparison with the literature data [36], we suggested that M5 and M6 may be sapogenin which formed by loss of all glycosidic units from protopanaxatriol saponins. The MS and MS2 spectra and possible metabolic pathways of 25-hydroxy-ginsenoside Rh1/F1 and protopanaxatriol in positive and negative ion mode are shown in Fig. 5a–d. M4 and M7 showed the molecular ion at m/z 697 [M−H+CH3COOH]− in MS spectra, and exhibited m/z 441, 423 and 405 in MS2 spectra, which hinted those maybe the metabolites of ginsenoside Re and ginsenoside Rg1, by losing of one glucose molecular and/or one rhamnose molecular. By comparison with literature data [35, 37], we suggested that both of them were 20(S) (R)-ginsenoside Rh1/ginsenoside F1. M8 showed a molecular ion at m/z 798 [M−H]− in MS spectra, and exhibited m/z 717 [M−H−SO3]− in MS2 spectra, which was consistent with the fragmentation of salvianolic acid B sulfates. In accordance with the literature data on the characteristic of MS/MS [36], M8 was identified as salvianolic acid B sulfates. M9 showed a molecular ion at m/z 783 [M−H]− in MS spectra, and exhibited m/z 621 [M−H−Glu]− and 459 [M−H−2Glu]− in MS2 spectra. The results showed the same fragmentation pathway as the metabolite of ginsenoside Rb1 and ginsenoside Rd. By comparison with literature data [38, 39], M9 was suggested as ginsenoside Rg3.
Fig. 5.
EIC (a), MS (b) and MS2 (c) spectra of 25-hydroxy-ginsenoside Rh1/F1 and protopanaxatriol and the possible metabolic biotransformation pathways (d)
Conclusion
By analyzing the constituents in rat serum of FTZ based on UPLC–MS technique and serum pharmacochemistry approach, a method for rapid analysis of the potential effective constituents in a Chinese Medicine formula FTZ have been established. In this study, 27 of the prototype constituents and 9 of the metabolites in rat blood after oral administration of FTZ were identified by the UPLC/Q–TOF system, which enhanced the speed and targeting of bioactive constituents analysis.
These results indicated that most of the alkaloids, ginsenosides, and pentacyclic triterpenes could be observed in rat blood through oral administration of FTZ. Meanwhile the salvianolic acid analogues could be converted into metabolites, such as salvianolic acid B sulfates. Our present work on the comprehensive analysis of the FTZ constituents in rat serum suggest that the serum pharmacochemistry study using UPLC–Q–TOF technique offer a rapid and reliable approach for the identification of potential bioactive compositions for complex herb prescriptions. Systemic pharmacokinetic investigation of the constituents in rat serum after oral administration of FTZ is warranted for better understanding the pharmacokinetic basis of the health benefits of FTZ.
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of P. R. China (No. 30973913 et No. 81102502), Major projects of Chinese National Science and Technology (2009zx09103-407), Projects of combining production, teaching and research of Guangdong Province and Ministry of Education (No. 2009B 090 300349). We appreciate Miss Nazia Ali for editing the manuscript.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.Guo J, Bei WJ, Hu YM, Tang CP, He W, Liu XB, Huang LH, Cao Y, Hu XG, Zhong XL, Cao L. J Ethnopharmacol. 2011;135(2):299–307. doi: 10.1016/j.jep.2011.03.012. [DOI] [PubMed] [Google Scholar]
- 2.Tang FT, Guo J, He W, Wang LJ, Luo DS, Bei WJ (2011) Chin J Integr Med 1(Epub ahead of print) [DOI] [PubMed]
- 3.Wu HY, Bei WJ, Guo J. J Geriatr Cardiol. 2009;6:119–125. [Google Scholar]
- 4.Tang FT, Cao Y, Wang TQ, Wang LJ, Guo J, Zhou XS, Xu SW, Liu WH, Liu PQ, Huang HQ. Eur J Pharmacol. 2011;650(1):275–284. doi: 10.1016/j.ejphar.2010.07.038. [DOI] [PubMed] [Google Scholar]
- 5.Tang CP, Jiang T, Guo J, Wei YP, Yang CY, Chen FC. J Chin Med Mater. 2010;33:1285–1289. [PubMed] [Google Scholar]
- 6.Guo J, Bei WJ, Tang CP, Hu YM, Chen FC, Huang GB, Luo DS. J Chin Med Mater. 2009;32:582–585. [Google Scholar]
- 7.Cao J, Qi LW, Chen J, Yi L, Li P, Ren MT, Li YJ. Biomed Chromatogr. 2009;23:397–405. doi: 10.1002/bmc.1130. [DOI] [PubMed] [Google Scholar]
- 8.Chan ECY, Yap SL, Lau AJ, Leow PC, Toh DF, Koh Hwee-Ling. Rapid Commun Mass Spectrom. 2007;21:519–528. doi: 10.1002/rcm.2864. [DOI] [PubMed] [Google Scholar]
- 9.Wang DW, Liu ZQ, Guo MQ, Liu SY. J Mass Spectrom. 2004;39:1356–1365. doi: 10.1002/jms.727. [DOI] [PubMed] [Google Scholar]
- 10.Wang XJ. J Chin Mater Med. 2006;31:789–792. [PubMed] [Google Scholar]
- 11.Wang X, Sun H, Fan Y, Li L, Makino T, Kano Y. Biol Pharm Bull. 2005;28(6):1106–1108. doi: 10.1248/bpb.28.1106. [DOI] [PubMed] [Google Scholar]
- 12.Sun H, Lv H, Zhang Y, Wang X, Bi K, Cao H. J Sep Sci. 2007;30(18):3202–3206. doi: 10.1002/jssc.200700251. [DOI] [PubMed] [Google Scholar]
- 13.Wang X, Lv H, Sun H, Jiang X, Wu Z, Sun W, Wang P, Liu L, Bi K. J Sep Sci. 2008;31(1):9–15. doi: 10.1002/jssc.200700376. [DOI] [PubMed] [Google Scholar]
- 14.Lv H, Sun H, Wang X, Sun W, Jiao G, Zhou D, Zhao L, Cao H, Zhang G. J Sep Sci. 2008;31(4):659–666. doi: 10.1002/jssc.200700596. [DOI] [PubMed] [Google Scholar]
- 15.Wang XJ, Sun WJ, Sun H, Lv HT, Wu ZM, Wang P, Liu L, Cao HX. J Pharm Biomed Anal. 2008;46:477–490. doi: 10.1016/j.jpba.2007.11.014. [DOI] [PubMed] [Google Scholar]
- 16.Zhou DY, Xu Q, Xue XX, Zhang FF, Liang XM. J Pharm Biomed Anal. 2006;42:441. doi: 10.1016/j.jpba.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 17.Lai CM, Li SP, Yu H, Wan JB, Kan KW, Wang YT. J Pharm Biomed Anal. 2006;40:669. doi: 10.1016/j.jpba.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 18.Liu JH, Wang X, Cai SQ, Komatsu K, Namba T. J Chin Pharm Sci. 2004;4:225–236. [Google Scholar]
- 19.Wang SP, Liu L, Wang LL, Jiang P, Zhang JQ, Zhang WD, Liu RH. Rapid Commun Mass Spectrom. 2010;24:1650. doi: 10.1002/rcm.4561. [DOI] [PubMed] [Google Scholar]
- 20.Liu SY, Cui M, Liu ZQ, Song FR. J Am Soc Mass Spectrom. 2004;15:133. doi: 10.1016/j.jasms.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 21.Zhou L, Kang J, Fan L, Ma XC, Zhao HY, Han H, Wang BR, Guo DA. J Pharm Biomed Anal. 2008;47:42. doi: 10.1016/j.jpba.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 22.Cardoso SM, Guyot S, Marnet N, Lopes-da-Silva JA, Renard CM, Coimbra MA. J Sci Food Agric. 2005;85:25. doi: 10.1002/jsfa.1925. [DOI] [Google Scholar]
- 23.Liu AH, Guo H, Ye M, Lin YH, Sun JH, Xu M, Guo DA. J Chromatogr A. 2007;1161:170. doi: 10.1016/j.chroma.2007.05.081. [DOI] [PubMed] [Google Scholar]
- 24.Xu M, Guo H, Han J, Sun SF, Liu AH, Wang BR, Ma XC, Liu P, Qiao X, Zhang ZC, De-an Guo DA. J Chromatogr A. 2007;858:184–198. doi: 10.1016/j.chroma.2006.10.073. [DOI] [PubMed] [Google Scholar]
- 25.Deevanhxay P, Suzuki M, Maeshibu N, Li H, Tanaka K, Hirose S. J Pharm Biomed Anal. 2009;50:417–418. doi: 10.1016/j.jpba.2009.05.023. [DOI] [PubMed] [Google Scholar]
- 26.Wang C, Wang SW, Fan GR, Zou HF. Anal Bioanal Chem. 2010;396:1736. doi: 10.1007/s00216-009-3409-1. [DOI] [PubMed] [Google Scholar]
- 27.Chen JH, Wang FM, Liu J, Lee Sen-Chun F, Wang XR, Yang HH. Anal Chim Acta. 2008;613:189. doi: 10.1016/j.aca.2008.02.060. [DOI] [PubMed] [Google Scholar]
- 28.Kinuko I, Masataka M, Takao Y, Takashi T, Dong-Ung L, Wolfgang W. J Nat Prod. 2001;7:896–898. [Google Scholar]
- 29.Sun JH, Yang M, Wang XM, Xu M, Liu AH, Guo DA. J Pharm Biomed Anal. 2007;44:564. doi: 10.1016/j.jpba.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 30.Zhu ZY, Zhang H, Zhao L, Dong X, Li X, Chai YF, Zhang GQ. Rapid Commun Mass Spectrom. 2007;21:1855–1865. doi: 10.1002/rcm.3023. [DOI] [PubMed] [Google Scholar]
- 31.Song JZ, Li SL, Zhou Y, Qiao CF, Chen SL, Xu HX. J Pharm Biomed Anal. 2010;53:279–286. doi: 10.1016/j.jpba.2010.03.022. [DOI] [PubMed] [Google Scholar]
- 32.Concannon S, Ramachandran VN, Smyth WF. Rapid Commun Mass Spectrom. 2000;14:1161–1162. doi: 10.1002/1097-0231(20001215)14:23<2260::AID-RCM161>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 33.Hu MY, Su GH, Sze Cho-Wing S, Ye WC, Tong Y. Biomed Chromatogr. 2010;24:443. doi: 10.1002/bmc.1311. [DOI] [PubMed] [Google Scholar]
- 34.Zhu MM, Han FM, Chen HX, Peng ZH, Chen Y. Rapid Commun Mass Spectrom. 2007;21:2019–2022. doi: 10.1002/rcm.3057. [DOI] [PubMed] [Google Scholar]
- 35.Chen GT, Gao HY, Song Y, Yang M, Guo DA, Wu LJ. Mod Chin Med. 2008;11:37–40. [Google Scholar]
- 36.Wei YJ, Li P, Shu B, Song Y. Chin J Anal Chem. 2007;1:13–18. [Google Scholar]
- 37.Chen GT, Yang M, Guo DA. Chin J Chin Mater Med. 2009;12:1541–1542. [Google Scholar]
- 38.Yang L, Xu SJ, Zeng X, Liu YM, Deng SG, Wu ZF, Ou RM. Acta Pharm Sinica. 2006;8:742–746. [PubMed] [Google Scholar]
- 39.Yang L, Xu SJ, Zeng X, Wu ZF, Deng YH, Liu YM, Deng SG, Ou RM. Chem J Chin Univ. 2006;27:1042–1044. [Google Scholar]