Termination codon readthrough is used as a mechanism of expression of a growing number of viral and cellular proteins, but in many cases, the mRNA signals that promote readthrough are poorly characterized. Here, the authors investigated the readthrough signal of Colorado tick fever virus (CTFV) segment 9 RNA (Seg-9). Seg-9 encodes a 36-kDa protein VP9 and by readthrough of a UGA stop codon, a 65-kDa product, VP9′. Using a reporter system, the authors defined the minimal sequence requirements for readthrough and confirmed activity in both mammalian and insect cell-free translation systems, and in transfected mammalian cells. Mutational analysis revealed that readthrough was UGA-specific and that the local sequence context around the UGA influenced readthrough efficiency.
Keywords: ribosome, translation, stop codon, RNA structure, readthrough, CTFV
Abstract
Termination codon readthrough is utilized as a mechanism of expression of a growing number of viral and cellular proteins, but in many cases the mRNA signals that promote readthrough are poorly characterized. Here, we investigated the readthrough signal of Colorado tick fever virus (CTFV) segment 9 RNA (Seg-9). CTFV is the type-species of the genus Coltivirus within the family Reoviridae and is a tick-borne, double-stranded, segmented RNA virus. Seg-9 encodes a 36-kDa protein VP9, and by readthrough of a UGA stop codon, a 65-kDa product, VP9′. Using a reporter system, we defined the minimal sequence requirements for readthrough and confirmed activity in both mammalian and insect cell-free translation systems, and in transfected mammalian cells. Mutational analysis revealed that readthrough was UGA specific, and that the local sequence context around the UGA influenced readthrough efficiency. Readthrough was also dependent upon a stable RNA stem–loop structure beginning eight bases downstream from the UGA codon. Mutational analysis of this stem–loop revealed a requirement for the stem region but not for substructures identified within the loop. Unexpectedly, we were unable to detect a ribosomal pause during translation of the CTFV signal, suggesting that the mechanism of readthrough, at least at this site, is unlikely to be dependent upon RNA secondary-structure induced ribosomal pausing at the recoded stop codon.
INTRODUCTION
Translation termination begins when a stop codon enters the decoding center of the A-site of the small ribosomal subunit. Stop codons are recognized by protein release factors, which subsequently promote hydrolysis of the peptidyl–tRNA linkage in the peptidyl transferase center of the large subunit (Petry et al. 2008; Loh and Song 2010). In eukaryotes, termination requires the collective action of two release factors, eRF1, in part a structural mimic of an A-site tRNA (Song et al. 2000; Laurberg et al. 2008), and eRF3, which stimulates the process. While eRF1 binds to and recognizes all three stop codons, the formation of an eRF1–eRF3 complex and the eRF3-associated GTPase activity are required for termination in vivo (for review, see von der Haar and Tuite 2007). Termination is generally a highly efficient process, but is influenced by the nature of the stop codon present (UAA, UAG, or UGA) and its flanking nucleotides, especially the immediately adjacent 3′ base (for review, see Harrell et al. 2002). Some termination codon contexts are noticeably “leaky” (e.g., UGAC) (McCaughan et al. 1995); that is, they allow “readthrough” at frequencies ranging from 0.3% to 5% (Bertram et al. 2001). In readthrough, the stop codon is misread as a sense codon (it can be said to have been “recoded”) and decoded by a near-cognate or suppressor tRNA, with translation continuing to the next termination codon. Readthrough is exploited in the expression of several viral and cellular genes, where it is referred to as “programmed” readthrough (for review, see Namy et al. 2004; Dreher and Miller 2006; von der Haar and Tuite 2007; Namy and Rousset 2010). Readthrough allows the production of a C-terminally extended polypeptide at a defined frequency, and in viruses it is often used to express replicases.
Until recently the body of experimental and comparative analysis had indicated that most plant, and many animal virus readthrough signals required only limited flanking information (Harrell et al. 2002). For example, in the alphavirus Sindbis virus, expression of the viral polymerase (nsp4) by readthrough of a UGA codon was thought to require only a single cytosine residue immediately downstream from the stop codon (Li and Rice 1993). Similarly, in the tobamovirus tobacco mosaic virus (TMV), expression by readthrough of the p183 RNA-dependent RNA polymerase is believed to involve only the six bases following the stop (UAG CAA UUA) (Skuzeski et al. 1991). Indeed, eukaryotic readthrough signals have been classified into three main groups based on either a (perceived) sole requirement for local sequence context (Types I and II), or the additional involvement of a 3′ structural element (Type III) (Beier and Grimm 2001; Harrell et al. 2002). In Type I, (tobamovirus-like), the six bases 3′ of the stop codon usually conform to the motif UAG-CAA-UYA, and in Type II (alphavirus-like), the stimulatory context is generally UGA-CGG or UGA-CUA. Type III signals are exemplified by that of the retrovirus Moloney murine leukemia virus (MuLV), which possesses a 3′ stimulatory RNA structure in the form of an RNA pseudoknot located eight nucleotides downstream from the gag UAG codon (Wills et al. 1991; Feng et al. 1992; Alam et al. 1999; for review, see Brierley et al. 2007). Recent work, however, has suggested that mRNA secondary structures are likely to be more generally involved in readthrough than had previously been thought (Firth et al. 2011). In particular, phylogenetic and experimental analysis suggests that the definition of the Type II motif should (in general, though perhaps not ubiquitously) be modified to include a 3′ RNA structural component. A functional 3′ stimulatory RNA stem–loop has now been identified in several alphaviruses, including Sindbis virus, and is predicted for several plant viruses with Type II organization (Firth et al. 2011). Secondary structure–dependent readthrough signals present an additional level of complexity that could promote readthrough to higher levels or offer the possibility of regulation (Robinson and Cooley 1997; Orlova et al. 2003).
In this study we describe an investigation of the readthrough signal of Colorado tick fever virus (CTFV) segment 9 RNA (Seg-9) (Fig. 1). CTFV is the type-species of the genus Coltivirus within the family Reoviridae and is a tick-borne, double-stranded, segmented RNA virus. Coltiviruses have been isolated from rodents, humans, and varieties of Ixodidae ticks. CTFV is endemic in northwestern America and is responsible for Colorado tick fever, a disease characterized in humans by symptoms including fever, chills, headaches, and malaise (Mohd Jaafar et al. 2009). CTFV RNA Seg-9 encodes a 36-kDa protein VP9 and by readthrough of a UGA stop codon, a 65-kDa product, VP9′ (Mohd Jaafar et al. 2004). The readthrough product has previously been detected in virus-infected cells, but the signal has not been characterized beyond computer-aided RNA folding, which revealed the possibility of an RNA secondary structure downstream from the recoded UGA (Mohd Jaafar et al. 2004). Here, we used a readthrough reporter construct to investigate the CTFV readthrough signal. In addition to defining the key elements of local context, we identified and characterized a stable RNA hairpin located eight bases downstream from the UGA codon. Surprisingly, we were unable to detect a ribosomal pause during translation of the CTFV signal, suggesting that the mechanism of readthrough, at least at this site, is unlikely to be dependent upon RNA secondary-structure induced ribosomal pausing at the recoded stop codon.
FIGURE 1.
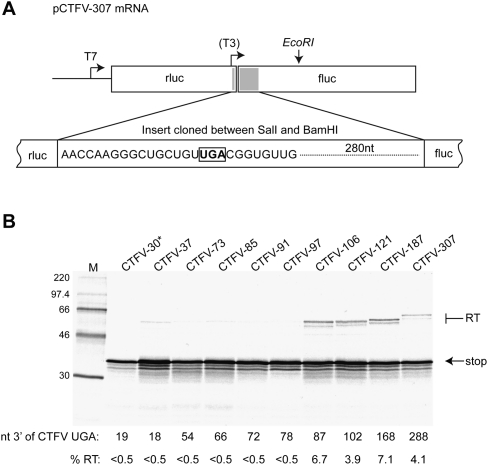
Characterization of the minimal sequence requirements for CTFV readthrough. (A) Schematic of the pCTFV-307 reporter mRNA. A portion of the CTFV Seg-9 sequence (indicated as a gray box) encompassing the VP9 UGA stop codon (16 nt upstream of and 208 nt downstream from the UGA) was cloned into the SalI and BamHI sites of the p2luc reporter plasmid. Transcripts for in vitro translation were generated using T7 RNA polymerase and EcoRI-cut pCTFV-307 and derivatives. The location of the T3 promoter present in the structure mapping construct pCTFV-121-T3 is indicated. (B) Deletion analysis of the CTFV readthrough signal. A series of reporter plasmid variants were prepared with stepwise, in-frame deletions from the 3′ end of the inserted viral sequence. Plasmids were linearized with EcoRI and run-off transcripts translated in Flexi RRL at a final RNA concentration of 50 μg/mL in the presence of [35S]methionine and 140 mM added KCl. The products were resolved by 12% SDS-PAGE and visualized by autoradiography. Molecular size markers were also run on the gel (M). The number of nucleotides 3′ of the CTFV UGA is shown below the gel. Products derived from termination at the CTFV UGA (stop) or following readthrough (RT) are indicated by arrows. The readthrough efficiency of each mRNA is indicated (%RT). The asterisk (CTFV-30) highlights that this mRNA has a shorter stretch upstream of the UGA (11 nt) in comparison to the other mRNAs (16 nt).
RESULTS
Defining the 3′ boundary of the CTFV readthrough signal
The first open reading frame (ORF) of the CTFV Seg-9 mRNA (nt 41–1051) ends in an opal (UGA) stop codon and encodes VP9, a structural protein of 337 amino acids; the second (nt 1052–1846) is accessed by readthrough of the UGA stop signal and encodes an extended, 602-amino acid product, VP9′. To determine the minimal 3′ region required for readthrough in CTFV, a 307-bp fragment of viral cDNA was cloned between the SalI and BamHI sites of the dual-luciferase reporter vector p2luc (Grentzmann et al. 1998) to generate plasmid CTFV-307 (Fig. 1). The cloned fragment contained 16 bp upstream of and 288 bp downstream from the VP9 UGA stop codon, and was expected to contain all of the sequences necessary for readthrough based on other known viral readthrough signals (Namy and Rousset 2010). CTFV-307 contains the Renilla (rluc) and Firefly luciferase (fluc) ORFs in the configuration 5′ rluc-VP9-UGA-VP9′-fluc 3′, thus ribosomes that terminate at the CTFV UGA codon express rluc, and those that readthrough, an rluc-fluc fusion protein. CTFV-307 contains a T7 RNA polymerase promoter upstream of rluc, allowing synthetic mRNAs to be generated to investigate readthrough in in vitro translation reactions, and an SV40 promoter to facilitate examination of readthrough in transfected tissue culture cells.
The translation of in vitro synthesized EcoRI-cut CTFV-307 mRNA was carried out in Flexi rabbit reticulocyte lysate (FlexiRRL) supplemented with 140 mM KCl (see Materials and Methods) and gave products of the expected sizes (rlucVP9 “stop” product, ∼37 kDa, rlucVp9-VP9′fluc readthrough product, ∼70 kDa) (Fig. 1B). Following quantification, and taking into account the methionine content of the two proteins, the readthrough efficiency was calculated to be 4.1%; thus, approximately one ribosome in 25 reads through the Seg-9 VP9 UGA codon. In order to determine the minimal 3′ sequence requirement for readthrough, further reporter plasmids were prepared that contained progressively less viral information at the 3′ end. As can be seen in Figure 1B, the readthrough product was synthesized efficiently when ≥87 nt of CTFV sequence was present downstream from the stop codon, but was sharply reduced (to <0.5%) upon removal of a further 9 nt (CTFV-97). These data indicate that the CTFV readthrough signal is contained within a 106-nt stretch, of which 87 nt are present immediately downstream from the recoded UGA. Longer inserts were also tested (up to 469 nt, comprising over 60% of the downstream ORF) and the readthrough efficiencies were in the region from 4% to 6% (data not shown). Thus, it seems unlikely that additional mRNA elements are present outside of the 106-nt region identified above (and this is supported by phylogenetic analysis) (see Firth et al. 2011).
RNA secondary structure probing of the CTFV readthrough signal
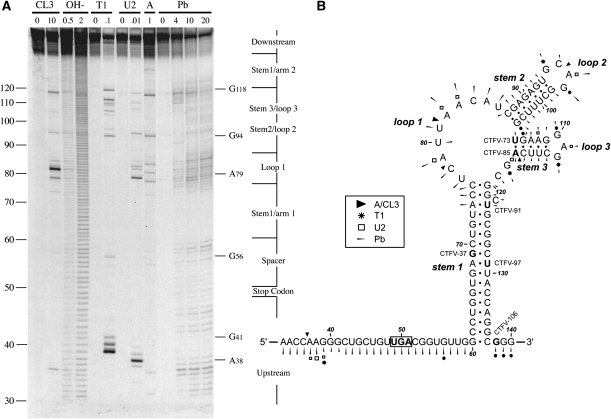
In their study of readthrough in CTFV, Mohd Jaafar et al. (2004) proposed the potential involvement of a stem–loop structure predicted by mfold, as shown in Figure 2. We used enzymatic and chemical RNA structure probing to investigate whether this configuration formed in the mRNA. Plasmid CTFV-121-T3 was linearized with BamHI, T3 run-off transcripts synthesized, and the 153-nt long RNA end-labeled with [γ−33P]ATP. The labeled transcripts were subjected to limited chemical and enzymatic RNA structure probing prior to analysis on 10% denaturing polyacrylamide gels. The enzymatic probes used were RNases CL3, T1, and U2, which preferentially cleave single-stranded C, G, and A residues respectively, and RNase A, which targets single-stranded CpA and UpA. A chemical probe was also used, lead acetate, which shows general specificity for single stranded regions. A representative stucture mapping gel is shown in Figure 2A; and in Figure 2B the data are mapped onto the mfold prediction. The bases are numbered such that +1 represents the first base of the transcript and the recoded UGA thus corresponding to bases 49–51.
FIGURE 2.
Structure probing of the CTFV readthrough signal. (A) RNA derived by transcription of pCTFV-121-T3/BamHI with T3 RNA polymerase was 5′ end-labeled with [γ-33P]ATP and subjected to limited RNase or chemical cleavage using structure-specific probes. Sites of cleavage were identified by comparison with a ladder of bands created by limited alkaline hydrolysis of the RNA (OH-; RNA heated to 100°C for 0.5 or 2 min) and the position of known RNase U2 and T1 cuts, determined empirically. Products were analyzed on a 10% acrylamide/7M urea gel containing formamide. Enzymatic structure probing was with RNases CL3, T1, U2, and A. Uniquely cleaved nucleotides were identified by their absence in untreated control lanes (0). The number of units of enzyme added to each reaction is indicated. Chemical structure probing was with lead acetate (Pb; mM concentration in reaction). (B) The sequence of the probed CTFV RNA and the inferred secondary structure. The sensitivity of bases in the CTFV readthrough region to the various probes is shown for an mfold prediction. The first base of the transcript is numbered 1. The reactivies of the T1 (asterisk), U2 (open square), A and CL3 (black triangle) probes are marked. Lead cleavages are indicated by thin arrows. The size of the symbols is approximately proportional to the intensity of cleavage at that site. Also indicated is the location of the 3′ edge of the truncated versions of CTFV-307 (labeled as in Fig. 1), with the last viral base in each truncation emboldened.
The structure mapping data were highly supportive of the stem–loop structure of Figure 2B. In the gel of Figure 2A, few single-stranded enzymatic cleavages were seen in the predicted stems, but several cuts were seen in the loops, including A79, A82, and A83 in loop 1, and A96 in loop 2 (RNase U2); G94 in loop 2 and G110 and G112 in loop 3 (RNase T1); C78, C84 in loop 1, and C95 in loop 2 (RNase CL3); and C78, U81, C84 in loop 1, and C95 in loop 2 (RNase A). Strong protection from lead acetate cleavage was observed in two main regions, G60–G72 and G124–C137, strongly supporting the existence of the long helix (stem 1) in the mfold of Figure 2B. In further support, no enzymatic cleavages were observed in this region. While the predicted base pairs at the top of stem 1 (U73–C76 with G120–G124) did show limited accessibility to lead acetate, and weak cleavage of U73 was observed with RNases A and CL3, this is likely to be a consequence of the proposed unpaired base (C122) in the 3′ arm of stem 1 and the weak U73–G124 pair in close proximity. It seems likely, therefore, that the top of stem 1 may “breathe.” Protection against lead acetate cleavage was also evident for stem 2 and stem 3, although not as strong as that seen for stem 1. This suggests that stems 2 and 3 may be less stable, and consistent with this, they have a greater proportion of AU and GU base pairs. Indeed, we noticed some cleavage of stem 3 with single-stranded enzymatic probes (e.g., weak RNase T1 cleavage of G106 and G109; strong RNase A cleavage of C116).
The pattern of cleavages observed is highly consistent with the mfold prediction of a stable stem–loop structure with a pair of less-stable hairpins within the major loop. Taking the deletion analysis into account (Fig. 1), it is likely that the identified structure has a key role in readthrough. The end-point of the last functional deletion mutant (CTFV-106) was 87 nt downstream from the UAG, corresponding to base G138 in the structure mapping RNA, which lies immediately downstream from the 3′ arm of stem 1 (Fig. 2). The next deletion mutant (CTFV-97) showed greatly reduced activity in readthrough and consistent with this, would lack most of the 3′ arm of stem 1. The cleavage pattern flanking the major stem was largely consistent with single-stranded RNA conformation (e.g., the pentamer AAGGG at positions 37–41 was highly reactive), but there were noticeably fewer enzymatic cleavages around the recoded UGA and immediately 3′ of this. It is possible that this region is inaccessible to the enzymes, perhaps due to steric hindrance, since it is accessible to the chemical probe.
Mutational analysis of the stimulatory stem–loop structure
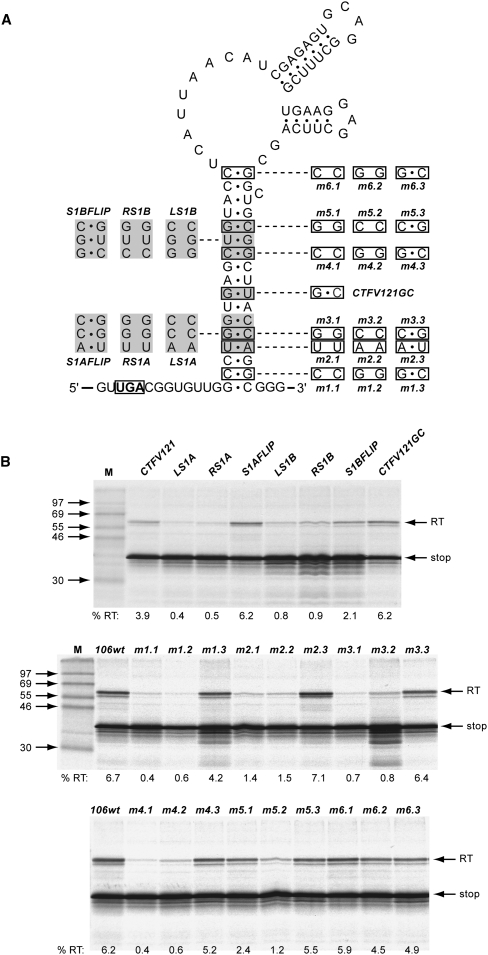
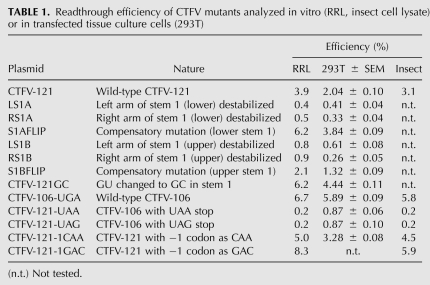
To investigate the involvement of the stem–loop structure and its integral hairpins in readthrough, we introduced a series of mutations into this region and assessed readthrough in RRL. Initially, we focused on stem 1, creating individual complementary mutations within each arm of the stem that should destabilize the structure, and double mutations with changes in both arms of the stem that should be compensatory and allow the stem to reform (Fig. 3). The first set of mutations were created in CTFV-121 and involved changes to three consecutive base pairs in two blocks, one in the lower region of stem 1 (LS1A, RS1A, S1Aflip), the other in the upper region (LS1B, RS1B, S1Bflip). As introduction of the RS1A mutation would generate an in-frame stop codon in the right arm of stem 1, it was necessary in RS1a and S1Aflip to simultaneously change the central GU pair to GC. As a control, therefore, a version of CTFV-121 was created that also included this change, CTFV-121-GC. In this mRNA, readthrough was found to be increased somewhat (to 6.2%) in comparison to the wild-type sequence (3.9%), probably a consequence of increased stability of this stem. The effect on readthrough of the destabilizing complementary changes in the lower and upper regions of stem 1 were very similar, with readthrough reduced dramatically. In mRNAs with the compensatory double mutations, readthrough was either fully restored (S1Aflip) or restored close to the wild-type value (S1Bflip). A similar pattern of readthrough was observed in assays in transfected 293T-tissue culture cells (Table 1). Thus, the formation of stem 1 is critical in the promotion of efficient readthrough. To investigate this further, a series of six complementary and compensatory mutations were made to individual base pairs along the length of stem 1. The pattern of readthrough observed for these constructs was similar to the block mutations above, in that destabilization of stem 1 reduced readthrough, and reformation of the base pair restored readthrough in all cases (Fig. 3B). It was noticeable, however, that the inhibitory effect of the stem mutations showed some polarity, with individual destabilization mutations being highly inhibitory to readthrough when in the lower and central region of stem 1 (2, 4, 5, and 11 bp), yet, less so toward the top of the stem (13 and 17 bp).
FIGURE 3.
The role of the CTFV stimulatory RNA stem 1 region in readthrough. (A) Complementary and compensatory changes to base pairs within stem 1 were prepared, targeting either blocks of three consecutive base pairs (based on CTFV-121 and shown to the left of the main stem) or single base pairs (based on CTFV-106 and shown to the right of the main stem). In CTFV121GC, the central GU pair was changed to GC. This mutation was also present in RS1A and S1AFLIP. (B) Plasmids containing the mutations detailed in A were linearized, transcribed, translated, and analyzed according to the legend of Figure 1.
TABLE 1.
Readthrough efficiency of CTFV mutants analyzed in vitro (RRL, insect cell lysate) or in transfected tissue culture cells (293T)
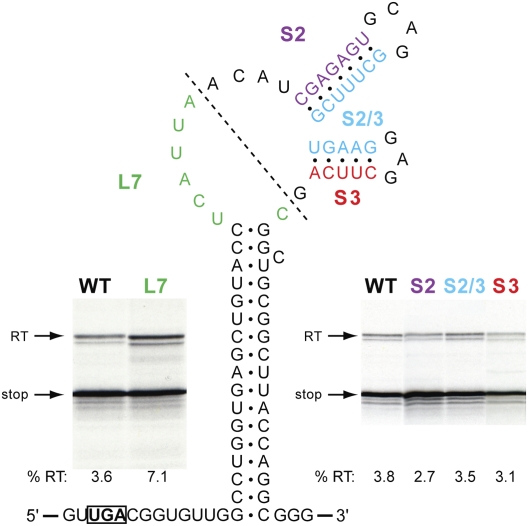
We next investigated the potential involvement in readthrough of the integral hairpins (stem–loops 2 and 3), through destabilizing complementary changes in either the 5′ arm of stem 2 (S2), the 3′ arm of stem 2 plus the 5′ arm of stem 3 (S2/3), or the 3′ arm of stem 3 (S3) (Fig. 4). A further mRNA was also generated (L7), in which the integral hairpins were deleted, leaving only seven bases in the loop of stem 1. From this analysis, it is clear that the substructures in the main loops are not involved in readthrough. Destabilization of the two stems, individually (S2, S3) or together (S2/3), had very little effect on readthrough, and reduction of the size of the main loop by deletion of the integral hairpins, in fact, stimulated readthrough somewhat. The latter observation could be a consequence of an improved stability of stem 1.
FIGURE 4.
Investigating the role of the CTFV stimulatory RNA loop region in readthrough. Bases within arm 1 of stem 2 (S2, purple), arm 2 of stem 2 and arm 1 of stem 3 (S2/3, blue), or arm 2 of stem 3 (S3, red) were changed to the complementary Watson–Crick bases (e.g., in S2, CGAGAGU was replaced by GCUCUCA) and readthrough measured as detailed in the legend to Figure 1. A deletion mutation was also tested (L7, green), in which most of the main loop was removed (boundaries of the deletion indicated by a dotted line), leaving the seven nucleotides highlighted in green.
The observed approximately twofold reduction in in vitro readthrough efficiency of CTFV-121 in comparison to CTFV-106 was recapitulated in tissue culture cells and in an insect cell lysate in vitro translation system (see Table 1 and below). We do not know why this is the case, but a possible explanation is that some competition for folding of the main stem occurs in the presence of the additional 3′ viral sequences of CTFV-121. A modest inhibitory effect was also noted for CTFV-307, albeit not for CTFV-187. The stem–loop is predicted to fold independently (in all of the constructs with ≥87 nt downstream from the UGA) and there is no obvious potential for long-range interactions between the loop and downstream CTFV sequences. We suspect that in some mRNAs, sequences 3′ of the stem–loop can affect stem–loop stability through stacking effects, but further work will be required to confirm this hypothesis.
Local context analysis of VP9 stop codon readthrough
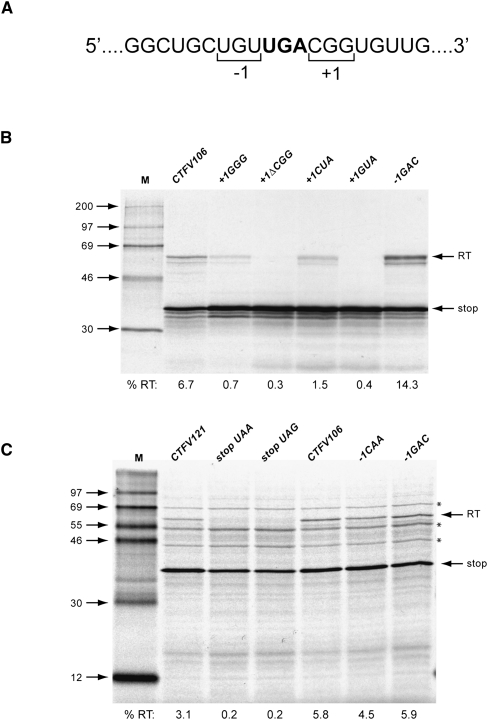
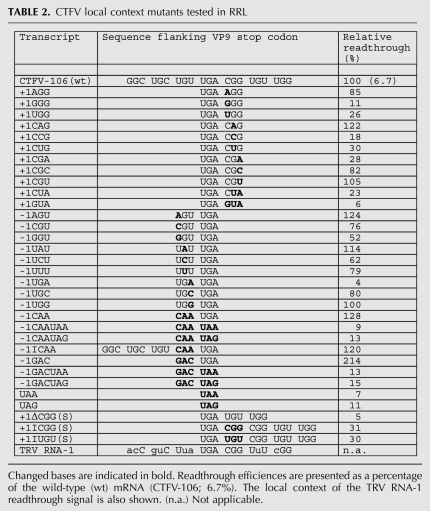
The mRNA sequence immediately 3′ of the stop codon (5′-UGACGGUGU-3′) is similar to that found in a number of Type II readthrough signals, suggesting that the specific sequence context of the VP9 UGA may also contribute to readthrough. We tested three aspects of local sequence context; the importance of the codons immediately 3′ (CGG) or 5′ (UGU) of the UGA, the effect of replacing the UGA codon with alternative stop codons (UAA or UAG), and the consequence of increasing or decreasing the spacing between UGA and stimulatory stem–loop structure. We began by introducing individual point mutations into the CGG triplet and measured readthrough in RRL (Fig. 5; Table 2). Consistent with an involvement in readthrough, most of the mutations reduced readthrough frequency (especially GGG and CCG), albeit some triplets were tolerated (e.g., CAG, CGC, CGU). In a separate experiment, the CGG triplet was replaced by CUA, the codon found in the +1 position (with UGA codon denoted as zero) of the Sindbis virus readthrough signal, or by GUA as a control. With CUA, readthrough was reduced about fourfold, and virtually abolished with GUA. These experiments confirm the importance of the CGG codon and reveal that simple replacement of this codon by the +1 CUA codon present in the Sindbis virus signal (Type II) does not fully restore function. This may be due to inappropriate additional flanking elements or the presence of the stimulatory stem–loop. Mutational analysis of the UGU codon immediately upstream of the UGA (the −1 codon) revealed that this triplet contributes less to readthrough, with a reduction of no more than 50% with any mutation (excepting the change to UGA, which introduced an additional in-frame stop codon and abolished readthrough) and some instances where readthrough was stimulated a little. Replacing the UGU codon with the −1 codon from the readthrough signal of either the alphavirus Venezuelan equine encephalitis virus (VEEV; CAA) or the retrovirus MuLV (GAC) also leads to a stimulation of readthrough, up to twofold with the MuLV-1 codon GAC. How this effect is mediated is uncertain, but it could reflect changes in the mRNA primary sequence, the amino acid encoded by the −1 codon, or the identity of the tRNA decoding the −1 codon (Mottagui-Tabar et al. 1998).
FIGURE 5.
Contribution of local sequence context to CTFV readthrough. (A) RNA sequence flanking the recoded UGA codon (bold) with the triplets encoding the amino acid to the 5′ (−1) or 3′ (+1) of the UGA bracketed. (B) Mutations were introduced into CTFV-106 in the vicinity of the UGA and tested in readthrough assays. Plasmids were linearized, transcribed, translated, and analyzed according to the legend of Figure 1. The gel in this panel shows a selection of the mutants. Further mutants are detailed in Table 2. In the mutations shown in this panel, the +1 CGG codon was replaced by GGG, CUA, GUA, or deleted (ΔCGG), and the −1 UGU codon replaced by GAC. (C) A selection of CTFV RNAs were translated in an insect cell lysate expression system. The asterisked bands are present in unprogrammed translations and are the products of endogenous mRNAs present in the lysate. Further details are provided in Table 1.
TABLE 2.
CTFV local context mutants tested in RRL
Suppression of the alternative stop codons UAA or UAG within the CTFV context was tested in 293T cells and in RRL (Table 1). In 293T cells, readthrough of UAA and UAG occurred about three times less efficiently, but in RRL, readthrough was more dramatically reduced (∼20-fold). We also investigated (in RRL) the effect of including the stimulatory −1 codons CAA or GAC, but this did not greatly promote readthrough of UAA or UAG (Table 2). Readthrough of UAG in the context of the MuLV readthrough signal has been described previously in RRL (Feng et al. 1989), suggesting that the CTFV readthrough signal must contain an element(s) that precludes decoding of UAG (see Discussion).
The necessity for an appropriate (8 nt) spacing between the stop codon and stimulatory RNA was tested by deleting or inserting a codon within the spacer region. Deletion of the +1 codon CGG reduced readthrough about 20-fold, whereas addition of a codon at this position (either CGG or UGU) reduced readthrough about threefold (Table 2). While interpretation of these data is potentially confounded by local context effects, the simplest conclusion is that extending or shortening the spacer has a detrimental effect on readthrough, consistent with the requirement for an appropriate spacer length.
Readthrough in an insect cell lysate
CTFV is an arbovirus whose major vector is the tick Dermacentor andersoni. It was of interest, therefore, to assess readthrough in the commercially available (Promega) in vitro translation system derived from insect cells (prepared from the Spodoptera frugiperda Sf21 cell line), which may be a more relevant cell type. As can be seen in Figure 5C, the pattern of readthrough observed in the insect cell lysate with the test mRNAs was in complete accordance with that seen in RRL. This indicates that insect ribosomes are able to recognize the stimulatory elements of the CTFV readthrough signal and supports the biological relevance of the data acquired in RRL. As can be seen in Figure 5, no readthrough was observed when the UGA stop codon was replaced by UAA or UAG; thus, insect cells do not appear to contain a suppressor tRNA capable of reading UAA or UAG in the context of the CTFV signal.
Absence of ribosomal pausing at the CTFV readthrough site
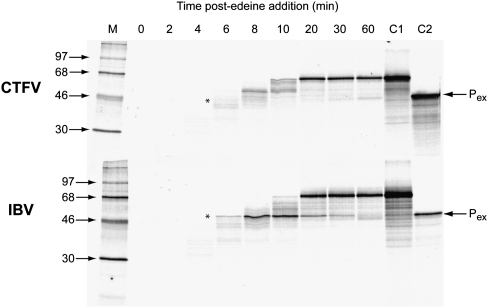
One possible role of the stimulatory hairpin is to induce ribosomal pausing over the stop codon, increasing the time for the action of a suppressor tRNA. Such RNA structure-induced ribosomal pausing has been extensively documented at sites of ribosomal frameshifting (for review, see Brierley et al. 2007). To test this possibility, pausing was assayed using an established time course assay (Somogyi et al. 1993; Kontos et al. 2001) and an mRNA derived from plasmid pPS0-CTFV-121-CGA, which contains the CTFV-121 readthrough signal cloned in-frame within the influenza PB1 gene, and the UGA codon replaced by CGA. The extent of pausing was estimated by comparing the levels of a translational intermediate corresponding to pausing at the stem–loop with that of full-length polypeptide produced during a time course of translation in RRL. To facilitate detection of intermediates corresponding to ribosomal pausing, the standard translation reaction was modified in two ways. First, the reactions were carried out at 26°C rather than 30°C, since the general reduction in the rate of translation at the lower temperature creates a longer “window” for recognition of translational intermediates. Second, in order to simplify the pattern of intermediates observed, translation was synchronized by the addition of edeine, a potent inhibitor of initiation (Szer and Kurylo-Borowska 1970), 3 min after the start of the reaction. As a positive control, an RNA containing the ribosomal frameshifting signal of the coronavirus infectious bronchitis virus (IBV) (pPS1a) (Kontos et al. 2001) was also assayed. As can be seen in Figure 6, both mRNAs specified the synthesis of a full-length product of ∼68 kDa. In the case of the control mRNA derived from pPS1a, a transient translational intermediate was seen (asterisk), whose size was consistent with it being derived from pausing at the IBV RNA pseudoknot. Indeed, identification of this polypeptide as a pseudoknot-induced product was further strengthened by the observation that it comigrated with the translation product of transcripts from pPS0 digested with XhoI, which cleaves the plasmid at the position of the inserted pseudoknot sequence of pPS1a. However, a pause product was completely absent in translations of pPS0-CTFV-121-CAG, indicating that the stimulatory activity in readthrough of the CTFV stem–loop does not seem to be mediated through the induction of ribosomal pausing.
FIGURE 6.
The CTFV stem–loop stimulatory RNA does not induce ribosomal pausing. Messenger RNAs derived from AvaII-cut pPS0-CTFV-106-CGA were translated in RRL for 3 min at 26°C prior to addition of edeine to 5 μM. Aliquots were removed at various times post-edeine addition, translation stopped, and products resolved on a 10% SDS–polyacrylamide gel. As a positive control, the experiment was also carried out with an mRNA derived from AvaII-cut pPS1a, which contains the frameshift-promoting pseudoknot of the coronavirus IBV at the equivalent position in the mRNA as the CTFV stimulatory RNA in pPS0-CTFV-106-CGA. The expected size of the pausing product (Pex; shown by an asterisk) was marked by translating RNA produced from XhoI-cleaved pPS0 (C2). Control C1 represents the translation product produced from the mRNAs in the absence of added edeine.
DISCUSSION
Stop codon readthrough has been documented in the expression of several viral and cellular genes (for review, see Namy and Rousset 2010). It is an elegant way to produce two proteins from a single mRNA, and in a defined ratio, with the potential for modulating this ratio through trans-acting factors. In gammaretroviruses, targeting of the viral replicase enzymes (derived from Pol) into virions is achieved by virtue of their expression as a C-terminal extension of Gag, following readthrough. Regarding CTFV, the role of readthrough is not fully understood, but the potential benefits are apparent. Viruses of the family Reoviridae have segmented, double-stranded RNA genomes that are synthesized within the protective space of the assembling capsid, which imposes a constraint on genome segment number. Readthrough could well be an adaptation to allow the synthesis of an additional protein without exceeding the maximum number of segments. The importance of maintaining a defined ratio of VP9 (a structural component of the viron) and VP9′ (a cytoplasmic protein of unknown function) remains to be determined. There is evidence that the levels of VP9′ diminish late in the virus replication cycle (Mohd Jaafar et al. 2004), but whether this reflects virus-controlled modulation of readthrough or is a consequence of diminished host cell viability remains to be determined. The idea that the efficiency of readthrough can be modulated is not without precedent. In MuLV, the viral reverse transcriptase itself can bind to and sequester eRF1, promoting readthrough and stimulating Gag-Pol synthesis to levels necessary for efficient replication of the virus (Orlova et al. 2003). Expression of the readthrough product of the Drosophila Kel gene also appear to be regulated—in this case during embryonic development—but the molecular basis is not understood (Robinson and Cooley 1997).
In the present study, we show that efficient readthrough in CTFV requires the presence of a downstream RNA stem–loop stimulator, supporting the view that many Type II readthrough signals have such stimulatory elements (Firth et al. 2011). To date, the best-studied readthrough stimulatory RNA structure is the MuLV pseudoknot (Wills et al. 1991; Feng et al. 1992; Alam et al. 1999). In vitro assays have revealed a requirement for certain evolutionarily conserved primary sequence elements within the MuLV signal, including several bases in loop 3 of the pseudoknot and within the 8-base spacer separating the recoded UAG and pseudoknot (Wills et al. 1994; Alam et al. 1999). The requirement for particular nucleotides in this pseudoknot does not seem to reflect an explicit structural role (Alam et al. 1999), indicating that they could be involved in interactions with a component(s) of the translation apparatus or other cytoplasmic factor(s). In contrast to the MuLV signal, the CTFV stimulatory RNA is a stem–loop and has no obvious primary nucleotide sequence requirements. Furthermore, there is no requirement for the loop substructures, and the upper portion of stem 1 is probably also dispensible for readthrough. Given the obvious differences between the MuLV and CTFV signals, an important question is how such disparate structures promote readthrough. The stimulatory RNAs presumably act to modulate the rate at which the stop codon is recognized by eRF1–eRF3 in competition with suppressor tRNA, but whether this is achieved by the same mechanism is debatable. In principle, cis-acting RNA structures could (1) modulate ribosome activity directly through mRNA–protein or mRNA–rRNA interactions; (2) interfere with release-factor function through steric hindrance, sequestration, or modulation of other proteins involved in termination (e.g., Gross et al. 2007); (3) recruit other factors that modulate release-factor function, e.g., through phosphorylation/dephosphorylation (Le Roy et al. 2005), or (4) pause ribosomes in the act of decoding the stop codon, perhaps by providing a barrier to unwinding by a ribosome-associated helicase (Takyar et al. 2005; Qu et al. 2011). In the latter scenario, stalling may bias the system toward readthrough passively, simply providing more time for suppressor tRNA binding, or actively, by introducing tension into the mRNA as the structure is unwound, stalling the elongation cycle in a state where release factor access or stop codon recognition is reduced. Effects of mRNA tension on ribosome function have already been proposed to play a role in the related phenomenon, ribosomal frameshifting (Plant and Dinman 2005; Namy et al. 2006).
Presently, the evidence favors the view that readthrough can be induced through numerous pathways. Aminoglycoside candidates for therapeutic intervention in diseases arising from pathogenic nonsense mutations are believed to stimulate readthrough through direct binding to, and modulation of ribosome function (for review, see Pokrovskaya et al. 2010). Readthrough can also be stimulated by sequestration or removal of release factors, or other factors involved in termination (Orlova et al. 2003; Gross et al. 2007; Bolger et al. 2008; Namy et al. 2008; Khoshnevis et al. 2010; for review, see Namy and Rousset 2010). We do not know how stimulatory RNAs promote readthrough, but it seems likely that their function is mediated by direct interaction with the ribosome. Consistent with this, in both CTFV and MuLV, the nucleotide spacing between the recoded stop codon and downstream stimulatory RNA is of a length (8 nt) consistent with an interaction with the ribosome at the time that decoding of the stop codon is taking place. What is intriguing, however, is that the CTFV stimulatory RNA is unable to promote ribosomal pausing in vitro, despite the presence of a long helix of considerable predicted stability (ΔG = −33 kcal/mol). This is the first time that readthrough signal-induced pausing has been investigated, so it is premature to suggest that this will hold true for other structured readthrough signals. Nevertheless, it does provide support for the view, together with other evidence (Wills et al. 1994), that readthrough stimulatory RNAs differ in their mechanism of action from frameshift-promoting structures, since all functional frameshift sites analyzed to date have been shown to be capable of inducing ribosomal pausing (although there is debate about how closely frameshifting efficiency and the extent of pausing are correlated) (Kontos et al. 2001).
The stem–loop is an important element of the CTFV readthrough signal, but additional elements of local sequence context flanking the UGA stop codon are also required. Given the similarity between the primary sequence in this region of CTFV (5′-UGACGGUGU-3′) and certain other Type II readthrough signals (e.g., tobacco rattle tobravirus RNA-1 [TRV]: 5′-UGACGGUUU-3′), this is perhaps unsurprising. Systematic studies have revealed that local context effects (as opposed to secondary structural effects) can extend as far as 9 nt downstream from the stop codon (Namy et al. 2001; Harrell et al. 2002). How these contexts promote readthrough is not known, but conceivably they could act by binding to rRNA. A complementarity between the TMV signal and a region of helix 16 of 18S rRNA has been noted (Namy et al. 2001); if such mRNA:rRNA interactions occur, they could modulate the decoding process through effects on the binding or activity of the release factor complex or near-cognate tRNAs. In support of this, it is known that many rRNA mutants affect termination, presumably through similar routes (Arkov and Murgola 1999). Bases immediately 5′ of the stop codon can also affect readthrough, and this is true for CTFV. It is not clear whether this is through the identity of the tRNA in the P-site (and/or its encoded amino acid) and its subsequent influence on the termination reaction (for review, see Bertram et al. 2001), or some effect of the bases themselves on the structure of the mRNA in the decoding site (Tork et al. 2004).
An interesting aspect of the CTFV signal is that readthrough appears to be restricted to UGA, both in RRL, insect cell lysates, and transfected mammalian cells. Suppression of both UGA and UAG (but to a much lesser extent, UAA) in the context of the MuLV signal is known to occur efficiently in RRL (Feng et al. 1989) and in a mammalian cell line (COS-7) (R Ramarao and I Brierley, unpubl.); thus, some feature of the CTFV signal must prevent the action of a UAG suppressor tRNA. Selective suppression of the UGA stop codon in RRL has also been observed for the Sindbis virus readthrough signal (Li and Rice 1993). A similar analysis of TRV also revealed specificity for UGA, albeit readthrough in the tRNA-depleted wheat germ system used was dependent upon the addition of a specific UGA suppressor, tRNATrp (CmCA). (Urban et al. 1996). Interestingly, the mRNAs used in the Sindbis and TRV experiments lacked the recently identified stem–loop stimulators (Firth et al. 2011); thus, it may be that the specificity of stop codon suppression is mediated through local context effects. Paradoxically, studies of the Type I readthrough signal of TMV, where readthrough is promoted solely by local sequence context, have revealed a general lack of stop codon specificity (Skuzeski et al. 1991; Zerfass and Beier 1992; Harrell et al. 2002). Thus, further work will be required to untangle the mechanism of selective stop codon suppression. We have considered the possibility that the UGA specificity of the CTFV signal could be accounted for if the stop codon was recognized by the selenocysteine insertion machinery (and that the VP9′ fusion protein is a selenoprotein). However, this seems unlikely. There is no obvious selenocysteine insertion element within the 3′ UTR of CTFV Seg-9 and, in addition, RRL is limited in the protein SBP2 necessary for efficient insertion of Sec tRNA[Ser]Sec in response to UGA (see Fixsen and Howard 2010).
RNA folding programs suggest the formation of stable RNA secondary structures immediately downstream from the stop codon of a number of the experimentally verified readthrough signals and some of the many candidates that are predicted from database screening programs (Namy et al. 2003; Sato et al. 2003; Williams et al. 2004) and from comparative genomics (Lin et al. 2007) (although there has yet to be an exhaustive survey). Given the frequent presence of extended 3′ structural elements at readthrough sites, the challenge will be to determine how they function.
MATERIALS AND METHODS
Construction of plasmids
A cDNA clone of CTFV Seg-9 in plasmid pcDNA3 was the kind gift of Dr. Houssam Attoui (Institute for Animal Health, Pirbright, UK). A 307-bp PCR fragment (generated using Pfu polymerase [Stratagene]) spanning the region 16 bp upstream of the VP9 UGA and 288 bp downstream was subcloned from this plasmid into BamHI and SalI-cut plasmid p2luc (Grentzmann et al. 1998) to generate the readthrough reporter plasmid CTFV-307. Related derivatives with 3′ truncations of increasing size were prepared by PCR amplification from CTFV-307 using primers tailed with BamHI and SalI restriction enzyme sites and ligated into appropriately cleaved p2luc. A schematic description of the organization of p2luc-CTFV constructs is shown in Figure 1. Plasmid pPS0-CTFV-106-CGA, used in the study of ribosomal pausing, was prepared by PCR amplication of the 106-bp of viral sequence from CTFV-106 using primers tailed with XhoI and PvuII and cloning into XhoI/PvuII-cut plasmid pPS0 (Kontos et al. 2001). Subsequently, the VP9 UGA codon was replaced with CGA by site-directed mutagenesis, performed using the Quikchange II site-directed mutagenesis kit (Stratagene) according to manufacturer's instructions. For measurement of readthrough in insect cell lysates, cDNAs were prepared from CTFV-plasmids by PCR and subcloned into AsiSI/EcoRI-cut insect expression vector pF25A ICE T7 (Promega). The PCR products included sequence information from the start codon of the upstream rluc reporter to the unique EcoRI site within the downstream fluc reporter gene. All plasmid sequences were confirmed by dideoxy sequencing (using the facility at the Department of Biochemistry, University of Cambridge).
In vitro transcription and translation
CTFV reporter plasmids were linearized with EcoRI and capped run-off transcripts generated using T7 RNA polymerase as described (Girnary et al. 2007). Messenger RNAs were recovered by a single extraction with phenol/chloroform (1:1 v/v), followed by ethanol precipitation. Remaining unincorporated nucleotides were removed by gel filtration through a NucAway spin column (Ambion). The eluate was concentrated by ethanol precipitation, the mRNA resuspended in water, checked for integrity by agarose gel electrophoresis, and quantified by spectrophotometry.
Messenger RNAs were translated in Flexi rabbit reticulocyte lysate (FlexiRRL, Promega) programmed with 50 μg/mL of template mRNA. Typical reactions were of 10 μL and composed of 60% (v/v) FlexiRRL, 20 μM amino acids (lacking methionine), 500 μM MgOAc, 2 mM DTT, 5 U of RNAse inhibitor (RNAguard, GE Healthcare Life Sciences), 140 mM KCl, and 0.2 MBq [35S]methionine. Reactions were incubated for 1 h at 30°C and stopped by the addition of an equal volume of 10 mM EDTA, 100 μg/mL of RNase A, followed by incubation at room temperature for 20 min. Samples were prepared for SDS-PAGE by the addition of 10 vol of 2× Laemmli's sample buffer (Laemmli 1970), boiled for 3 min, and resolved on 12% SDS–polyacrylamide gels. Dried gels were exposed to a Cyclone Plus Storage Phosphor Screen (Perkin Elmer), the screen scanned using a Typhoon TRIO Variable Mode Imager (GE Healthcare) in storage phosphor autoradiography mode, and bands were quantified using ImageQuantTL software (GE Healthcare). The calculations of readthrough efficiency take into account the differential methionine content of the various products.
Translations in insect cell lysates were carried out using a coupled TnT T7 insect cell expression system (Promega). Reactions comprised 80% (v/v) TnT T7 ICE master mix, 100 μg/mL of plasmid DNA, and 14.8 MBq/mL of [35S]methionine (PerkinElmer, >1000 Ci/mMole). Following incubation at 30°C for 4 h, reactions were stopped and samples prepared and analyzed as above.
Readthrough assays in tissue culture
293T cells were maintained in Dulbecco's modification of Eagle's medium supplemented with 10% (v/v) fetal calf serum. Plasmids were transfected using a commercial liposome method (FuGene 6, Roche). Cells were seeded in dishes of a 24-well plate and grown for 18–24 h until 80% confluency was reached. Transfection mixtures (containing plasmid DNA, serum-free medium [Optimem; Gibco-BRL] and FuGene) were set up as recommended by the manufacturer and added directly (dropwise) to the tissue-culture cell growth medium. The cells were harvested 36–48 h post-transfection, and reporter gene expression determined using a dual luciferase assay system kit (Promega). Each data point represents the mean value (±SEM) from six separate transfections.
RNA structure mapping
Plasmid CTFV-121 was modified by site-directed mutagenesis to include a T3 RNA polymerase promoter 30 bp upstream of the CTFV insert generating plasmid CTFV-121-T3. RNA for structure mapping was prepared by in vitro transcription of BamHI-digested pCTFV-121-T3 using T3 RNA polymerase. Transcription reactions were carried out on a 200-μL scale essentially as described (Girnary et al. 2007). Structure mapping was performed using a 5′ end-labeling procedure as described previously (Manktelow et al. 2005; Girnary et al. 2007). All probing reactions were performed in a final volume of 50 μL and contained ∼40,000 cpm 5′ [33P] end-labeled transcript, 10 μg of Escherichia coli rRNA, and the relevant enzymatic or chemical probe. Further details are provided in the legend to Figure 2.
Ribosomal pausing assays
Plasmid pPS0-CTFV-121-CGA was linearized with AvaII prior to transcription with T7 RNA polymerase and translation in Flexi RRL. Translations were allowed to proceed for 3 min at 26°C in the presence of [35S]methionine prior to the addition of edeine to a final concentration of 5 μM. Aliquots of 1.5 μL were withdrawn from the translation mixture at specified intervals, mixed with an equal volume of RNase A (100 μg/mL) in 10 mM EDTA (pH 7.5), and incubated for 20 min at 26°C prior to preparation for analysis on SDS–12% polyacrylamide gels. Pausing assays were also carried out with an mRNA derived from AvaII-cut plasmid pPS1a (Kontos et al. 2001) as a positive control. A size marker indicating the expected position of a CTFV hairpin-induced ribosomal pause product was prepared by translating XhoI-cut pPS0 mRNA at 26°C for 1 h.
ACKNOWLEDGMENTS
This work was supported by a grant from the Biotechnology and Biological Sciences Research Council, U.K. (I.B., T.D.K.B.). We thank Dr. Houssam Attoui for providing the CTFV plasmid and Ruoyi Sun for her help in the early part of this work.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.030338.111.
REFERENCES
- Alam SL, Wills NM, Ingram JA, Atkins JF, Gesteland RF 1999. Structural studies of the RNA pseudoknot required for readthrough of the gag-termination codon of murine leukemia virus. J Mol Biol 288: 837–852 [DOI] [PubMed] [Google Scholar]
- Arkov AL, Murgola EJ 1999. Ribosomal RNAs in translation termination: facts and hypotheses. Biochemistry (Mosc) 64: 1354–1359 [PubMed] [Google Scholar]
- Beier H, Grimm M 2001. Misreading of termination codons in eukaryotes by natural nonsense suppressor tRNAs. Nucleic Acids Res 29: 4767–4782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram G, Innes S, Minella O, Richardson JP, Stansfield I 2001. Endless possibilities: translation termination and stop codon recognition. Microbiology 147: 255–269 [DOI] [PubMed] [Google Scholar]
- Bolger TA, Folkmann AW, Tran EJ, Wente SR 2008. The mRNA export factor Gle1 and inositol hexakisphosphate regulate distinct stages of translation. Cell 134: 624–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I, Pennell S, Gilbert RJ 2007. Viral RNA pseudoknots: versatile motifs in gene expression and replication. Nat Rev Microbiol 5: 598–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher TW, Miller WA 2006. Translational control in positive strand RNA plant viruses. Virology 344: 185–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng YX, Levin JG, Hatfield DL, Schaefer TS, Gorelick RJ, Rein A 1989. Suppression of UAA and UGA termination codons in mutant murine leukemia viruses. J Virol 63: 2870–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng YX, Yuan H, Rein A, Levin JG 1992. Bipartite signal for read-through suppression in murine leukemia virus mRNA: an eight-nucleotide purine-rich sequence immediately downstream of the gag termination codon followed by an RNA pseudoknot. J Virol 66: 5127–5132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth AE, Wills NM, Gesteland RF, Atkins JF 2011. Stimulation of stop codon readthrough: frequent presence of an extended 3′ RNA structural element. Nucleic Acids Res 39: 6679–6691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fixsen SM, Howard MT 2010. Processive selenocysteine incorporation during synthesis of eukaryotic selenoproteins. J Mol Biol 399: 385–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girnary R, King L, Robinson L, Elston R, Brierley I 2007. Structure-function analysis of the ribosomal frameshifting signal of two human immunodeficiency virus type 1 isolates with increased resistance to viral protease inhibitors. J Gen Virol 88: 226–235 [DOI] [PubMed] [Google Scholar]
- Grentzmann G, Ingram JA, Kelly PJ, Gesteland RF, Atkins JF 1998. A dual-luciferase reporter system for studying recoding signals. RNA 4: 479–486 [PMC free article] [PubMed] [Google Scholar]
- Gross T, Siepmann A, Sturm D, Windgassen M, Scarcelli JJ, Seedorf M, Cole CN, Krebber H 2007. The DEAD-box RNA helicase Dbp5 functions in translation termination. Science 315: 646–649 [DOI] [PubMed] [Google Scholar]
- Harrell L, Melcher U, Atkins JF 2002. Predominance of six different hexanucleotide recoding signals 3′ of read-through stop codons. Nucleic Acids Res 30: 2011–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoshnevis S, Gross T, Rotte C, Baierlein C, Ficner R, Krebber H 2010. The iron-sulphur protein RNase L inhibitor functions in translation termination. EMBO Rep 11: 214–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontos H, Napthine S, Brierley I 2001. Ribosomal pausing at a frameshifter RNA pseudoknot is sensitive to reading phase but shows little correlation with frameshift efficiency. Mol Cell Biol 21: 8657–8670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Laurberg M, Asahara H, Korostelev A, Zhu J, Trakhanov S, Noller HF 2008. Structural basis for translation termination on the 70S ribosome. Nature 454: 852–857 [DOI] [PubMed] [Google Scholar]
- Le Roy F, Salehzada T, Bisbal C, Dougherty JP, Peltz SW 2005. A newly discovered function for RNase L in regulating translation termination. Nat Struct Mol Biol 12: 505–512 [DOI] [PubMed] [Google Scholar]
- Li G, Rice CM 1993. The signal for translational readthrough of a UGA codon in Sindbis virus RNA involves a single cytidine residue immediately downstream of the termination codon. J Virol 67: 5062–5067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MF, Carlson JW, Crosby MA, Matthews BB, Yu C, Park S, Wan KH, Schroeder AJ, Gramates LS, St Pierre SE, et al. 2007. Revisiting the protein-coding gene catalog of Drosophila melanogaster using 12 fly genomes. Genome Res 17: 1823–1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh PG, Song H 2010. Structural and mechanistic insights into translation termination. Curr Opin Struct Biol 20: 98–103 [DOI] [PubMed] [Google Scholar]
- Manktelow E, Shigemoto K, Brierley I 2005. Characterization of the frameshift signal of Edr, a mammalian example of programmed -1 ribosomal frameshifting. Nucleic Acids Res 33: 1553–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaughan KK, Brown CM, Dalphin ME, Berry MJ, Tate WP 1995. Translational termination efficiency in mammals is influenced by the base following the stop codon. Proc Natl Acad Sci 92: 5431–5435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohd Jaafar F, Attoui H, De Micco P, De Lamballerie X 2004. Termination and read-through proteins encoded by genome segment 9 of Colorado tick fever virus. J Gen Virol 85: 2237–2244 [DOI] [PubMed] [Google Scholar]
- Mohd Jaafar F, Mertens P, Attoui H 2009. Coltiviruses and seadornaviruses. In eLS. Wiley, Chichester, UK: doi: 10.1002/9780470015902.a0021553 [Google Scholar]
- Mottagui-Tabar S, Tuite MF, Isaksson LA 1998. The influence of 5′ codon context on translation termination in Saccharomyces cerevisiae. Eur J Biochem 257: 249–254 [DOI] [PubMed] [Google Scholar]
- Namy O, Rousset J-P 2010. Specification of standard amino acids by stop codons. In Recoding: Expansion of decoding rules enriches gene expression (ed. Atkins JF, Gesteland RF), pp 79–100 Springer Science and Business Media, New York [Google Scholar]
- Namy O, Hatin I, Rousset JP 2001. Impact of the six nucleotides downstream of the stop codon on translation termination. EMBO Rep 2: 787–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namy O, Duchateau-Nguyen G, Hatin I, Hermann-Le Denmat S, Termier M, Rousset JP 2003. Identification of stop codon readthrough genes in Saccharomyces cerevisiae. Nucleic Acids Res 31: 2289–2296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namy O, Rousset JP, Napthine S, Brierley I 2004. Reprogrammed genetic decoding in cellular gene expression. Mol Cell 13: 157–168 [DOI] [PubMed] [Google Scholar]
- Namy O, Moran SJ, Stuart DI, Gilbert RJ, Brierley I 2006. A mechanical explanation of RNA pseudoknot function in programmed ribosomal frameshifting. Nature 441: 244–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namy O, Galopier A, Martini C, Matsufuji S, Fabret C, Rousset JP 2008. Epigenetic control of polyamines by the prion [PSI+]. Nat Cell Biol 10: 1069–1075 [DOI] [PubMed] [Google Scholar]
- Orlova M, Yueh A, Leung J, Goff SP 2003. Reverse transcriptase of Moloney murine leukemia virus binds to eukaryotic release factor 1 to modulate suppression of translational termination. Cell 115: 319–331 [DOI] [PubMed] [Google Scholar]
- Petry S, Weixlbaumer A, Ramakrishnan V 2008. The termination of translation. Curr Opin Struct Biol 18: 70–77 [DOI] [PubMed] [Google Scholar]
- Plant EP, Dinman JD 2005. Torsional restraint: a new twist on frameshifting pseudoknots. Nucleic Acids Res 33: 1825–1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokrovskaya V, Nudelman I, Kandasamy J, Baasov T 2010. Aminoglycosides redesign strategies for improved antibiotics and compounds for treatment of human genetic diseases. Methods Enzymol 478: 437–462 [DOI] [PubMed] [Google Scholar]
- Qu X, Wen JD, Lancaster L, Noller HF, Bustamante C, Tinoco I Jr 2011. The ribosome uses two active mechanisms to unwind messenger RNA during translation. Nature 475: 118–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DN, Cooley L 1997. Examination of the function of two kelch proteins generated by stop codon suppression. Development 124: 1405–1417 [DOI] [PubMed] [Google Scholar]
- Sato M, Umeki H, Saito R, Kanai A, Tomita M 2003. Computational analysis of stop codon readthrough in D. melanogaster. Bioinformatics 19: 1371–1380 [DOI] [PubMed] [Google Scholar]
- Skuzeski JM, Nichols LM, Gesteland RF, Atkins JF 1991. The signal for a leaky UAG stop codon in several plant viruses includes the two downstream codons. J Mol Biol 218: 365–373 [DOI] [PubMed] [Google Scholar]
- Somogyi P, Jenner AJ, Brierley I, Inglis SC 1993. Ribosomal pausing during translation of an RNA pseudoknot. Mol Cell Biol 13: 6931–6940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Mugnier P, Das AK, Webb HM, Evans DR, Tuite MF, Hemmings BA, Barford D 2000. The crystal structure of human eukaryotic release factor eRF1-mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell 100: 311–321 [DOI] [PubMed] [Google Scholar]
- Szer W, Kurylo-Borowska Z 1970. Effect of edeine on aminoacyl-tRNA binding to ribosomes and its relationship to ribosomal binding sites. Biochim Biophys Acta 224: 477–486 [DOI] [PubMed] [Google Scholar]
- Takyar S, Hickerson RP, Noller HF 2005. mRNA helicase activity of the ribosome. Cell 120: 49–58 [DOI] [PubMed] [Google Scholar]
- Tork S, Hatin I, Rousset J-P, Fabret C 2004. The major 5′ determinant in stop codon read-through involves two adjacent adenines. Nucleic Acids Res 32: 415–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban C, Zerfass K, Fingerhut C, Beier H 1996. UGA suppression by tRNACmCATrp occurs in diverse virus RNAs due to a limited influence of the codon context. Nucleic Acids Res 24: 3424–3430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Haar T, Tuite MF 2007. Regulated translational bypass of stop codons in yeast. Trends Microbiol 15: 78–86 [DOI] [PubMed] [Google Scholar]
- Williams I, Richardson J, Starkey A, Stansfield I 2004. Genome-wide prediction of stop codon readthrough during translation in the yeast Saccharomyces cerevisiae. Nucleic Acids Res 32: 6605–6616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills NM, Gesteland RF, Atkins JF 1991. Evidence that a downstream pseudoknot is required for translational read-through of the Moloney murine leukemia virus gag stop codon. Proc Natl Acad Sci 88: 6991–6995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills NM, Gesteland RF, Atkins JF 1994. Pseudoknot-dependent read-through of retroviral gag termination codons: importance of sequences in the spacer and loop 2. EMBO J 13: 4137–4144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerfass K, Beier H 1992. Pseudouridine in the anticodon G psi A of plant cytoplasmic tRNA(Tyr) is required for UAG and UAA suppression in the TMV-specific context. Nucleic Acids Res 20: 5911–5918 [DOI] [PMC free article] [PubMed] [Google Scholar]