Over the last years, the microRNA (miRNA) pathway has emerged as a key component of the regulatory network of pluripotency. Although clearly distinct states of pluripotency have been described in vivo and ex vivo, differences in miRNA expression profiles associated with the developmental modulation of pluripotency have not been extensively studied. Here, the authors performed deep sequencing to profile miRNA expression in naive (embryonic stem cell [ESC]) and primed (epiblast stem cell [EpiSC]) pluripotent stem cells derived from mouse embryos of identical genetic background. This analysis showed that differentiation priming of EpiSC compared to ESC is evidenced by changes in miRNA expression. These dynamic changes in miRNAs signature are likely to reflect both redundant and specific roles of miRNAs in the fine-tuning of pluripotency during development.
Keywords: deep sequencing, embryonic stem cells, epiblast stem cells, microRNA, pluripotent stem cells
Abstract
Over the last years, the microRNA (miRNA) pathway has emerged as a key component of the regulatory network of pluripotency. Although clearly distinct states of pluripotency have been described in vivo and ex vivo, differences in miRNA expression profiles associated with the developmental modulation of pluripotency have not been extensively studied so far. Here, we performed deep sequencing to profile miRNA expression in naive (embryonic stem cell [ESC]) and primed (epiblast stem cell [EpiSC]) pluripotent stem cells derived from mouse embryos of identical genetic background. We developed a graphical representation method allowing the rapid identification of miRNAs with an atypical profile including mirtrons, a small nucleolar RNA (snoRNA)-derived miRNA, and miRNAs whose biogenesis may differ between ESC and EpiSC. Comparison of mature miRNA profiles revealed that ESCs and EpiSCs exhibit very different miRNA signatures with one third of miRNAs being differentially expressed between the two cell types. Notably, differential expression of several clusters, including miR290-295, miR17-92, miR302/367, and a large repetitive cluster on chromosome 2, was observed. Our analysis also showed that differentiation priming of EpiSC compared to ESC is evidenced by changes in miRNA expression. These dynamic changes in miRNAs signature are likely to reflect both redundant and specific roles of miRNAs in the fine-tuning of pluripotency during development.
INTRODUCTION
In the embryo, pluripotent cells are present for several days from preimplantation (16 cells) to post-implantation (pregastrulation) stage. Pluripotent cells have been derived by explanting cells from embryos at different stages of development under various growth conditions. These embryo-derived pluripotent stem cells (PSCs) are important paradigms for the study of pluripotency, self-renewal, and differentiation. PSCs can be classified into two distinct states, naive and primed, which are believed to represent successive snapshots of pluripotency as embryonic development proceeds (Nichols and Smith 2009). Naive PSCs share many properties with the inner cell mass of the blastocyst, while the primed PSCs resemble epiblast cells of a more advanced, pregastrulating stage embryo. In the mouse, the naive and primed states can be stabilized ex vivo, represented by embryonic stem cells (mESCs) and epiblast stem cells (EpiSCs), respectively. Culture conditions for the two types of cells differ, reflecting the use of different signaling pathways to maintain pluripotency and self-renewal: mESC maintenance is dependent on LIF and BMP4, or combined inhibition of Gsk3 and the MAP kinase pathway (Ying et al. 2003, 2008); EpiSCs maintenance requires both Activin and FGF2 (Brons et al. 2007; Greber et al. 2010). In humans, ESCs have been derived from blastocysts and, until recently, were regarded as the human equivalent of mESCs. However, based on multiple characteristics such as flat morphology, dependence on growth factors, or X-chromosome inactivation, hESCs (and human induced pluripotent stem cell [iPSCs]) are closer to mouse EpiSCs than to mESCs and, as such, more likely correspond to the primed rather than the naive state of pluripotency (Tesar et al. 2007; Stadtfeld and Hochedlinger 2010). Interestingly, mESCs and EpiSCs are inter-convertible, and the molecular and epigenetic determinants that convert one cell type into another has been investigated (Chenoweth et al. 2010). Of note, whereas the switch from the naive to the primed state is easily achievable in culture, the reverse is more demanding in terms of epigenetic reprogramming (Bao et al. 2009; Guo et al. 2009).
In recent years, the links between the microRNAs (miRNAs) pathway and pluripotency have been explored mainly using mouse ESCs as a model system. The ESC miRNA signature is characterized by a high abundance of miRNAs sharing a 5′-proximal AAGUGC motif and belonging essentially to three clusters, miR-290-295, miR17-92, and miR-302/367 (Houbaviy et al. 2003; Marson et al. 2008; Ciaudo et al. 2009; Svoboda and Flemr 2010). Expression of most microRNAs of these clusters is down-regulated upon differentiation. Studies of Dcgr8-deficient mESC lines showed that miRNAs have essential roles in proliferation and differentiation. ES cells have a peculiar cell cycle with a short G1 phase, and members of the three abundant clusters, referred to as ESCC (for ESC-specific cell cycle regulating), were shown to have redundant roles in the establishment and the maintenance of the ESC cell cycle profile through down-regulation of inhibitors of G1/S progression (Wang et al. 2008). Promoters of many miRNAs are bound in mESCs by core pluripotency factors such as OCT4, SOX2, and NANOG (Marson et al. 2008). Reciprocally, several miRNAs that are up-regulated during ESC differentiation have been shown to directly target mRNAs coding for pluripotency factors (Sox2, Nanog, Oct4, and Klf4) (Tay et al. 2008). Of note, mature miRNAs from the let7 family, which are present at low levels in ESCs but accumulate upon differentiation (Viswanathan et al. 2008), can antagonize ESCC miRNAs in promoting ESCs' self-renewal, leading to the proposal that the balance between these two types of miRNAs is crucial for the transition from self-renewal to differentiation (Melton et al. 2010). Accordingly, forced expression of miR291-3p, miR294 and miR-295, which all belong to the ESCC family, or pan-inhibition of let7 miRNAs, improve the reprogramming efficiency of MEFS into induced pluripotent stem cells (iPSCs) by Oct4, Klf4, and Sox2 (Judson et al. 2009; Melton et al. 2010). More recently, it was shown that modulation of expression of a few other miRNAs can affect the reprogramming efficiency (Li et al. 2011; Liao et al. 2011; Yang et al. 2011). Strikingly, expression of the miR302/367 cluster was shown to be sufficient to drive efficient reprogramming of murine and human somatic cells to a primed or naive pluripotent state in the absence of exogenous transcription factors (Anokye-Danso et al. 2011).
The naive and primed pluripotent states can be easily discriminated according to various criteria. However, the changes in miRNA expression profiles that are associated with this developmental modulation of pluripotency are largely unknown. Although miRNA profiling has been previously reported for either naive (mESCs) or primed (hESCs) PSCs, accurate comparison between the two types of stem cells has so far been hampered by different parameters such as the multiplicity of techniques used and differences in miRNA repertoires between rodents and primates. Recently, one group has reported the profiling of miRNAs in both ESCs and EpiSCs and shown that the two types of cells cluster separately; however, no detailed comparison has been provided (Chou et al. 2008). In the present study, we used Illumina deep sequencing to profile miRNA expression in mouse ESCs and EpiSCs. All the cell lines used in this study were derived from the same genetic background, thus the differences identified by our analysis must be related to differences in pluripotency states.
RESULTS AND DISCUSSION
A graphical representation method of deep sequencing data sets allows the accurate identification of atypical miRNAs
Two ESC and three EpiSC lines derived from (C57Bl6xDBA2)F1 embryos were used in this study. These lines were characterized and shown to be bona fide naive and primed PSCs, respectively (see Materials and Methods). To profile miRNA expression, we performed high-throughput Illumina sequencing of 18–30-nt small RNA libraries from three EpiSC and two ESC lines. Sequencing of each of the five libraries yielded between 4,859,714 to 9,413,373 small RNA reads that matched the Mus musculus genome (mm9), falling into the various RNA classes depicted in Supplemental Table S1. In total, we identified ∼17.5 million reads (14.4 million from EpiSC and 3.1 million from ESC) that matched 608 out of the 672 miRNA stem–loop sequences annotated in miRBase r16 (Supplemental Material, File A).
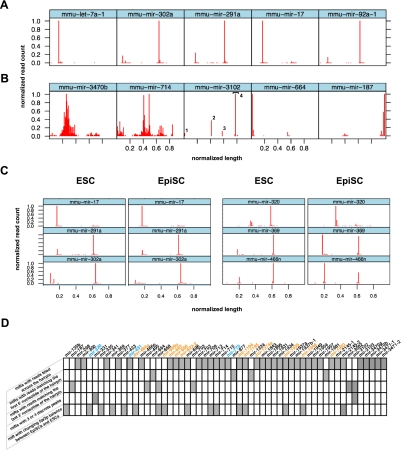
To annotate the miRNA sequences obtained from this study, we first aggregated the read data sets from the five libraries and, for each miRNA, we plotted the number of reads against their 5′ position in the miRNA stem–loop sequence available in miRBase (r16). Using this representation, comparisons of miR read counts profiles was limited by great variations of miRNA total read counts, as well as by variations of lengths of the miRNA stem–loop reference sequences available in miRBase (for an example, see Supplemental Fig. SA). To facilitate comparisons, we, therefore, normalized the read count plots as follows. For each of the 481 miRNAs with more than 29 sequence reads, the number of reads matching any position in a miRNA stem–loop was normalized to the highest number of reads observed at one position for that miRNA. These normalized read counts were then plotted against their 5′ offset normalized to the miRNA stem–loop sequence length (Supplemental Fig. SA). Displaying these plots in a high density lattice allowed rapid and global visualization of miRNA reads in the sequencing data sets (Supplemental Fig. SB). As expected, the vast majority of miRNAs with a significant number of reads generated two discrete peaks in the 5′ and 3′ halves of the miRNA stem–loop sequence, respectively (Fig. 1A; Supplemental Fig. SB). A major read count peak corresponded to the mature miRNA, species whereas a minor, or barely detectable peak, corresponded to the mature miRNA* species. Notably, 38 miRNAs (Fig. 1B; Supplemental Fig. SB) exhibited a nontypical profile with a wider distribution of sequence reads across one or both halves of the stem–loop sequence, suggesting that reads corresponding to these miRNAs were not generated through the canonical Drosha/Dicer biogenesis pathway. These miRNAs have a nontypical stem–loop structure with an abnormally long or short stem and a reduced loop. Altogether, these observations indicate that these small RNAs are probably not generated from bona fide miRNAs. In line with this, seven out of the 38 atypical miRNAs we identified have been recently removed from the last release of miRBase (r17).
FIGURE 1.
Graphical representation of miRNA reads from mouse ESCs and EpiSCs. Standardized graphical representation (normalized number of reads plotted along the normalized length of the stem–loop sequence) allows the rapid discrimination of miRNAs with expected profiles (A) from miRNAs with atypical profiles (B) including wide distribution of reads (mir-3470b and mir-714), putative Dicer double cut processing (mir-3102; the 4 peaks are numbered), and major reads count peak at one extremity of the stem–loop sequence (mir-664 and mir-187). (C) Side-by-side comparison of lattices obtained from ESCs and EpiSCs data sets allows efficient visualization of miRNAs with similar (mir-17, mir-291a, mir-302a) and different (mir-320, mir-369, mir-466n) processing in the two types of PSCs. All lattices display the 5′ offset of reads except for mir-187, for which 3′ offset is represented. (D) Summary of the 56 noncanonical miRNAs identified by our analysis. Differentially expressed miRNAs are indicated with color codes: orange for those more highly expressed in ESCs and blue for those more highly expressed in EpiSCs.
The lattice representation was also very convenient for the rapid visualization of miRNAs with read count peaks at unusual offsets in the stem–loop precursor sequence. For instance, the miR-3102 lattice revealed an unusual profile with four read count peaks (Fig. 1B). This is consistent with the recent report that the stem of the miR-3102 precursor is cleaved twice by Dicer, giving rise to two adjacent miRNA:miRNA* duplexes (Chiang et al. 2010). Four miRNAs (mir-664, mir-877, mir-1224, and mir-1981) showed a major read count peak at the first nucleotide of the precursor stem–loop sequence (pre-miR) (Fig. 1B; Supplemental Fig. SB). The mirtron nature of three of these (mir-877, mir-1224, and mir-1981) explains this atypical read count distribution since the first nucleotide of their pre-miR sequence corresponds to the first nucleotide of an intron. The fourth miRNA, mir-664, was recently annotated as an endogenous small-hairpin RNA (Chiang et al. 2010), raising the possibility that reads mapping upstream of the miRBase stem–loop sequence had been missed by our analysis. When we extended the 5′ end of the mir-664 stem–loop sequence by 20 nt, we were, indeed, able to recover 58 additional reads from our data sets. Closer examination revealed that miR-664 is embedded in the snora36b small nucleolar RNA (snoRNA) transcript (Supplemental Fig. SC). Interestingly, small RNAs derived from snoRNA-encoding loci have been described in various eukaryotes (Taft et al. 2009), suggesting a link between the RNA silencing and snoRNA-mediated RNA processing pathways. When read counts were plotted against their 3′ rather than their 5′ relative offset in the miRNA stem–loop sequence, six miRNAs (mir-3082, -3102, -668, -495, -187, and mir-702) with 3′ read ends mapping to the last nucleotide of the miRBase stem–loop reference were easily identified on the corresponding lattices (Supplemental Fig. SD). Such a distribution was expected for the three mirtrons, mir-3082, mir-3102, and mir-702, as well as for mir-495, for which the sequences flanking the stem–loop in miRBase are shortened as it belongs to a dense cluster of miRNAs. No additional read matches from the sequencing data sets were recovered by 3′ extension of the two remaining mir-668 and mir-187 stem–loop sequences.
Finally, we aggregated the sequencing data sets from the two ESC libraries and from the three EpiSC libraries, respectively, and we generated side-by-side, easy-to-compare lattices of read count miRNA profiles for both cell types (Fig. 1C; Supplemental Fig. SE). In most cases, miRNA reads mapped to the same positions and with similar relative occurrences in the stem–loop precursor sequences, suggesting that miRNA biogenesis does not vary significantly between ESCs and EpiSCs. However, we could observe a clear change in the balance between the strands of the miRNA:miRNA* duplexes for a subset of 16 miRNAs (Fig. 1C; Supplemental Fig. SE). For example, reads mapping to the 3′ part of the mir-369 stem–loop (miR-369-3p) were about four times more abundant than reads mapping to its 5′ part in ESCs, whereas this ratio was about 1:1 in EpiSCs. Similarly, we observed an inversion of this ratio for miR-320, from 1:4 in ESCs to 10:1 in EpiSCs (Fig. 1C). Interestingly, biogenesis of miR-320 has been found to be DGCR8-independent in mouse ES cells and Babiarz and collaborators suggested that the prominence of miR-320 3′ reads reflects the direct transcription of miR-320 as a short hairpin whose 5′ arm is poorly cloned in small RNA libraries (Babiarz et al. 2008). Further studies will be necessary to establish whether these inversions of miRNA:miRNA* ratios do, indeed, reflect differences in biogenesis between ESCs and EpiSCs.
In conclusion, our graphical analysis of the read allowed us to identify 56 out of the 608 miRNAs stem–loop precursors present in our data sets with a singular distribution (Fig. 1D). We nevertheless reasoned that putative regulatory functions of these potentially noncanonical miRNAs deserve future studies and, therefore, we kept these miRNAs under the scope of our expression profiling analysis.
ESCs and EpiSCs exhibit a different miRNA expression profile
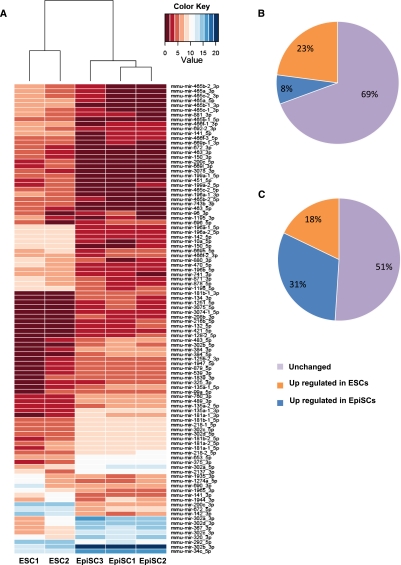
To profile expression of mature miRNAs in ESCs and EpiSCs, we counted the number of sequence reads matching either the 5′ or the 3′ arm of the miRNA stem–loop precursor in each aggregated library and for each miRNA. Using this procedure (see Materials and Methods), count values were attributed to miRNA 5p and miRNA 3p species, irrespective of whether these species are annotated in miRBase as miRNA or miRNA* species (Supplemental Material, Files B–D). We first performed unsupervised hierarchical clustering of our data sets and observed that the three EpiSC samples clustered closely together and could be clearly discriminated from ES cells on the basis of their miRNAs signature (Fig. 2A). We then searched for differentially expressed miRNAs between ESCs and EpiSCs samples. With an adjusted P-value ≤ 0.05, 302 out of the 987 mature miRNAs present in our data sets were found to be differentially expressed (30.5%) (Fig. 2B; Supplemental Material, File E). Among them, 226 mature miRNAs were more highly expressed in ESCs, while 76 mature miRNAs were more highly expressed in EpiSCs (Fig. 2B). Although fewer mature miRNAs were found to be more highly expressed in EpiSCs compared to ESCs, they represent a higher proportion of the number of miRNAs reads present in our data sets (Fig. 2C).
FIGURE 2.
ESC and EpiSC differ in their miRNA expression profiles. (A) Hierarchical clustering of different ESC and EpiSC lines miRNA expression profiles. Heat map shows the top 100 most variable miRNAs. The color coding indicates the relative abundance of miRNA species in the data set. (B,C) Pie chart distribution of number of mature miRNAs (B) and corresponding mean number of reads (C) found to be more highly expressed either in ESCs or in EpiSC, or unchanged.
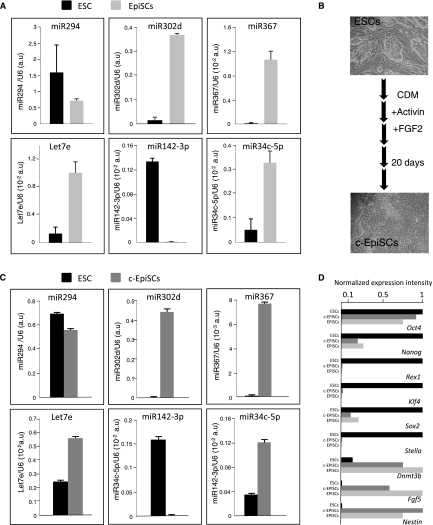
To validate this differential miRNA expression pattern, we performed real-time RT-PCR to assay for the expression of six representative miRNAs: miR-302d, miR34c, miR-367, and let7e that were found to be more highly expressed in EpiSCs compared to ESCs and miR-294 and miR-142-3p that were found to be more highly expressed in ESCs from deep sequencing data. Differential expressions of the representative miRNAs measured by real time RT-PCR were consistent with those computed from the small RNA sequencing data sets (Fig. 3A). We extended this analysis to six additional cell lines (two ESCs and four EpiSCs) of different genetic backgrounds and confirmed differential expression for five out of the six representative miRNAs (Supplemental Fig. SF). When transferred into EpiSC culture conditions, ESCs can rapidly differentiate into EpiSCs (Guo et al. 2009). We thus maintained ESCs under EpiSCs culture conditions and observed a rapid conversion as judged by morphology and marker expression (Fig. 3B,D). Importantly, this conversion was accompanied by a marked increase in miR-302d, miR34c, miR-367, and let7e and a decrease in miR-142-3p expression levels. miR-294 expression was moderately affected in this setting. Altogether, these data indicate that ESCs and EpiSCs differ in their miRNA expression profile.
FIGURE 3.
Differential expression of miRNAs between ESC and EpiSC cell lines. (A) Expression of four miRNAs more highly expressed in EpiSCs (miR-367, -34c-5p, -302d, let7e) and two more highly expressed in ESCs (miR-294 and -142-3p) was monitored on the two ESCs and the three EpiSCs lines using real-time RT-PCR. For each miRNA, the values are normalized using U6 rRNA as reference. Error bars represent the standard error of the mean. (B) Schematic representation of the ESCs to EpiSCs conversion procedure. (C) Real time RT-PCR expression analysis of the same miRNAs before and after conversion. (D) Pluripotency and differentiation markers expression profile in ESCs, c-EpiSCs, and EpiSCs. For each marker, expression level measured by real-time RT-PCR was arbitrarily set to 1 in either ESCs, c-EpiSCs, or EpiSCs.
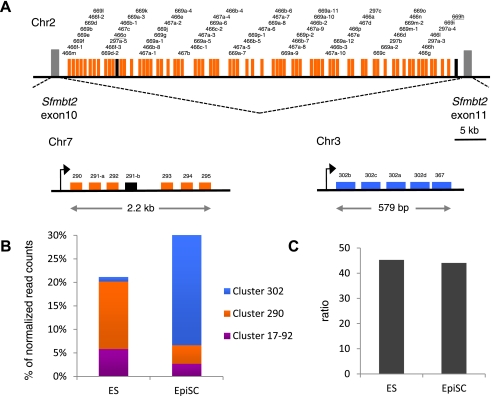
We noticed that many differentially expressed mature miRNAs from our data sets were generated from genomically clustered miRNA stem–loop sequences. Indeed, 62% of them (187/302) mapped to eight of the eleven mouse loci containing at least five miRNAs (Supplemental Material, File F). Of note, miRNAs more highly expressed in ESCs and those more highly expressed in EpiSCs mapped to separate clusters. Moreover, for six of these clusters, including the miR290-295 and miR302/367 clusters and a huge cluster of repetitive miRNAs, miR297/466/467/699, mapping on chromosome 2 (Fig. 4A), all or most of the members of the cluster showed coordinated changes in their expression levels. These observations suggest that the difference in transcriptional regulation of a limited number of primary miRNA transcripts may account for a large part of the differences in mature miRNAs' signature between ESC and EpiSC. For example, reads mapping to 67 out of the 69 miRNA stem–loop sequences of the large chromosome 2 cluster, miR297/466/467/699, represent more than half (123/226) of the mature miRNAs that were found more highly expressed in ESCs (Fig. 4A; Supplemental Material, File F).
FIGURE 4.
ESC and EpiSC express different clusters of pluripotency-associated miRNAS. (A) Schematic representation of the genomic organization of miR290-295 cluster on chromosome 7, miR302/367 cluster on chromosome 3, and a large repetitive cluster on chromosome 2 mapping to intron 10 of the imprinted Sfmbt2 gene. Differentially expressed miRNAs are indicated with color codes: orange for those more highly expressed in ESCs, blue for those more highly expressed in EpiSCs, black for miRNAs that are not differentially expressed. (B) Relative expression of miRNAS from miR17-92, miR290-295, and miR302/367 clusters in ESCs and EpiSCs. (C) Ratio of miRNAS from miR17-92, miR290-295, and miR302/367 clusters over let7 miRNAs.
ESCs and EpiSCs express different set of pluripotency-associated miRNAs
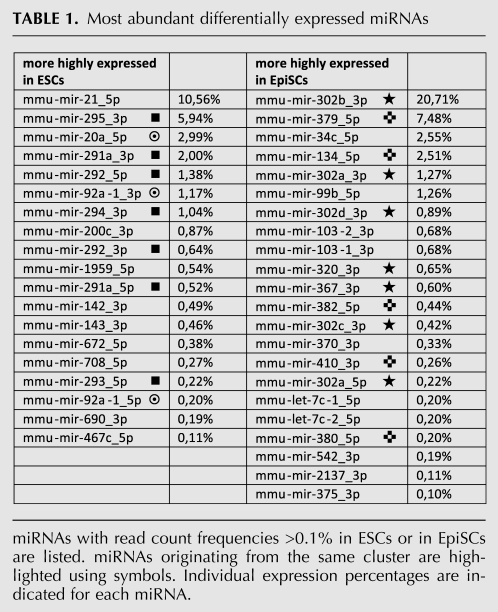
Several miRNAs previously associated with pluripotency were found to be differentially expressed between ESCs and EpiSCs. Table 1 shows the list of the most abundant differentially expressed mature miRNAs (for complete list, see Supplemental Material, File E). Seven out of the 22 most abundant miRNAs more highly expressed in EpiSCs mapped to the miR302/367 cluster. In contrast, members of the miR290-295 and miR17-92 clusters were found to be more highly expressed in ESCs. Strikingly, when we looked at the relative contribution of these clusters in ESC and EpiSC miRNA signatures, we found that miRNAs were essentially produced from the miR290-295 and miR17-92 clusters in ESCs, while the vast majority of miRNA in EpiSCs came from the miR302/367 cluster (Fig. 4B). Interestingly, it was recently reported that inner cells of mouse blastocysts and mESCs display similar expression of miRNAs derived from the miR290-295 cluster (Tang et al. 2010). Moreover, during mouse development, the miR-302 transcript and mature miR-302a can hardly be detected at the blastocyst stage, while their expression is robust and largely confined to pluripotent epiblast at E6.5 and E7.5 (Card et al. 2008). These findings suggest that PSCs retain ex vivo miRNA signature characteristics of the developmental stage from which they are derived. Members of the miR290-295, miR17-92, and miR302/367 clusters have been shown to have redundant contributions to cell cycle regulation and capacity for self-renewal of PSCs, as well as to promoting reprogramming of somatic nuclei to pluripotency (Wang et al. 2008; Judson et al. 2009; Anokye-Danso et al. 2011). miRNAs from the three clusters are related in sequence, notably in their 5′ region. In particular, the AAGUGCU seed sequence is common to several of them, suggesting that they may target identical mRNAs (Ciaudo et al. 2009; Svoboda and Flemr 2010; Anokye-Danso et al. 2011). However, other seed sequences are represented in these clusters, and the exact contribution of each member is not yet clearly understood. Within the miR302/367 cluster, all miRNAs except miR367 share a common AAGUGCU seed sequence. The recent demonstration that miR367 expression is critically required for miR302/367-mediated reprogramming of somatic cells to iPS (Anokye-Danso et al. 2011) clearly suggests that miRNAs regulate pluripotency through more than the dominant AAGUGCU seed.
TABLE 1.
Most abundant differentially expressed miRNAs
miRNA expression profiling revealed notable differences in miRNA expression between mouse and human ESCs, although miRNA signatures of both type of cells are dominated by miRNAs that share similar seed sequences (Suh et al. 2004; Bar et al. 2008; Laurent et al. 2008; Goff et al. 2009; Stadler et al. 2010). Thus, miRNAs from the conserved miR302/367 cluster represent the most abundant class of miRNAs in hESCs, while being far less abundant in mESCs. Our observation that the EpiSCs miRNA profile differs from that of mESCs and is characterized by the overrepresentation of miRNAs from the miR302/367 cluster is in accordance with the proposal that hESCs correspond to the primed rather than the naive state of pluripotency. Therefore, previously reported differences in miRNA expression profiles between mESCs and hESCs may simply rely on differences in their differentiation states rather than on a divergence in miRNA repertoire usage between human and rodent. Nevertheless, nonconserved miRNAs are expressed in rodent and primate PSCs and may have species-specific roles in the regulation of pluripotency. For example, it is known that expression of the large C19MC primate-specific cluster is up-regulated in undifferentiated hESCs (Laurent et al. 2008), while the large mouse-specific repetitive cluster on chromosome 2 is essentially expressed in PSCs and gonads (this study; Laurent et al. 2008). Participation of these two clusters in the regulation of pluripotency remains to be established. Both clusters localize to imprinted genomic regions, a characteristic shared by another large cluster, the eutherian-specific miR-379/miR-410 cluster mapping to the imprinted Dlk1-Dio3 locus. Interestingly, absent or reduced expression of this cluster was observed in partially reprogrammed mouse iPSCs, while normal expression identified iPSC clones with full developmental potential including the capacity to generate full-term mice through tetraploid complementation (Liu et al. 2010; Stadtfeld et al. 2010). Nine miRNAs mapping to the Dlk1-Dio3 cluster (miR-134, miR-370, miR-376a, miR-379, miR-382, miR-410, miR-431, miR-433, miR-543) were found to be more highly expressed in EpiSCs, indicating that differences in the expression of this cluster enable different states of pluripotency to be distinguished.
Both miR290-295 and miR302/367 clusters are transcribed as a single polycistronic primary transcript (Houbaviy et al. 2003; Card et al. 2008), the expression of which is coupled to the core pluripotency transcription factor network (Card et al. 2008; Judson et al. 2009). Indeed, their promoter regions are bound by OCT4, SOX2, and NANOG in mESCs and hESCs. Our analysis suggests that their transcriptional regulation differs between naive and primed PSCs. Further work will be necessary to establish whether this is due to differences in the activity of the core pluripotency network, to epigenetic regulation, or to expression of other transcription factors. In addition to their redundant function in the regulation of pluripotency, these miRNAs may also have specific roles in the fine-tuning of gene expression at two developmental stages. Recently, in hESCs, miR-302a has been shown to collaborate with OCT4 to silence NR2F2, an early activator of neural gene expression (Rosa and Brivanlou 2011). It is tempting to speculate that coordinated expression of lineage specification genes, together with their miRNAs-mediated post-transcriptional silencing, may be a general and important feature of primed pluripotency, ensuring a rapid switch from pluripotency to commitment in response to differentiation clues.
miRNA signature unravels the primed state of EpiSCs
Among the miRNAs up-regulated in ESCs, we noted the presence of several miRNAs (miR-200c, miR-141, and miR-205) involved in the regulation of the epithelial to mesenchymal transition (Gregory et al. 2008). During ESC differentiation, members of the miR-200 family were shown to be down-regulated and forced expression of miR-200c, and miR-141 during the differentiation process can stall differentiated ESCs into an EpiSCs-like state (Gill et al. 2011). Interestingly, Activin signaling, which is constitutively activated under EpiSC culture conditions, also plays a role in the maintenance of the epithelial phenotype and may compensate for the lower expression of miR-200c, miR-141, and miR-205 in EpiSCs. It is noteworthy that, although Cdh1 (E-cadherin) is strongly expressed in both types of cells, the expression of Cdh2 (N-cadherin) is higher in EpiSCs compared to ESCs and has been shown to be inversely correlated to the expression of miR-200 (Samavarchi-Tehrani et al. 2010; Gill et al. 2011).
Among the miRNAs up-regulated in EpiSCs, we also noted the presence of several let7 miRNAs. Reads corresponding to all let7 miRNAs represent 0.8% of the total number of reads in ESCs, and this percentage increases up to 1.2% in EpiSCs. Although the low proportion in both cell types is in accordance with their undifferentiated states, the increase of let-7 miRNA expression was proportional to that of miRNAs from the miR302/367, miR290-295, and miR17-92 clusters (Fig. 4C), consistent with the idea that balanced expression between the two categories of miRNAs may be critical for maintenance of pluripotency. Indeed, let-7 miRNAs have been proposed to counteract the activity of miRNAs promoting pluripotency (Melton et al. 2010). Yet, as it is known that EpiSCs exhibit higher expression of different markers of early germ layers compared to ESCs (Tesar et al. 2007), the increase in let-7 miRNAs expression might also reflect a move toward differentiation priming in EpiSCs.
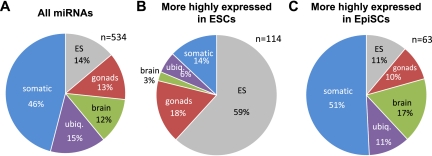
To explore in a more comprehensive manner the possibility that EpiSC miRNome may be primed to differentiation, we annotated the miRNAs differentially expressed between ESCs and EpiSCs based on a recent miRNA-expression profiling performed in multiple embryonic, fetal, and adult mouse tissues (Chiang et al. 2010). According to their major trend of expression, we classified 534 miRNAs analyzed by these authors into five categories: ESC, Gonads, Brain, Somatic, and Ubiquitous (see Materials and Methods; Fig. 5A). Importantly, the annotation profiles obtained for miRNAs more highly expressed in ESCs or in EpiSCs were significantly different from each other (Fig. 5B,C). A majority of miRNAs more highly expressed in ESCs was classified as ESC (59%) or Gonads (18%). In contrast, only a minority of miRNAs more highly expressed in EpiSCs was annotated as ESC and Gonads, the vast majority of them being ubiquitously expressed or expressed in differentiated tissues (79%). Seventeen percent of these miRNAs have prominent expression in brain, reminiscent of the neural differentiation default pathway of the epiblast in the absence of Nodal, or human ESCs in the absence of Activin signaling (Vallier et al. 2004; Camus et al. 2006). Altogether, these data suggest that miRNA signatures provide an important sensor for the primed status of EpiSCs compared to the naive status of ESCs.
FIGURE 5.
Pie charts of miRNAs annotated according to their main trend of expression. (A) Classification of 534 miRNAs according to data published by Chiang et al. (2010). (B) Classification of miRNAs more highly expressed in ESCs (n = 114 annotated). (C) Classification of miRNAs more highly expressed in EpiSCs (n = 63 annotated). The distributions in A vs. B and B vs. C were found to be significantly different (χ2 test, P < 10−5).
CONCLUSION
The present study has established, for the first time, a deep profiling of miRNA expression in EpiSCs and its comparison with ESCs. Dynamic changes in the miRNA signatures unraveled by our analysis reflect both redundant and specific aspects of miRNA-mediated regulation of pluripotency during development. We also show that this miRNA signature can enable PSC states to be discriminated, thereby constituting an important predictive indicator for use of PSCs in basic research and medical applications.
MATERIALS AND METHODS
Cell lines and culture conditions
The two ESC lines used, ESC1 and ESC2, have been described elsewhere (Zhao et al. 2007; Liu et al. 2010). Importantly, they have been tested positively both for germ line transmission and tetraploid complementation. The three EpiSC lines, named EpiSC1 (8.73), EpiSC2 (9.73), and EpiSC3 (3.83), were also derived and characterized previously (Maruotti et al. 2010). All cell lines used for deep sequencing were from the same (C57Bl/6xDBA/2)F1 genetic background. Additional cell lines used for real-time RT-PCR were: PGK (129xPGK background) (Penny et al. 1996) and LF2 ESCs (129/Ola background) (Nichols et al. 1990); FTPas1-2 [(129/SvxC57Bl/6)F1 background] and FT129/4-5 (129/Sv background) EpiSCs recently derived in Alice Jouneau's laboratory. ES cell lines were cultured in Dulbecco's Modified Eagle Media containing 15% fetal calf serum, 1000 U/mL LIF, 0.1 mM 2-mercaptoethanol, 0.05 mg/mL of streptomycin, and 50 U/mL of penicillin in the presence (ESC1 and ESC2) or the absence (PGK and LF2) of mitomycin-C treated mouse embryonic fibroblasts. For conversion, trypsin-dissociated feeder free (129/SvxC57Bl/6)F1 ESCs were cultured on serum-coated plates in chemically defined medium (CDM) (Brons et al. 2007) supplemented with FGF2 (12 ng/mL, R&D) and Activin A (20 ng/mL, R&D). After a few days, cells were passaged as clumps using collagenase and replated in the same conditions. This first passage was rapidly followed by the appearance of flattened colonies. After five passages (overall 20 d of culture), a homogenous population of cells with the morphology of EpiSCs was obtained, from which RNA was extracted.
Preparation of the small RNA libraries and high-throughput sequencing
Feeders were removed from the ES cultures prior to RNA extraction to avoid contamination with fibroblast miRNAs. Total RNA was extracted using Trizol (Invitrogen) and submitted to Bioanalyser for quality assessment. Small RNAs from EpiSC and ES cells were cloned using the DGE-Small RNA Sample Prep Kit and the Small RNA Sample Prep v1.5 Conversion Kit from Illumina, following the manufacturer's instructions. Libraries were sequenced using the Illumina Genome Analyzer II.
Informatic analysis of small RNA libraries
Sequence databases
M. musculus miRNA stem–loop sequences were extracted from miRBase r16. Ribosomal and tRNA sequences were obtained from the Silva database (http://www.arb-silva.de/) and from the Genomic tRNA Database (http://gtrnadb.ucsc.edu/Mmusc/mm9-tRNAs.fa), respectively. Sequences of repeated elements were obtained by extracting Rodentia annotated sequences from Repbase (http://www.girinst.org/repbase/). Messenger RNA and mitochondrial sequences were retrieved from ftp://ftp.ensembl.org/pub/release-56/fasta/mus_musculus/cdna/Mus_musculus.NCBIM37.56.cdna.all.fa.gz and ftp://ftp.ensembl.org/pub/release-56/fasta/mus_musculus/dna/Mus_musculus.NCBIM37.56.dna_rm.chromosome.MT.fa.gz, respectively. SnoRNA sequences were retrieved from Ensembl NCBIM37 using BioMart (http://www.ensembl.org/biomart/martview). Intronic sequences were extracted using a BioMart request for coordinates of all M. musculus exons and subsequent extraction of sequences excluded by these coordinates. We also used Biomart to extract the coordinates of all genes from the M. musculus genome (NCBIM37.56) for which corresponding mRNAs were identified. The reference library of inter-genic sequences was generated by extracting the sequences that are not covered by these genes.
Annotations of sequence reads
Sequence reads were first trimmed from the adapter sequence and matched against (1) the miRBase r16 miRNA stem–loop sequences using novoalign v2.06.09 (http://www.novocraft.com/). Reads matching these sequences with zero or one mismatch were retained for subsequent analysis, whereas unmatched reads were rematched to (2) the rRNA and tRNA sequences. Reads matching these sequences with zero or one mismatch were counted, whereas unmatched reads were rematched to (3) snoRNAs. This procedure was further iteratively applied to (4) Rodentia repeated sequences, (5) introns, (6) mRNAs, (7) intergenic sequences, and (8) mitochondrial sequences to produce the annotations in Supplemental Table S1.
miRNA read mapping and expression profiling
Using a Perl script, we parsed the Novoalign output files to count and map the miRNA reads to the miRNA stem–loop sequences (Supplemental Material, File A). To further assign read counts to either the 5p or 3p miRNAs, each miRNA stem–loop sequence was iteratively split between −20 and +20 nt relative to the middle of the miRNA stem–loop sequence. For each split position, the numbers of reads mapping entirely to the upstream and downstream substrings were computed and stored, whereas reads mapping across the split position were discarded. Then, the split position for which the sum of the upstream and downstream reads was the nearest of the total number of reads mapped to the miRNA stem–loop sequence was retained (marked by parentheses in Supplemental Material, Files C,D), and the upstream and downstream read counts were assigned to the 5p and 3p miRNAs, respectively. Using this algorithm, miRNA5p and miRNA3p read counts were unambiguously assigned in most cases, independently of miRBase annotations for miRNA and miRNA* species. Note that, in the case of noncanonical miRNAs with reads tilled across the stem–loop precursor, the procedure leads to arbitrarily assigning read counts to miRNA5p and miRNA3p species which may not be biologically relevant; however, these counts still reflect the expression level of the miRNA stem–loop precursor.
Expression profiling of mature miRNAs was performed using the hit tables generated as described above (Supplemental Material, Files A,B) and the DEseq R package (Anders and Huber 2010). Unsupervised hierarchical clustering of the 100 miRNAs with highest variances in the data sets was performed after sample normalization using the gplots R package and its heatmap.2 function.
Trellis graphs
To generate the miR read count profiles, we generated tabulated data frames in which each record contained (1) the identifier of the miRNA stem–loop sequence, (2) the normalized offset relative to the miRNA stem–loop sequence length, (3) the frequency of reads matching at this offset expressed as a fraction of the highest number of reads matching at a unique offset in the miRNA stem–loop sequence, and (4) the total number of reads matching the miRNA stem–loop sequence. We processed these data frames with the lattice R package (Sarkar 2008) using the xyplot function and the miRNA stem–loop identifier as a conditioning variable. To avoid representation distortions linked to low sampling, miRNAs with a total number of read matches <30 were excluded from this analysis.
Real-time PCR
Real-time PCR reagents for miRNAs and control U6 RNA were from Qiagen. The RT reactions were performed using 1 μg total RNA and the miScript Reverse Transcription Kit from Qiagen following the manufacturer's instructions. After the RT reactions, the cDNA products (final volume 20 μl) were diluted five times, and 2μl of the diluted cDNAs was used for the PCR reaction using the QuantiTect SYBR Green PCR Master Mix and the miScript Universal Primer from Qiagen. The PCR reaction was conducted at 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 30 sec on a LightCycler 480 real-time PCR machine (Roche). Differences between samples and controls were calculated based on the 2-ΔΔCT method (Schmittgen and Livak 2008). Real-time RT-PCRs for mRNAs were performed as described in Ciaudo et al. (2009) using Rrm2 primers as a control. A list of all primers used in this study is provided as Supplemental Table S2. All real-time PCR reactions have been done in triplicate.
Clustering and annotations of differentially expressed miRNAs
Clusters of miRNAs were defined using miRBase annotations as groups of miRNAs whose inter-distances do not exceed 10 kb. Recently, Chiang and colleagues established the relative miRNA expression profile in various mouse tissues and developmental stages (ovary, testis, brain, newborn, ESC, E7.5, E9.5, and E12.5 embryos) (Chiang et al. 2010). We used these data (except E7.5, which was considered as ambiguous since pluripotent cells can persist at this stage depending on the advancement of development) to classify miRNAs expressed in our data set into five categories according to their major trend of expression. Each miRNA was annotated as either somatic (newborn, E12.5, and E9.5), gonads (ovary and testis), brain, or ESC when the number of reads exceeded 50% of the total for the corresponding category. miRNAs expressed in all categories were annotated as ubiquitous.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We are grateful to Joern Toedling and Marie-Agnès Dillies for helpful discussions on bioinformatics analyses. This work was supported in part by the Institut Pasteur/Institut National de la Recherche Agronomique joint grant PTR 331 and the Centre National de la Recherche Scientifique. Work from E.H. and C.C. was supported by the ANR and EU Project FP7-CP-IP-242129-SyBoSS and the EpiGeneSys Network of Excellence. C.C. received a fellowship from the Federation of European Biochemical Societies.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.028878.111.
REFERENCES
- Anders S, Huber W 2010. Differential expression analysis for sequence count data. Genome Biol 11: R106 doi: 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anokye-Danso F, Trivedi CM, Juhr D, Gupta M, Cui Z, Tian Y, Zhang Y, Yang W, Gruber PJ, Epstein JA, et al. 2011. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell 8: 376–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R 2008. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev 22: 2773–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S, Tang F, Li X, Hayashi K, Gillich A, Lao K, Surani MA 2009. Epigenetic reversion of post-implantation epiblast to pluripotent embryonic stem cells. Nature 461: 1292–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar M, Wyman SK, Fritz BR, Qi J, Garg KS, Parkin RK, Kroh EM, Bendoraite A, Mitchell PS, Nelson AM, et al. 2008. MicroRNA discovery and profiling in human embryonic stem cells by deep sequencing of small RNA libraries. Stem Cells 26: 2496–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, et al. 2007. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 448: 191–195 [DOI] [PubMed] [Google Scholar]
- Camus A, Perea-Gomez A, Moreau A, Collignon J 2006. Absence of Nodal signaling promotes precocious neural differentiation in the mouse embryo. Dev Biol 295: 743–755 [DOI] [PubMed] [Google Scholar]
- Card DA, Hebbar PB, Li L, Trotter KW, Komatsu Y, Mishina Y, Archer TK 2008. Oct4/Sox2-regulated miR-302 targets cyclin D1 in human embryonic stem cells. Mol Cell Biol 28: 6426–6438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenoweth JG, McKay RD, Tesar PJ 2010. Epiblast stem cells contribute new insight into pluripotency and gastrulation. Dev Growth Differ 52: 293–301 [DOI] [PubMed] [Google Scholar]
- Chiang HR, Schoenfeld LW, Ruby JG, Auyeung VC, Spies N, Baek D, Johnston WK, Russ C, Luo S, Babiarz JE, et al. 2010. Mammalian microRNAs: Experimental evaluation of novel and previously annotated genes. Genes Dev 24: 992–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou YF, Chen HH, Eijpe M, Yabuuchi A, Chenoweth JG, Tesar P, Lu J, McKay RD, Geijsen N 2008. The growth factor environment defines distinct pluripotent ground states in novel blastocyst-derived stem cells. Cell 135: 449–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciaudo C, Servant N, Cognat V, Sarazin A, Kieffer E, Viville S, Colot V, Barillot E, Heard E, Voinnet O 2009. Highly dynamic and sex-specific expression of microRNAs during early ES cell differentiation. PLoS Genet 5: e1000620 doi: 10.1371/journal.pgen.1000620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill JG, Langer EM, Lindsley RC, Cai M, Murphy TL, Kyba M, Murphy KM 2011. Snail and the microRNA-200 family act in opposition to regulate epithelial-to-mesenchymal transition and germ layer fate restriction in differentiating ESCs. Stem Cells 29: 764–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff LA, Davila J, Swerdel MR, Moore JC, Cohen RI, Wu H, Sun YE, Hart RP 2009. Ago2 immunoprecipitation identifies predicted microRNAs in human embryonic stem cells and neural precursors. PLoS ONE 4: e7192 doi: 10.1371/journal.pone.0007192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber B, Wu G, Bernemann C, Joo JY, Han DW, Ko K, Tapia N, Sabour D, Sterneckert J, Tesar P, et al. 2010. Conserved and divergent roles of FGF signaling in mouse epiblast stem cells and human embryonic stem cells. Cell Stem Cell 6: 215–226 [DOI] [PubMed] [Google Scholar]
- Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ 2008. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 10: 593–601 [DOI] [PubMed] [Google Scholar]
- Guo G, Yang J, Nichols J, Hall JS, Eyres I, Mansfield W, Smith A 2009. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development 136: 1063–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbaviy HB, Murray MF, Sharp PA 2003. Embryonic stem cell-specific microRNAs. Dev Cell 5: 351–358 [DOI] [PubMed] [Google Scholar]
- Judson RL, Babiarz JE, Venere M, Blelloch R 2009. Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat Biotechnol 27: 459–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent LC, Chen J, Ulitsky I, Mueller FJ, Lu C, Shamir R, Fan JB, Loring JF 2008. Comprehensive microRNA profiling reveals a unique human embryonic stem cell signature dominated by a single seed sequence. Stem Cells 26: 1506–1516 [DOI] [PubMed] [Google Scholar]
- Li Z, Yang CS, Nakashima K, Rana TM 2011. Small RNA-mediated regulation of iPS cell generation. EMBO J 30: 823–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao B, Bao X, Liu L, Feng S, Zovoilis A, Liu W, Xue Y, Cai J, Guo X, Qin B, et al. 2011. MicroRNA cluster 302-367 enhances somatic cell reprogramming by accelerating a mesenchymal-to-epithelial transition. J Biol Chem 286: 17359–17364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Luo GZ, Yang W, Zhao X, Zheng Q, Lv Z, Li W, Wu HJ, Wang L, Wang XJ, et al. 2010. Activation of the imprinted Dlk1-Dio3 region correlates with pluripotency levels of mouse stem cells. J Biol Chem 285: 19483–19490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. 2008. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 134: 521–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruotti J, Dai XP, Brochard V, Jouneau L, Liu J, Bonnet-Garnier A, Jammes H, Vallier L, Brons IG, Pedersen R, et al. 2010. Nuclear transfer-derived epiblast stem cells are transcriptionally and epigenetically distinguishable from their fertilized-derived counterparts. Stem Cells 28: 743–752 [DOI] [PubMed] [Google Scholar]
- Melton C, Judson RL, Blelloch R 2010. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature 463: 621–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, Smith A 2009. Naive and primed pluripotent states. Cell Stem Cell 4: 487–492 [DOI] [PubMed] [Google Scholar]
- Nichols J, Evans EP, Smith AG 1990. Establishment of germ-line-competent embryonic stem (ES) cells using differentiation inhibiting activity. Development 110: 1341–1348 [DOI] [PubMed] [Google Scholar]
- Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N 1996. Requirement for Xist in X chromosome inactivation. Nature 379: 131–137 [DOI] [PubMed] [Google Scholar]
- Rosa A, Brivanlou AH 2011. A regulatory circuitry comprised of miR-302 and the transcription factors OCT4 and NR2F2 regulates human embryonic stem cell differentiation. EMBO J 30: 237–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samavarchi-Tehrani P, Golipour A, David L, Sung HK, Beyer TA, Datti A, Woltjen K, Nagy A, Wrana JL 2010. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell 7: 64–77 [DOI] [PubMed] [Google Scholar]
- Sarkar D 2008. Lattice: Multivariate data visualization with R. Springer, New York [Google Scholar]
- Schmittgen TD, Livak KJ 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3: 1101–1108 [DOI] [PubMed] [Google Scholar]
- Stadler B, Ivanovska I, Mehta K, Song S, Nelson A, Tan Y, Mathieu J, Darby C, Blau CA, Ware C, et al. 2010. Characterization of microRNAs involved in embryonic stem cell states. Stem Cells Dev 19: 935–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Hochedlinger K 2010. Induced pluripotency: History, mechanisms, and applications. Genes Dev 24: 2239–2263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Apostolou E, Akutsu H, Fukuda A, Follett P, Natesan S, Kono T, Shioda T, Hochedlinger K 2010. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature 465: 175–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh MR, Lee Y, Kim JY, Kim SK, Moon SH, Lee JY, Cha KY, Chung HM, Yoon HS, Moon SY, et al. 2004. Human embryonic stem cells express a unique set of microRNAs. Dev Biol 270: 488–498 [DOI] [PubMed] [Google Scholar]
- Svoboda P, Flemr M 2010. The role of miRNAs and endogenous siRNAs in maternal-to-zygotic reprogramming and the establishment of pluripotency. EMBO Rep 11: 590–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft RJ, Glazov EA, Lassmann T, Hayashizaki Y, Carninci P, Mattick JS 2009. Small RNAs derived from snoRNAs. RNA 15: 1233–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F, Barbacioru C, Bao S, Lee C, Nordman E, Wang X, Lao K, Surani MA 2010. Tracing the derivation of embryonic stem cells from the inner cell mass by single-cell RNA-Seq analysis. Cell Stem Cell 6: 468–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I 2008. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature 455: 1124–1128 [DOI] [PubMed] [Google Scholar]
- Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD 2007. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 448: 196–199 [DOI] [PubMed] [Google Scholar]
- Vallier L, Reynolds D, Pedersen RA 2004. Nodal inhibits differentiation of human embryonic stem cells along the neuroectodermal default pathway. Dev Biol 275: 403–421 [DOI] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ, Gregory RI 2008. Selective blockade of microRNA processing by Lin28. Science 320: 97–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, Blelloch R 2008. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat Genet 40: 1478–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CS, Li Z, Rana TM 2011. microRNAs modulate iPS cell generation. RNA 17: 1451–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying QL, Nichols J, Chambers I, Smith A 2003. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell 115: 281–292 [DOI] [PubMed] [Google Scholar]
- Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A 2008. The ground state of embryonic stem cell self-renewal. Nature 453: 519–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Yao R, Hao J, Ding C, Fan Y, Dai X, Li W, Hai T, Liu Z, Yu Y, et al. 2007. Establishment of customized mouse stem cell lines by sequential nuclear transfer. Cell Res 17: 80–87 [DOI] [PubMed] [Google Scholar]