PIWI-interacting RNAs (piRNAs) are at the heart of the nucleic acid–based adaptive immune system against transposons in animal gonads. To date, how the piRNA pathway senses an element as a substrate and how de novo piRNA production is initiated remain elusive. Here, by using a GFP transgene, the authors screened and obtained clonal silkworm BmN4 cell lines, producing massively amplified GFP-derived piRNAs capable of silencing GFP in trans. In multiple independent piRNA-silenced cell lines, they detected a common transcript from an endogenous piRNA cluster, in which a part of the cluster is uniquely fused with an antisense GFP sequence. Bioinformatic analyses suggest that the fusion transcript is a source of GFP primary piRNAs. These data implicate a role for transcription from a piRNA cluster in initiating de novo piRNA production against a new insertion.
Keywords: piRNA, piRNA cluster, silkworm, trans-silencing, transposon
Abstract
PIWI-interacting RNAs (piRNAs) are at the heart of the nucleic acid–based adaptive immune system against transposons in animal gonads. To date, how the piRNA pathway senses an element as a substrate and how de novo piRNA production is initiated remain elusive. Here, by utilizing a GFP transgene, we screened and obtained clonal silkworm BmN4 cell lines producing massively amplified GFP-derived piRNAs capable of silencing GFP in trans. In multiple independent cell lines where GFP expression was silenced by the piRNA pathway, we detected a common transcript from an endogenous piRNA cluster, in which a part of the cluster is uniquely fused with an antisense GFP sequence. Bioinformatic analyses suggest that the fusion transcript is a source of GFP primary piRNAs. Our data implicate a role for transcription from a piRNA cluster in initiating de novo piRNA production against a new insertion.
INTRODUCTION
Controlling transposon activity is essential for host genomes, especially in germ-line cells, to ensure an adequate transmission of parental genomic information to the next generation (Girard and Hannon 2008; Klattenhoff and Theurkauf 2008; Ghildiyal and Zamore 2009; Malone and Hannon 2009). To achieve this, organisms have evolved an elegant defense system against transposons. In animal germ lines, PIWI proteins, a subclade of Argonaute family proteins, and associated PIWI-interacting RNAs (piRNAs) are at the center of this defense system. piRNAs are 23–30 nt small RNAs that act as sequence-specific guides for PIWI proteins that harbor target cleavage (slicer) activity. Mutations in the piRNA pathway-related genes cause derepression of transposons and defects in germ-line development.
The biogenesis of piRNAs markedly differs from that of siRNAs and microRNAs (miRNAs) in that it is Dicer independent (Vagin et al. 2006; Houwing et al. 2007). The piRNA biogenesis initiates with fragmentation of putative long, single-stranded piRNA precursors. Resulting RNA fragments, perhaps longer than mature piRNAs, are incorporated into a subset of PIWI proteins, Siwi in silkworm (Kawaoka et al. 2008a), with a specific nucleotide preference for uracil (1U) at the 5′ end of associated RNAs (Brennecke et al. 2007; Gunawardane et al. 2007; Girard and Hannon 2008; Klattenhoff and Theurkauf 2008; Ghildiyal and Zamore 2009; Kawaoka et al. 2009, 2011a; Malone and Hannon 2009). 3′ ends of PIWI-associated RNAs are further trimmed by a 3′ to 5′ exonuclease named Trimmer (Kawaoka et al. 2011a) to the mature length, followed by 2′-O-methylation catalyzed by Hen1 (Horwich et al. 2007; Houwing et al. 2007; Kirino and Mourelatos 2007; Ohara et al. 2007; Saito et al. 2007; Kawaoka et al. 2011a). This so-called primary processing pathway generates primary piRNAs with 1U bias. PIWI and primary piRNA complexes then cleave their complementary targets across from positions 10 and 11 from the guide piRNAs (Brennecke et al. 2007; Gunawardane et al. 2007). 3′ fragments of cleavage products are then incorporated into another subset of PIWI proteins, BmAgo3 in silkworm (Kawaoka et al. 2008a), which do not show a first-nucleotide bias, and again processed into mature secondary piRNAs with adenine at the position 10 (10A), overlapping with primary 1U piRNAs precisely by 10 nt (Brennecke et al. 2007; Gunawardane et al. 2007; Girard and Hannon 2008; Klattenhoff and Theurkauf 2008; Ghildiyal and Zamore 2009; Kawaoka et al. 2009, 2011a; Malone and Hannon 2009). These secondary 10A piRNAs can in turn generate secondary 1U piRNAs—in theory indistinguishable from primary 1U piRNAs—by cleaving their complementary targets. Such a cleavage-dependent piRNA biogenesis is called a ping-pong amplification cycle (Brennecke et al. 2007; Gunawardane et al. 2007). In insects, primary and secondary 1U piRNAs are often antisense to transposons, while secondary 10A piRNAs tend to be sense to transposons. The ping-pong amplification cycle is broadly conserved among species, including flies, mice, zebrafish, and silkworm (Aravin et al. 2008; Houwing et al. 2008; Kawaoka et al. 2009, 2011b).
Previous genetic studies in flies showed that integration of an exogenous cassette into a piRNA cluster in the telomeric-associated sequence on the X chromosome (X-TAS) results in production of piRNA-like 22–24 nt small RNAs and trans-silencing against the element (Ronsseray et al. 1991, 1996, 2003; Todeschini et al. 2010). These phenomena require piRNA pathway genes (Todeschini et al. 2010). Thus, it is likely that integration of an element into TAS generated de novo piRNAs to cause trans-silencing. However, the molecular nature of these de novo piRNAs is unknown and the underlying mechanism in which a piRNA cluster recognizes transposons to initiate de novo piRNA production remains unclear. To gain insights into these issues, we utilized a silkworm ovary-derived cell line, BmN4, which expresses PIWI/piRNA complexes endogenously (Kawaoka et al. 2009), and sought to see whether a GFP transgene can trigger de novo piRNA production.
RESULTS AND DISCUSSION
Establishment of clonal cell lines with various GFP expression levels
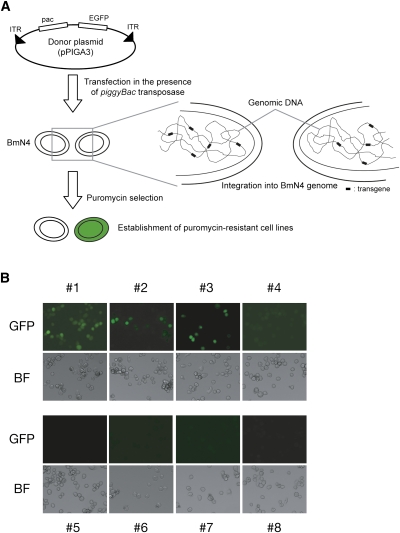
First, by using a piggyBac-based transposition system (Mitsutake and Kobayashi 2011), we introduced a GFP-containing cassette into the BmN4 genome (Fig. 1A; see Materials and Methods for details). In this system, a transgene between two inverted terminal repeats (ITRs), which are excised by the piggyBac transposase, is integrated into genomic TTAA loci. The cassette contained a puromycin-resistant gene, allowing us to isolate transformants by continuous puromycin treatment. We further semicloned eight puromycin-resistant cell lines (#1–#8). Microscopic analysis revealed that one of the eight lines (#1) highly expressed GFP, whereas five lines (#4–#8) showed no GFP expression. The #2 and #3 lines contained both GFP-positive and GFP-negative cells, indicating that these lines were composed of at least two types of cells (Fig. 1B). To test whether transposition occurred successfully, we performed inverse PCR, followed by sequencing analyses. Our data confirmed that each line contained at least one intact transgene with an intact inverted terminal repeat (Supplemental Table S1). Furthermore, by genomic PCR analysis, we detected the full-length GFP sequence in #1–#8 (Supplemental Fig. S1). Collectively, GFP expression was silenced in seven out of eight cell lines.
FIGURE 1.
Establishment of clonal cell lines with various GFP expression levels. (A) Schematic representation for a piggyBac-based transposition system. (B) Eight lines with various GFP expressions. Note that #2 and #3 did not seem to be genuine clonal lines. (BF) Bright field. See also Supplemental Figure S1.
De novo production of piRNAs deriving from a GFP transgene
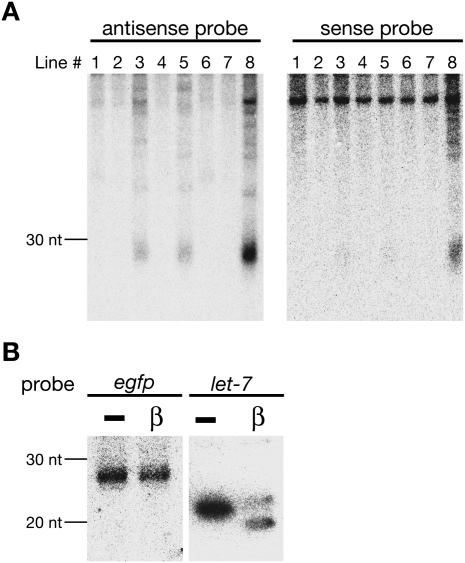
To examine the molecular mechanism that silences GFP expression in the series of BmN4 cell lines, we performed Northern blot analysis with internally 32P-radiolabeled GFP probes (Fig. 2A). We detected abundant 26–30 nt small RNAs, which precisely matched to the mature piRNA length in BmN4 cells, in #3, #5, and #8. #8 expressed both antisense and sense small RNAs deriving from GFP, whereas sense GFP piRNAs were hardly detected in #3 and #5 in this condition. In contrast to 26–30 nt small RNAs, we could not detect 21–22 nt GFP-derived small RNAs, which correspond to endogenous small interfering RNAs (endo-siRNAs) (Ghildiyal and Zamore 2009).
FIGURE 2.
De novo production of piRNAs deriving from a GFP transgene. (A) Total RNAs were subjected to Northern blot analysis with internally 32P-U-radiolabeled antisense or sense GFP probes. (B) Chemical characterization of the 3′ end of GFP-derived antisense piRNAs in #8. Northern blot analysis was performed against total RNAs treated with NaIO4-mediated oxidation, followed by a β-elimination reaction. let-7, an endogenous miRNA in BmN4 cells, served as a control.
The 3′ ends of piRNAs are 2′-O-methylated (Horwich et al. 2007; Houwing et al. 2007; Kirino and Mourelatos 2007; Ohara et al. 2007; Saito et al. 2007; Kurth and Mochizuki 2009; Kamminga et al. 2010). Modified 3′-ends are resistant to β-elimination, followed by oxidation, allowing us to investigate the 3′-end modification state of GFP-derived small RNAs. We found that GFP-derived 26–30 nt small RNAs were resistant to β-elimination reaction (Fig. 2B). Hence, we concluded that small RNAs present in #3, #5, and #8 were de novo-generated canonical piRNAs.
GFP-derived piRNAs mediate silencing in trans
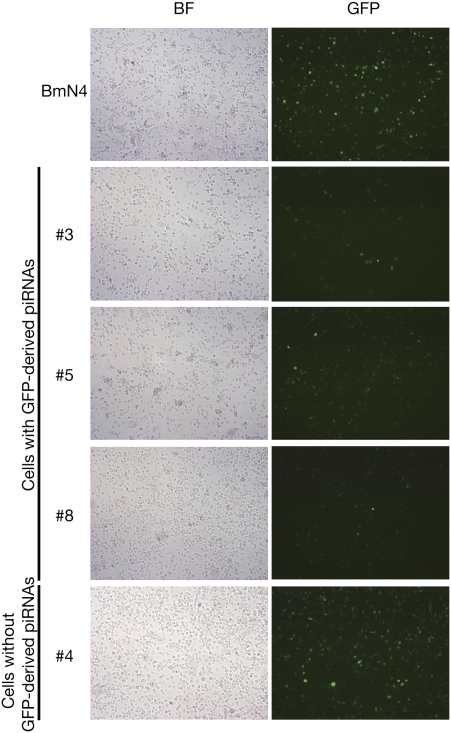
To know how GFP-derived piRNAs function, we transfected a pIZ vector-based GFP reporter (see Materials and Methods) into wild-type (WT) BmN4 cells, those cell lines expressing GFP-derived piRNAs (#3, #5, and #8), and #4, which expressed neither GFP-derived piRNAs nor GFP proteins (Fig. 3). When compared with WT BmN4, the three lines expressing GFP-derived piRNAs showed a marked trans-silencing activity for GFP expression (Fig. 3). On the other hand, #4 exhibited no trans-silencing activity. We quantitatively measured GFP expression in these conditions, and confirmed that #3, #5, and #8, but not #4, exhibited trans-silencing activity (Supplemental Fig. S2). Collectively, our data suggested that GFP-derived piRNAs act in trans to silence GFP expression, and that #4 may use a distinct mechanism that can act only in cis to achieve GFP silencing.
FIGURE 3.
GFP-derived piRNAs mediate silencing in trans. A pIZ vector-based GFP reporter was transfected into WT BmN4, and #3, #5, and #8, which expressed GFP piRNAs, and #4, which expressed neither GFP proteins nor GFP piRNAs. Trans-silencing activity was monitored based on GFP expression. (BF) Bright field. See also Supplemental Figure S2.
GFP-derived piRNAs are amplified through the ping-pong cycle
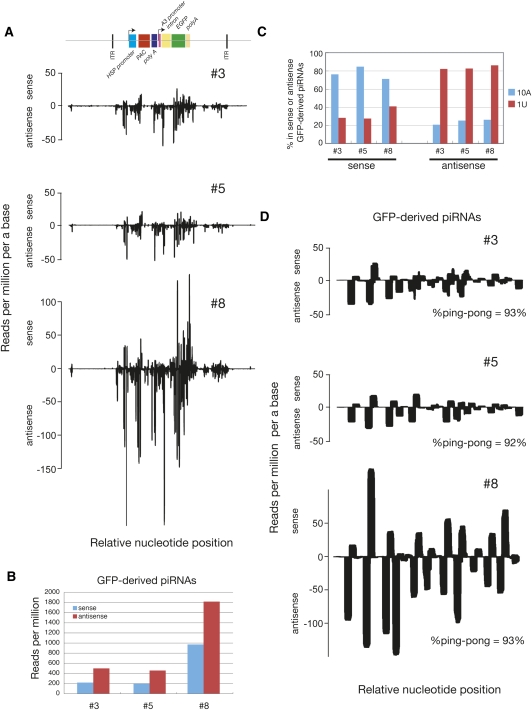
To determine the nature of GFP-derived piRNAs, we deeply sequenced piRNAs of #3, #5, and #8. Sequenced reads were mapped to the silkworm genome for normalization and mapped to an annotated transposon data set and the GFP-containing transgene cassette. The cassette contains two regulatory regions: puromycin-resistant gene (pac) under the control of heat shock protein 70 (hsp70) promoter and EGFP under actin3-derived promoter and an intronic region (Fig. 4A). We found that many piRNAs were mapped to the GFP-containing cassette, while the abundance of piRNAs matching PAC was much less (Fig. 4A), suggesting that puromycin selection might have eliminated the cells where the piRNA pathway had efficiently silenced PAC expression. piRNAs were also generated from backbone sequences, promoters, and the actin3 intronic region (Fig. 4A; Supplemental S3), but their abundance was generally much lower than exonic piRNAs (∼1/10-fold).
FIGURE 4.
GFP-derived piRNAs are amplified through the ping-pong cycle. (A) Density plot of pPIGA3-derived piRNAs. (B) Strand bias of GFP-derived piRNAs. (C) 1U and 10th A bias of GFP-derived piRNAs. (D) Density plot of GFP-derived piRNAs. Percentage of ping-pong participants of GFP-derived piRNAs are indicated (see Materials and Methods). Relative nucleotide position (x-axis) indicates the nucleotide position relative to the reference sequences in the same scale. See also Supplemental Figures S3 and S4.
Consistent with our Northern blot data (Fig. 2A), the relative expression of GFP-derived piRNAs was the strongest in #8, and GFP-derived piRNAs were antisense biased (∼2.3-fold more antisense piRNAs than sense piRNAs) (Fig. 4B). Antisense piRNAs from these three lines had a strong U bias at their 5′ ends (Fig. 4C). In contrast, the 10th position of sense GFP-derived piRNAs was highly enriched for A (Fig. 4C). These were also true for piRNAs matching other regions of the cassettes (data not shown). Visualization of GFP-derived piRNA mapping pattern and bioinformatic analyses showed that the perecentage of ping-pong participants observed in GFP-derived piRNAs was very high—more than 90% of antisense piRNAs overlapped with sense piRNAs precisely by 10 nt (Fig. 4D). Moreover, Siwi preferentially incorporated 1U antisense GFP-derived piRNA, while 10A sense GFP piRNA was enriched in BmAgo3 (Supplemental Fig. S4). These features are representative for PIWI-mediated cleavage-dependent piRNA production, suggesting that GFP-derived de novo primary antisense piRNAs cleave sense GFP mRNAs driven by actin3 promoter to produce sense piRNAs, forming a ping-pong amplification cycle against the newly introduced transgene.
Transcription of antisense GFP RNA from an endogenous piRNA cluster
How were GFP-derived primary piRNAs generated? Given that we detected antisense piRNAs from #3, #5, and #8, antisense GFP RNA must have been expressed under the control of an endogenous promoter(s). If so, such antisense transcription might have triggered de novo production of primary 1U piRNAs. To test this possibility, we performed strand-specific quantitative RT–PCR, and found that antisense transcription of GFP was not limited to #3, #5, and #8 (data not shown). Therefore, antisense transcription per se is not sufficient to cause piRNA-based transgene silencing.
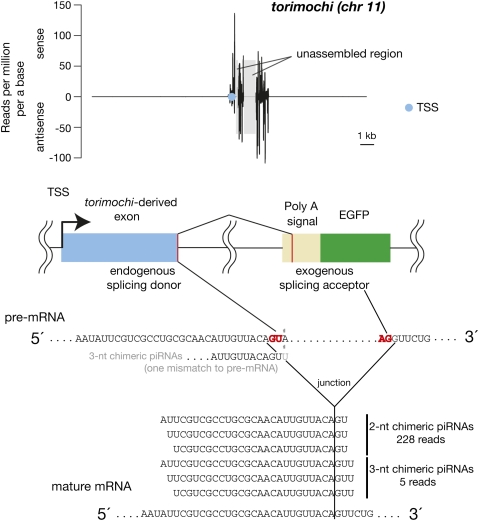
Next, we sought to identify the transcription start sites (TSSs) that transcribed antisense GFP RNAs in #1, which expresses GFP proteins abundantly; #3, #5, and #8, which show GFP silencing with GFP-derived piRNAs; and #2, #4, #6, and #7, which exhibit GFP silencing without expressing piRNAs. We identified 13, 48, and 62 redundant TSSs from each category, respectively. Interestingly, #3, #5, and #8 utilized one common TSS on chromosome 11 (chr 11) to generate antisense GFP RNA. This TSS is embedded within a large endogenous piRNA cluster, herein named as torimochi (birdlime in Japanese; our transgene was trapped in this locus) (14/62 TSSs). We found that an endogenous torimochi-related sequence was fused with antisense GFP RNAs (Fig. 5); an endogenous GU site in torimochi-derived exon acted as a splicing donor; and an exogenous AG site in the poly(A) signal served as a splicing acceptor to generate a fusion transcript. The torimochi-antisense GFP fusion was not detected in #1 (0/13), and #2, #4, #6, and #7 (0/48 in total). The region that served as an intron within the antisense poly(A) signal sequence generated much less piRNAs (Fig. 5; Supplemental Fig. S5A). These observations led us to hypothesize that antisense transcription from the torimochi locus is responsible for triggering de novo production of GFP-derived piRNAs.
FIGURE 5.
Transcription of antisense GFP RNA from torimochi, an endogenous piRNA cluster. (Blue circle) Transcription start site (TSS) embedded with an endogenous piRNA cluster named torimochi. In #3, #5, and #8, this TSS-expressed antisense GFP fused with the torimochi-derived sequence. The fusion occurred in a splicing-dependent mechanism. We identified two types of “chimeric” piRNAs, which covered the junction between exogenous and endogenous sequences. Total abundances of each chimeric piRNA in three libraries are indicated. Chimeric piRNAs were not found in the WT BmN4-derived library. Density of piRNAs was normalized as described previously (Brennecke et al. 2007; Kawaoka et al. 2008b). See also Supplemental Figure S5.
If fusion transcription from torimochi triggered de novo piRNA production, the transcript would produce a “chimeric” piRNA that matches both exogenous and endogenous sequences. Careful examination of piRNAs deriving from #3, #5, and #8 lines identified two types of chimeric piRNAs that were completely absent in the WT BmN4 cell line: 2-nt chimeric piRNAs and 3-nt chimeric piRNAs (Fig. 5; Supplemental Fig. S5B). The 2 or 3 nt at the 3′ end of the 2-nt or 3-nt chimeric piRNAs, respectively, matched exogenous sequence (corresponding to the antisense poly(A) signal sequence), and the remainder of the chimeric piRNAs corresponded to the endogenous torimochi sequence. The 3-nt chimeric piRNAs matched perfectly with the spliced mature mRNA, but not the pre-mRNA. Thus, 3-nt chimeric piRNAs were probably derived from mature mRNA after splicing, although we cannot exclude the possibility that 3-nt chimeric piRNAs were generated by the addition of U to 2-nt chimeric piRNAs by a terminal uridylyltransferase. For 2-nt chimeric piRNAs, the overlapping 2-nt was exactly the same as the 2-nt sequence from the GU site of pre-mRNAs, making it difficult to determine whether 2-nt chimeric piRNAs came from an endogenous or exogenous sequence. However, given that no 2-nt chimeric piRNAs were found in a WT BmN4 library (Supplemental Fig. S5B) and that intronic piRNAs are generally less abundant than exonic piRNAs (Supplemental Fig. S5A), at least a part of 2-nt chimeric piRNAs should be derived from mature torimochi-GFP fusion mRNAs exclusively expressing in #3, #5, and #8. Taken together, we envision that insertion into the torimochi cluster, followed by antisense transcription from endogenous torimochi-associated TSS, generated a torimochi-antisense GFP fusion transcript, which serves to produce de novo GFP-derived primary piRNAs. Although identification of ≥4-nt chimeric piRNAs would strengthen our model, we could not find such chimeric piRNAs from the current data set. More rigorous conclusions will be obtained by further studies with the aid of modified transgenes, which would generate different sets of chimeric piRNAs.
Detection of torimochi-GFP chimeric antisense piRNAs covering the splicing junction exclusively in #3, #5, and #8 (Fig. 5; Supplemental Fig. S5B) suggests that primary piRNA production can, in part, occur after splicing. This is consistent with previous studies showing that 3′ UTR of genic mRNAs can produce sense primary piRNAs (Aravin et al. 2007; Robine et al. 2009; Saito et al. 2009). However, we also detected piRNAs from the region that served as an intron within the antisense poly(A) signal sequence, albeit with lower abundance than exonic piRNAs (Fig. 5; Supplemental Fig. S5A). Therefore, it appears that primary piRNAs could be produced both before and after splicing. In contrast, considering that the ping-pong amplification cycle is thought to occur mainly in the cytoplasm, secondary piRNAs should be generated preferentially from spliced mRNAs. The fact that actin3 intronic piRNAs were much less abundant than exonic piRNAs, especially in sense orientation, supports this view (Fig. 4; Supplemental Fig. S3).
CONCLUSION
Here, we established multiple independent BmN4 cell lines that produce de novo piRNAs with significant ping-pong signatures against a nonrepetitive protein-coding sequence. In these cell lines, we detected a common transcript from an endogenous piRNA cluster, in which a part of an endogenous piRNA cluster, torimochi, is uniquely fused with an antisense GFP sequence. Identification of chimeric piRNAs, which covered the junction site between exogenous and endogenous sequences in the fusion transcript, confirmed that the fusion transcript is a source of de novo piRNAs. Collectively, we envision that de novo piRNA production is triggered when an exogenous sequence is trapped and transcribed from endogenous piRNA cluster loci, and that the “piRNA-generating signal” should be encoded in piRNA cluster loci. Transposon-controlling loci (e.g., flamenco/COM, X-TAS) are well characterized in flies (Ronsseray et al. 1991, 1998; Pelisson et al. 1994; Prud'homme et al. 1995; Roche and Rio 1998; Marin et al. 2000; Stuart et al. 2002; Desset et al. 2003; Brennecke et al. 2007). Our current study and Muerdter et al. (2012) collectively discovered a functional piRNA cluster in flies, silkworm, and mice, probing evolutionary conservation of such master loci that can produce de novo piRNAs. Interestingly, despite functional conservation to produce de novo piRNAs, sequence natures of these master loci are completely different among flies and silkworm. This raises a new question: What defines transposon-controlling loci in the eukaryotic genome?
MATERIALS AND METHODS
Cell culture
BmN4 cells were cultured at 27°C in IPL-41 (Thermo Scientific) or TC-100 (Applichem) supplemented with 10% fetal bovine serum.
Establishment of multiple clonal cell lines
BmN4 cells were transfected with pPIGA3 (GenBank accession no. AB593377), which encodes EGFP, in the presence of piggyBac transposase and Cellfectin reagent (Invitrogen) (Fig. 1A) as described in Mitsutake and Kobayashi (2011). The cells were further selected by puromycin treatment, and the resultant line was used as the parental line to establish clonal cell lines. The parental cells were diluted to three to four cells per well in a 96-well plate. Cell growth was monitored by microscopy to isolate a line where a single cell grew. We established eight lines, two of which (#2 and #3) were not genuine clonal lines. After establishment, cells were cultured without puromycin.
Inverse PCR and genomic PCR
A total of 500 ng of genomic DNA from BmN4 cells and #1–#8 lines were digested with PstI (TaKaRa Bio, Inc.) overnight at 37°C. Resulting fragments were circularized by T4 DNA ligase (TaKaRa Bio, Inc.) for 16 h at 16°C. Circularized DNA was subjected to PCR reaction with DhspF (5′-AATAGAGGCGCTTCGTCTAC-3′) and RLR (5′-CAGTGACACTTACCGCATTGACAAGCACGC-3′) for the 5′ ITR region, and PRF (5′-CCTCGATATACAGACCGATAAAAACACATGC-3′) and PRR (5′-AGTCAGTCCAGAAACAACTTTGGCACATATC-3′) for the 3′ ITR region. The amplified fragments were cloned and sequenced. Primers used for genomic PCR experiments were GFP-F (5′-GTCACTAAATAGATGGACGC-3′), GFP-R (5′-ATGTTTCAGGTTCAGGGGGA-3′), GAPDH-F (5′-TGTTGAGGGCTTGATGAC-3′), and GAPDH-R (5′-ACCTTACCCACAGCTTTG-3′).
Northern blot
Northern blot was performed as described previously (Kawaoka et al. 2009). Radiolabeled sense and antisense GFP probes were prepared by using MaxiScript kit (Ambion) and [α-32P]UTP. For the detection of let-7, a DNA probe (5′-TACTATACAACCTACTACCTCA-3′) was radiolabeled with T4 polynucleotide kinase (Takara Bio, Inc.) and [γ-32P]ATP.
Analysis of RNA 3′ ends
A NaIO4 reaction was performed by incubating total RNA in 100 μL of borax/boric acid buffer (pH 8.6) containing 25 mM NaIO4 for 30 min at room temperature. Ten μL of glycerol was added to quench unreacted NaIO4. β-elimination was then performed by adding 10 μL of 500 mM NaOH, followed by incubation for 90 min at 45°C. The resultant RNA was collected by ethanol precipitation.
Transfection for testing trans-silencing activity
pIZ/V5-His vector, which possesses the ie2 promoter of Orgyia pseudotsugata nucleopolyhedrovirus for the constitutive expression of a gene of interest in insect cell lines, was used for testing trans-silencing activity. The coding region of GFP was inserted into pIZ vector (pIZ/GFP). Next, 5 × 105 BmN4 cells were transfected with 1 μg of pIZ/GFP in the presence of 5 μL of FuGENE HD reagent (Roche). After transfection for 12 h, cells were cultured in normal medium for 60 h. GFP expression levels were monitored by using BX51 microscopy (Olympus). Plasmid transfection efficiency was measured by using a Luciferase (Luc)-expressing plasmid (a modified pGL3-basic vector in which a luciferase gene is driven by the baculovirus ie-1 promoter), which was used to normalize GFP expression. In short, protein concentration of each lysate was determined by using Coomassie Plus Protein Assay Reagent (PIERCE), and the luminescence from Luc expression was measured by ARVO (Wallac). Prior to the Western blot, the loading amount of each lysate was normalized according to the value of Luc luminescence/total protein concentration. GFP expressions were determined by Western blot with anti-GFP antibody (1:1000) as described elsewhere (Kawaoka et al. 2009).
Establishment of #8 lines stably expressing Flag-Siwi or Flag-BmAgo3
The #8 derivative lines stably expressing Flag-Siwi or Flag-BmAgo3 were generated as described previously (Kawaoka et al. 2009). In short, BmN4 cells were transfected with pIZ/Flag-Siwi or Flag-BmAgo3, and transformants were further selected by continuous zeocin selection.
Immunoprecipitation and quantitative PCR
Lysate from #8 lines expressing Flag-Siwi or Flag-BmAgo3 were subjected to immunoprecipitation with anti-Flag antibody. Small RNA fractions were extracted from the immunoprecipitates with the aid of a miRVana miRNA isolation kit (Ambion) according to the manufacturer's instruction. Reverse transcription and following qPCR were performed as described previously (Kawaoka et al. 2011c). Primers used in this experiment were 5′-TGAACTTCAGGGTCAGCTTGCCGTAGGT-3′ for antisense GFP piRNA-1, 5′-CTGAAGTTCATCTGCACCACCGGCAAGC-3′ for sense GFP piRNA-1, and 5′-TACTATACAACCTACTACCTCA-3′ for let-7.
piRNA library construction
Total RNA was prepared using Trizol reagent (Invitrogen) according to the manufacturer's protocol. The total RNA (10 μg) was loaded onto a 15% denaturing polyacrylamide gel containing 8 M urea, electrophoresed, and then stained with SYBRGold (Invitrogen). Signals were visualized using LAS-1000 film (Fujifilm). Small RNA libraries were constructed using a small RNA cloning kit (Takara). DNA sequencing was performed using the Solexa genetic analysis system (Illumina) (Kawaoka et al. 2009). One nanogram of the prepared cDNA was used for the sequencing reactions with the Illumina GA; 10,000–15,000 clusters were generated per “tile”; and 36 cycles of the sequencing reactions were performed. The protocols of the cluster generation and sequence reactions were performed according to the manufacturer's instructions.
Sequence analysis
Solexa sequencing generated reads of up to 36 nt in length. The 3′ adaptor sequences were identified and removed, allowing for up to two mismatches. Reads without adaptor sequences were discarded. Reads shorter than 23 nt or longer than 30 nt were excluded, resulting in reads of 23–30 nt. Alignment to the B. mori genome (International Silkworm Genome Consortium 2008), 121 annotated transposons, and 1668 ReAS clones were performed with SOAP2 (ver. 2.20), allowing no mismatch (Li et al. 2009). To compare the reads among different data sets, reads were expressed in reads per million (RPM) by normalizing to the total number of perfect genome mapping. In addition, we evaluated ping-pong significance by calculating the likelihood for average piRNA mapping to an element to have a complementary ping-pong partner (percentage of ping-pong participants) (Brennecke et al. 2008; Malone et al. 2009; Kawaoka et al. 2011b).
RACE
GeneRacer kit (Invitrogen) was used to identify transcription start sites that transcribed antisense GFP RNA according to the manufacturer's instructions. Primers used in this experiment were described as follows: asGFP_1st (5′-ACGGCAGCGTGCAGCTCGCC-3′) for first PCR, and asGFP_nest (5′-CCTGAGCAAAGACCCCAACG-3′) for nested PCR.
DATA DEPOSITION
piRNAs sequenced in this study are deposited in DRA000374 (DNA Data Bank of Japan; DDBJ).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
This work was supported in part by Special Coordination Funds for Promoting Science and Technology from the Ministry of Education, Culture, Sports, Science, and Technology of the Japanese Government (MEXT) (No. 22115502 to Su.K., No. 17018007 to T.S., and the Professional Program for Agricultural Bioinformatics); Scientific Research (B) (No. 18380044) to J.K.; the National Bio-Resource Project “Silkworm” of MEXT; and a Grant-in-Aid for Scientific Research on Innovative Areas (“Functional machinery for non-coding RNAs”) to Su.K. and Y.T. Sh.K. is a recipient of a fellowship from the Japan Society for the Promotion of Science.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.029777.111.
REFERENCES
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ 2007. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316: 744–747 [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Bourc'his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ 2008. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell 31: 785–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103 [DOI] [PubMed] [Google Scholar]
- Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, Hannon GJ 2008. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science 322: 1387–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desset S, Meignin C, Dastugue B, Vaury C 2003. COM, a heterochromatic locus governing the control of independent endogenous retroviruses from Drosophila melanogaster. Genetics 164: 501–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD 2009. Small silencing RNAs: an expanding universe. Nat Rev Genet 10: 94–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard A, Hannon GJ 2008. Conserved themes in small-RNA-mediated transposon control. Trends Cell Biol 18: 136–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC 2007. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315: 1587–1590 [DOI] [PubMed] [Google Scholar]
- Horwich MD, Li C, Matranga C, Vagin V, Farley G, Wang P, Zamore PD 2007. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr Biol 17: 1265–1272 [DOI] [PubMed] [Google Scholar]
- Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, et al. 2007. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell 129: 69–82 [DOI] [PubMed] [Google Scholar]
- Houwing S, Berezikov E, Ketting RF 2008. Zili is required for germ cell differentiation and meiosis in zebrafish. EMBO J 27: 2702–2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International Silkworm Genome Consortium 2008. The genome of a lepidopteran model insect, the silkworm Bombyx mori. Insect Biochem Mol Biol 38: 1035–1045 [DOI] [PubMed] [Google Scholar]
- Kawaoka S, Minami K, Katsuma S, Mita K, Shimada T 2008a. Developmentally synchronized expression of two Bombyx mori Piwi subfamily genes, SIWI and BmAGO3 in germ-line cells. Biochem Biophys Res Commun 367: 755–760 [DOI] [PubMed] [Google Scholar]
- Kawaoka S, Hayashi N, Katsuma S, Kishino H, Kohara Y, Mita K, Shimada T 2008b. Bombyx small RNAs: genomic defense system against transposons in the silkworm, Bombyx mori. Insect Biochem Mol Biol 38: 1058–1065 [DOI] [PubMed] [Google Scholar]
- Kawaoka S, Hayashi N, Suzuki Y, Abe H, Sugano S, Tomari Y, Shimada T, Katsuma S 2009. The Bombyx ovary-derived cell line endogenously expresses PIWI/PIWI-interacting RNA complexes. RNA 15: 1258–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaoka S, Izumi N, Katsuma S, Tomari Y 2011a. 3′ end formation of PIWI-interacting RNAs in vitro. Mol Cell 43: 1015–1022 [DOI] [PubMed] [Google Scholar]
- Kawaoka S, Arai Y, Kadota K, Suzuki Y, Hara K, Sugano S, Simizu K, Tomari Y, Shimada T, Katsuma S 2011b. Zygotic amplification of secondary piRNAs during the silkworm embryogenesis. RNA 17: 1401–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaoka SK, Kadota Y, Arai Y, Suzuki T, Fujii H, Abe Y, Yasukochi K, Mita S, Sugano K, Simizu Y, et al. 2011c. The silkworm W chromosome is a source of female-enriched piRNAs. RNA 17: 2144–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamminga LM, Luteijn MJ, den Broeder MJ, Redl S, Kaaij LJ, Roovers EF, Ladurner P, Berezikov E, Ketting RF 2010. Hen1 is required for oocyte development and piRNA stability in zebrafish. EMBO J 29: 3688–3700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirino Y, Mourelatos Z 2007. Mouse Piwi-interacting RNAs are 2′-O-methylated at their 3′ termini. Nat Struct Mol Biol 14: 347–348 [DOI] [PubMed] [Google Scholar]
- Klattenhoff C, Theurkauf W 2008. Biogenesis and germline functions of piRNAs. Development 135: 3–9 [DOI] [PubMed] [Google Scholar]
- Kurth HM, Mochizuki K 2009. 2′-O-methylation stabilizes Piwi-associated small RNAs and ensures DNA elimination in Tetrahymena. RNA 15: 675–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J 2009. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25: 1966–1967 [DOI] [PubMed] [Google Scholar]
- Malone CD, Hannon GJ 2009. Molecular evolution of piRNA and transposon control pathways in Drosophila. Cold Spring Harb Symp Quant Biol 74: 225–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ 2009. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137: 522–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin L, Lehmann M, Nouaud D, Izaabel H, Anxolabehere D, Ronsseray S 2000. P-Element repression in Drosophila melanogaster by a naturally occurring defective telomeric P copy. Genetics 155: 1841–1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsutake H, Kobayashi J 2011. piggyBac-mediated stable transformation of cultured Bombyx mori cells using in vitro synhesized transposase mRNA. J Insect Biotechnol Sericology 80: 15–24 [Google Scholar]
- Muerdter F, Olovnikov I, Molaro A, Rozhkov NV, Czech B, Gordon A, Hannon GJ, Aravin AA 2012. Production of artificial piRNAs in flies and mice. RNA 18: 42–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohara T, Sakaguchi Y, Suzuki T, Ueda H, Miyauchi K, Suzuki T 2007. The 3′ termini of mouse Piwi-interacting RNAs are 2′-O-methylated. Nat Struct Mol Biol 14: 349–350 [DOI] [PubMed] [Google Scholar]
- Pelisson A, Song SU, Prud'homme N, Smith PA, Bucheton A, Corces VG 1994. Gypsy transposition correlates with the production of a retroviral envelope-like protein under the tissue-specific control of the Drosophila flamenco gene. EMBO J 13: 4401–4411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prud'homme N, Gans M, Masson M, Terzian C, Bucheton A 1995. Flamenco, a gene controlling the gypsy retrovirus of Drosophila melanogaster. Genetics 139: 697–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robine N, Lau NC, Balla S, Jin Z, Okamura K, Kuramochi-Miyagawa S, Blower MD, Lai EC 2009. A broadly conserved pathway generates 3′UTR-directed primary piRNAs. Curr Biol 19: 2066–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche SE, Rio DC 1998. Trans-silencing by P elements inserted in subtelomeric heterochromatin involves the Drosophila Polycomb group gene, Enhancer of zeste. Genetics 149: 1839–1855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronsseray S, Lehmann M, Anxolabehere D 1991. The maternally inherited regulation of P elements in Drosophila melanogaster can be elicited by two P copies at cytological site 1A on the X chromosome. Genetics 129: 501–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronsseray S, Lehmann M, Nouaud D, Anxolabehere D 1996. The regulatory properties of autonomous subtelomeric P elements are sensitive to a Suppressor of variegation in Drosophila melanogaster. Genetics 143: 1663–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronsseray S, Marin L, Lehmann M, Anxolabehere D 1998. Repression of hybrid dysgenesis in Drosophila melanogaster by combinations of telomeric P-element reporters and naturally occurring P elements. Genetics 149: 1857–1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronsseray S, Josse T, Boivin A, Anxolabehere D 2003. Telomeric transgenes and trans-silencing in Drosophila. Genetica 117: 327–335 [DOI] [PubMed] [Google Scholar]
- Saito K, Sakaguchi Y, Suzuki T, Suzuki T, Siomi H, Siomi MC 2007. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi-interacting RNAs at their 3′ ends. Genes Dev 21: 1603–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Inagaki S, Mituyama T, Kawamura Y, Ono Y, Sakota E, Kotani H, Asai K, Siomi H, Siomi MC 2009. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature 461: 1296–1299 [DOI] [PubMed] [Google Scholar]
- Stuart JR, Haley KJ, Swedzinski D, Lockner S, Kocian PE, Merriman PJ, Simmons MJ 2002. Telomeric P elements associated with cytotype regulation of the P transposon family in Drosophila melanogaster. Genetics 162: 1641–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todeschini AL, Teysset L, Delmarre V, Ronsseray S 2010. The epigenetic trans-silencing effect in Drosophila involves maternally-transmitted small RNAs whose production depends on the piRNA pathway and HP1. PLoS ONE 5: e11032 doi: 10.1371/journal.pone.0011032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD 2006. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313: 320–324 [DOI] [PubMed] [Google Scholar]