RBFOX1 and RBFOX2 are alternative splicing factors that are predominantly expressed in the brain and skeletal muscle. They specifically bind the RNA element UGCAUG, and regulate alternative splicing positively or negatively in a position-dependent manner. The molecular basis for the position dependence of these and other splicing factors on alternative splicing of their targets is not known. The authors explored the mechanisms of RBFOX splicing activation and repression using an MS2-tethering assay and found that the Ala/Tyr/Gly-rich C-terminal domain is sufficient for exon activation when tethered to the downstream intron, whereas both the C-terminal domain and the central RRM are required for exon repression when tethered to the upstream intron.
Keywords: RBFOX, alternative splicing, hnRNP H1, TFG
Abstract
RBFOX1 and RBFOX2 are alternative splicing factors that are predominantly expressed in the brain and skeletal muscle. They specifically bind the RNA element UGCAUG, and regulate alternative splicing positively or negatively in a position-dependent manner. The molecular basis for the position dependence of these and other splicing factors on alternative splicing of their targets is not known. We explored the mechanisms of RBFOX splicing activation and repression using an MS2-tethering assay. We found that the Ala/Tyr/Gly-rich C-terminal domain is sufficient for exon activation when tethered to the downstream intron, whereas both the C-terminal domain and the central RRM are required for exon repression when tethered to the upstream intron. Using immunoprecipitation and mass spectrometry, we identified hnRNP H1, RALY, and TFG as proteins that specifically interact with the C-terminal domain of RBFOX1 and RBFOX2. RNA interference experiments showed that hnRNP H1 and TFG modulate the splicing activity of RBFOX1/2, whereas RALY had no effect. However, TFG is localized in the cytoplasm, and likely modulates alternative splicing indirectly.
INTRODUCTION
The RBFOX family splicing factors play important roles in tissue-specific alternative splicing regulation. There are three paralogs in mammals: RBFOX1—formerly known as A2BP1 (Ataxin-2 Binding Protein 1) or Fox-1—is specifically expressed in brain, skeletal muscle, and heart (Jin et al. 2003; Underwood et al. 2005); RBFOX2—formerly known as RBM9 (RNA-binding motif protein 9)—is more ubiquitously expressed (Underwood et al. 2005); RBFOX3 is less well studied, and was recently discovered to be the same as NeuN (neuronal nuclei), a marker of post-mitotic neuronal cells (Kim et al. 2009).
RBFOX proteins are characterized by a very highly conserved RNA-Recognition Motif (RRM) that is nearly invariant in human, mouse, zebrafish, fruitfly, and nematode (Kuroyanagi 2009). The central RRM is flanked by less-conserved N- and C-terminal domains unique to RBFOX proteins. Unlike many other splicing factors, which tend to have very degenerate binding sites, the RBFOX family of proteins specifically binds to a (U)GCAUG element. Both RBFOX1 and RBFOX2 genes express multiple isoforms via alternative promoters and alternative cassette exons (Nakahata and Kawamoto 2005; Underwood et al. 2005). Both N- and C-terminal domains of the protein isoforms are highly diversified, and some isoforms lack the second half of the RRM. Therefore, some of these isoforms are expected to have different activities or to lack alternative splicing functions (Nakahata and Kawamoto 2005). Alternative splicing of RBFOX1 is regulated during neuronal depolarization, allowing it to modulate the activity of its target genes (Lee et al. 2009).
RBFOX-family proteins regulate alternative splicing positively or negatively in a position-dependent manner. They usually promote exon inclusion when binding to the intron downstream from an alternative cassette exon, and exon skipping when binding to the upstream intron (Jin et al. 2003; Underwood et al. 2005; Ponthier et al. 2006; Zhou et al. 2007; Zhang et al. 2008; Tang et al. 2009; Yeo et al. 2009). Several target genes have been extensively studied by use of reporter minigenes such as mitochondrial ATP synthase γ-subunit (F1γ) (Jin et al. 2003), calcitonin/calcitonin-gene-related peptide (CGRP) (Zhou et al. 2007), CaV1.2 L-Type calcium channel (Tang et al. 2009), fibronectin (Jin et al. 2003), non-muscle myosin II heavy chain-B (NMHC-B) (Nakahata and Kawamoto 2005), epithelial cell-specific fibroblast growth factor receptor 2 (FGFR2) (Baraniak et al. 2006), and RBFOX1 and RBFOX2 themselves (Damianov and Black 2010). Global analysis was utilized more recently to evaluate the splicing regulatory networks of the RBFOX family of proteins. A combination of microarray, CLIP-seq, high-throughput RT–PCR, and computational analyses by different laboratories resulted in the identification of many endogenous target genes, and confirmed the so-called “RNA map,” meaning that the splicing outcome (activation or repression) depends in a predictable manner on the location of the UGCAUG element (Zhang et al. 2008; Venables et al. 2009; Yeo et al. 2009). Certain other splicing factors, such as Nova and PTB, also have a similar map with position-dependent activities (Ule et al. 2006; Llorian et al. 2010). The fact that different splicing factors with completely distinct binding sites have RNA maps with similar features suggests a convergence in the underlying molecular mechanisms of splicing regulation.
Despite the recent progress in the identification of endogenous targets, little is known about the molecular mechanisms by which RBFOX proteins—or other splicing factors—regulate alternative splicing positively or negatively in a context-dependent manner. Most studies to date have focused on the repressive effect. Using a calcitonin/CGRP minigene as a model, Zhou and colleagues showed that RBFOX1/2 prevent SF1 from binding to the branchpoint sequence in intron 3, and repress the formation of the spliceosomal E′ complex; the proteins also interfere with binding of Tra2β and SRSF6 (formerly SRp55) to ESEs (Exonic Splicing Enhancers) via a UGCAUG binding site in exon 4, and block the formation of the spliceosomal E complex (Zhou and Lou 2008). Fukumura et al. (2009) analyzed exon 9 of F1γ as a model system, and found that RBFOX1 represses this exon by inhibiting splicing of intron 9, which is U1-snRNP independent and U2-snRNP dependent. RBFOX2 interacts with hnRNP H1 and cooperates with it and with hnRNP F to repress exon IIIc of an FGFR2 minigene by antagonizing the binding of SRSF1 (formerly SF2/ASF) (Mauger et al. 2008). The C-terminal domain of RBFOX1 is critical for exon repression in an F1γ minigene (Fukumura et al. 2007). However, the potential involvement of this domain in alternative exon activation, or in repression of alternative exons flanked by U1-dependent introns, has not been determined. The only study to date that provided a mechanistic clue about splicing activation by Fox-1/2 reported an interaction in a yeast two-hybrid assay between RBFOX1 and the U1-snRNP-specific U1-C polypeptide (Ohkura et al. 2005).
Here we have started to systematically explore the mechanism of RBFOX1/2 regulation of alternative splicing. We used an MS2-tethering assay to evaluate the requirement of RBFOX1 domains in both exon activation and repression. We found that the C-terminal portion is the only fragment that is critical for cassette-exon activation when tethered to the downstream intron. In contrast, both the RRM and the C-terminal domain are required for exon repression when tethered to the upstream intron. We also used coimmunoprecipitation combined with mass spectrometry to identify proteins that interact with the RBFOX1/2 C-terminal domain, and evaluated their roles in RBFOX1/2 activity.
RESULTS
The C-terminal domain of RBFOX1 is sufficient for exon activation when tethered to the downstream intron
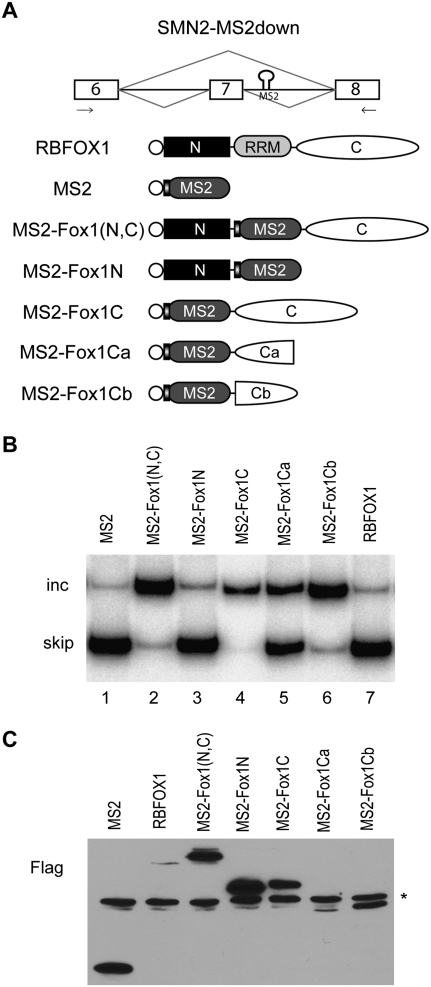
To elucidate the mechanisms of exon activation and repression by RBFOX1, we first used an MS2 tethering assay to determine whether the RRM and the flanking domains can be separated. We replaced the RRM with the bacteriophage MS2 coat protein, which specifically binds to a 21-nt RNA stem–loop (Carey et al. 1983). We inserted this RNA element in the intron downstream from exon 7 in a human SMN2 minigene (Fig. 1A; Hua et al. 2008). This minigene lacks natural RBFOX1 binding motifs. The alternative exon 7 was predominantly skipped when transiently cotranfected with MS2 protein or RBFOX1 alone into HeLa cells (Fig. 1B, lanes 1, 7). However, the MS2-Fox1(N,C) construct, in which the RBFOX1 RRM was replaced by MS2 coat protein, strongly induced exon 7 inclusion (Fig. 1B, lane 2). This result shows that RBFOX1 can enhance exon inclusion when tethered to the downstream intron, and that the RRM and the flanking domains can be separated. We next asked which domains are required for this effect. We only kept either the N- or C-terminal domain fused with MS2 coat protein, and cotransfected them with the SMN2 minigene. The C-terminal domain but not the N-terminal domain was sufficient to induce exon 7 inclusion (Fig. 1B, lanes 3, 4). We further subdivided the C-terminal domain into two similarly sized fragments. The C-terminal fragment, MS2-Fox1Cb, was fully functional in exon activation. The fragment proximal to the RRM, MS2-Fox1Ca, was less active, promoting exon inclusion to a lesser extent (Fig. 1B, lanes 5, 6). However, Western blotting showed that the expression level of MS2-Fox1Ca was lower than that of the other mutants (Fig. 1C). Therefore, its weaker activity in exon activation may simply reflect lower protein expression.
FIGURE 1.
The C-terminal domain of RBFOX1 is sufficient for exon activation when tethered to the downstream intron. (A) Diagrams of the modified SMN2 minigene and MS2-fused RBFOX1 constructs. An MS2 binding site was inserted in the intron downstream from exon 7. The primers for RT–PCR are indicated by arrows below the exons. RBFOX1 has one RRM (gray) flanked by N-terminal (black square) and C-terminal (white oval) domains. All of the protein constructs have a Flag epitope tag at the N terminus, indicated by an open circle. The MS2 module also has a T7 tag at its N terminus, represented by a vertical bar. (B) HeLa cells were cotransfected with the SMN2-MS2down reporter minigene and different protein constructs. RNA and proteins were extracted 48 h after transfection. Radioactive RT–PCR was performed to measure the changes in the alternative splicing isoforms. (C) Western blot analysis using Flag antibody was performed to show the expression of the protein mutants. (*) Nonspecific band.
Fox1N and Fox1Ca mutant proteins are mislocalized, but restoring nuclear localization is not sufficient to rescue their activity
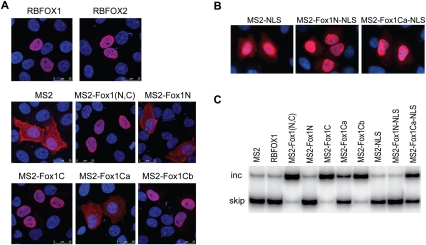
We used indirect immunofluorescence against the Flag tag to examine the localization of the different transiently expressed constructs. MS2 protein gave a diffuse pattern in both nucleus and cytoplasm, whereas MS2-Fox1(N,C) localized in the nucleus, as did the wild-type Fox-1 protein (Fig. 2A). As expected, the two mutant proteins that were active in promoting exon 7 inclusion, MS2-Fox1C and MS2-Fox1Cb, also localized in the nucleus. However, both MS2-Fox1N and MS2-Fox1Ca, which completely or partially lost the splicing activation potential, distributed in both the nucleus and the cytoplasm (Fig. 2A). This result is consistent with previous reports that the C-terminal region is critical for nuclear localization (Nakahata and Kawamoto 2005; Lee et al. 2009).
FIGURE 2.
Subcellular localization of the RBFOX1 mutant proteins. (A) Indirect immunofluorescence of HeLa cells transfected with the indicated constructs was performed using Flag antibody. (B) Indirect immunofluorescence of HeLa cells transfected with RBFOX1 constructs containing an SV40 NLS was carried out using Flag antibody. (C) HeLa cells were cotransfected with the SMN2-MS2down reporter minigene and the indicated RBFOX1 constructs. RNA was extracted 48 h after transfection and analyzed as in Figure 1B.
To determine whether the lack of splicing activity is caused by mislocalization, we forced MS2, MS2-Fox1N, and MS2-Fox1Ca to localize in the nucleus by fusing an SV40 nuclear localization signal (NLS) at the C terminus of the mutant proteins. Indirect immunofluorescence confirmed their nuclear localization (Fig. 2B). However, when cotransfected with the SMN2 minigene, Fox1N-NLS still failed to promote exon inclusion. Fox1Ca-NLS displayed slightly greater activity in exon activation, compared with the mutant without the NLS, but it was still not fully functional (Fig. 2C). However, this could again be due to its lower expression level; although we transfected more plasmid to try to increase its expression, this mutant protein failed to accumulate to the same level as the other proteins, suggesting that it is less stable. We conclude from these experiments that the N-terminal domain of RBFOX1 is not functional in enhancing alternative exon inclusion.
Both the RRM and the C-terminal domain are important for exon repression when tethered to the upstream intron
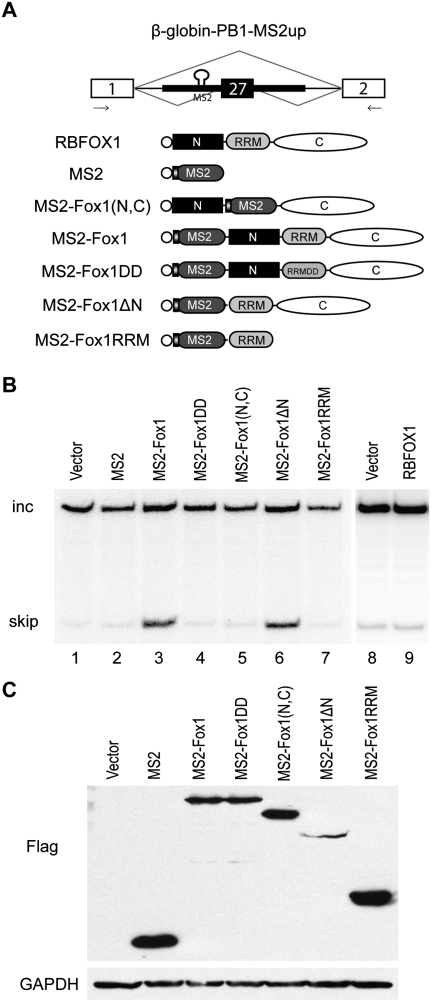
As mentioned above, RBFOX1 usually promotes exon skipping when binding to the upstream intron of alternative cassette exons. We again used the MS2-tethering assay to examine RBFOX1's activity in exon repression. We modified a β-globin PB1 minigene (Zhang et al. 2008) to address this question. We mutated the three natural RBFOX1 motifs in the upstream intron, as well as two motifs in the exon. We then inserted an MS2 binding site in the intron upstream of the alternative exon (Fig. 3A). The alternative exon was predominantly included when cotransfected with MS2 protein, or vector, or RBFOX alone (Fig. 3B, lanes 1, 2, 8, 9). But MS2 protein fused to the N terminus of RBFOX1 markedly enhanced exon skipping (Fig. 3B, lane 3). Like all canonical RRMs, the RBFOX1 RRM utilizes the β-sheet to bind nucleic acids; two exposed phenylalanine residues in the RNP (ribonucleoprotein) submotifs are essential for binding to RNA by intercalating with single-stranded bases (Auweter et al. 2006). We mutated the two phenylalanines to aspartic acids, which was reported to eliminate RNA binding (Mayeda et al. 1994; Auweter et al. 2006), and tested whether the resulting protein can still enhance exon skipping when tethered via MS2. To our surprise, the 2 amino acid substitution completely abolished the exon repression activity, even though the protein is still tethered to the minigene transcript through MS2 (Fig. 3B, lane 4). Similarly, the MS2-Fox1(N,C) mutant, which lacks the entire RRM, failed to promote exon skipping (Fig. 3B, lane 5), although the same protein enhanced exon inclusion when tethered to the downstream intron (see above). We infer that the RRM, including specific RNA interacting residues, is also involved in protein–protein interactions or nonspecific RNA–protein interactions, which are essential for exon repression, but not for exon activation.
FIGURE 3.
Both the RRM and the C-terminal domain are required for exon repression when tethered to the upstream intron. (A) Diagrams of the modified β-globin-PB1 minigene and MS2-fused RBFOX1 constructs. Three natural RBFOX1/2 binding sites in the upstream intron and one natural binding site in the alternative exon were mutated, and an MS2-binding site was inserted in the upstream intron. The primers for RT–PCR are indicated by arrows below the exons. All of the protein mutants have a Flag epitope tag, which is indicated by an open circle. The RRM with two Phe-to-Asp mutations is designated RRMDD. (B) HeLa cells were cotransfected with the β-globin-PB1-MS2up reporter minigene and different RBFOX1 constructs. RNA and protein were extracted 48 h after transfection. Radioactive RT–PCR was performed to detect the changes of the alternative splicing isoforms. (C) Western blot analysis using Flag antibody was performed to show the expression of the protein mutants.
To test whether the N-terminal domain is necessary for the repressive effect, we deleted it and kept the RRM and the C-terminal domain after the MS2 tag. This mutant was fully functional in inducing exon skipping, similar to the full-length protein (Fig. 3B, lane 6). Therefore, the N-terminal domain is not required for exon repression, while the RRM is indispensable. When the RRM alone was fused to MS2, it could not promote exon skipping (Fig. 3B, lane 7). We conclude that both the RRM and C-terminal domain are important for exon repression.
Coimmunoprecipitation and mass spectrometry identify Fox-1C interacting proteins
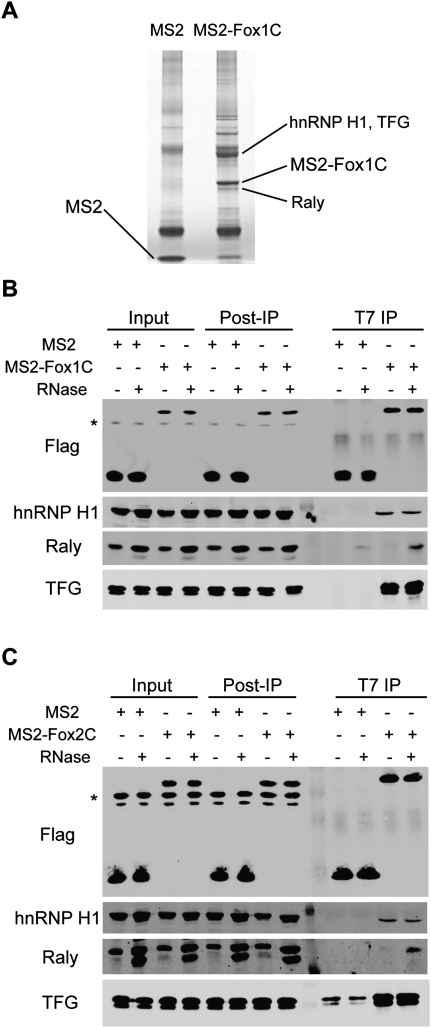
To explore the mechanisms responsible for exon activation by the C-terminal fragment of RBFOX1, we performed coimmunoprecipitation (IP) and mass spectrometry to identify proteins that interact with MS2-Fox1C. We used MS2 as a negative control. As both constructs have a T7 epitope tag between the N-terminal Flag tag and MS2, we used T7 monoclonal antibody cross-linked to Protein G magnetic beads for the IPs. The immunoprecipitated proteins were eluted, separated by SDS-PAGE, and stained with Coomassie Blue (Fig. 4A). We cut out eight prominent gel bands that were only observed in the MS2-Fox1C sample and corresponding gel slices at the same positions of the MS2 sample. Proteins were in-gel digested with trypsin overnight at room temperature. The tryptic peptides were extracted from the gel and analyzed using LC-MS/MS. Multiple protein candidates were identified, including several RNA-binding proteins (Supplemental Table S1). We focused on three candidate proteins: hnRNP H1, RALY, and TFG, which had a high number of unique peptides and high coverage and/or appeared most likely to play roles in alternative splicing based on their known properties.
FIGURE 4.
The C-terminal domains of RBFOX1 and RBFOX 2 interact specifically with hnRNP H1, RALY, and TFG. (A) HeLa cells were transfected with MS2 or MS2-Fox1C. The proteins were immunoprecipitated from whole-cell lysates using Protein G Dynabeads coated with T7 monoclonal antibody. The immunoprecipitated proteins were separated on a gradient SDS–polyacrylamide gel and stained with Coomassie Blue. (B,C) IP using T7 antibody cross-linked to Protein G Dynabeads and Western blotting with Flag, hnRNP H1, RALY, and TFG antibodies. IPs were performed either with (+) or without (−) nuclease treatment. Whole-cell lysates, post-IP lysates, and IPs were blotted with the indicated antibodies. (B) HeLa cells transfected with MS2 or MS2-Fox1C. (C) HeLa cells transfected with MS2 or MS2-Fox2C. (*) Nonspecific bands.
To confirm the protein interactions identified by mass spectrometry, we repeated the co-IP using T7 antibody, followed by Western blotting. To exclude the possibility that the observed interactions are actually mediated by binding to the same RNA, rather than reflecting protein–protein contacts, we also treated the cell lysates with a ribonuclease/benzonase cocktail before the IP. As both MS2 and MS2-Fox1C have a Flag tag at the N terminus, we used Flag antibody for Western blotting to verify the IP efficiency. TFG interacted strongly with Fox1C, independently of RNA and/or DNA (Fig. 4B). RALY gave a stronger signal when the lysate was pretreated with nuclease, in total, post-IP and IP samples. We have observed this with other RNA-binding proteins, and hypothesize that these proteins are probably associated with large RNP complexes, forming aggregates that precipitate with cell debris or are otherwise poorly accessible to the antibodies; nuclease treatment presumably increases the solubility and accessibility of the proteins. Irrespective of nuclease treatment, all three proteins—hnRNP H1, RALY, and TFG—interacted specifically with Fox1C (Fig. 4B; Supplemental Table S1). All three proteins also interacted with full-length RBFOX1, as seen by mass spectrometry analysis, confirming the interaction specificity.
We carried out similar IP-Western experiments with RBFOX2's C-terminal domain, to investigate whether these two paralogs have similar interacting partners. It was already reported that RBFOX2 interacts with hnRNP H1 (Mauger et al. 2008). Our data confirm the previous study, and further show that RALY and TFG interact with the RBFOX2 C-terminal domain (Fig. 4C). These results indicate that RBFOX1 and RBFOX2 have similar interacting partners, consistent with their similar functions in alternative splicing (Zhang et al. 2008).
Testing the involvement of hnRNP H/F, RALY, and TFG in RBFOX2-mediated alternative splicing
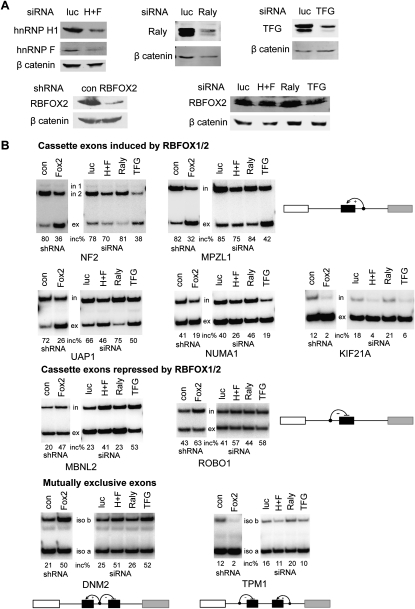
We carried out a siRNA knockdown experiment to test the functional significance of the observed protein–protein interactions. As hnRNP H1 and hnRNP F are 78% identical and appear to have redundant functions (Caputi and Zahler 2001), we knocked down both proteins simultaneously. We transfected siRNAs against luciferase, hnRNP H/F, RALY, or TFG into HeLa cells, and extracted both total RNA and protein after 72 h. All of the proteins were efficiently knocked down without significant changes in endogenous RBFOX2 levels (Fig. 5A). We tested selected RBFOX1/2 targets with cassette exons that are either induced or repressed by RBFOX1/2 up-regulation, as well as targets with mutually exclusive exons. These include both new and previously reported targets (Zhang et al. 2008). We also included HeLa cells with stable shRNA knockdown of RBFOX2 as a positive control (Zhang et al. 2008).
FIGURE 5.
Knockdown of hnRNP H/F and TFG showed modest inhibition on alternative splicing of RBFOX2 endogenous targets. HeLa cells were transfected with the indicated siRNAs. After 72 h, total proteins and RNAs were extracted. HeLa cells with stable knockdown of RBFOX2 using shRNA were analyzed in parallel as the positive control. (A) Western blotting analysis using the indicated antibodies. (B) Radioactive RT–PCR of RBFOX1/2 endogenous targets. The targets are classified into three groups: cassette exons induced by RBFOX1/2, cassette exons repressed by RBFOX1/2, and mutually exclusive exons. A representative diagram is shown for each category. The quantified exon-inclusion level is indicated below each panel.
Knockdown of hnRNP H1 and TFG resulted in similar slight changes in alternative splicing of several RBFOX endogenous targets, compared with RBFOX2 knockdown, though not all of the targets were affected; this was the case for all three types of alternative splicing patterns (Fig. 5B). This result suggests that hnRNP H1 and TFG assist the function of RBFOX in both exon activation and exon repression, through their interaction with the C-terminal domain. In contrast, knockdown of RALY had no effect on alternative splicing of any of the RBFOX endogenous targets tested (Fig. 5B).
TFG is involved in alternative splicing regulation of RBFOX targets
TFG (TRK-fused gene) is a chromosomal translocation partner of NTRK1 (a nerve-growth-factor receptor gene) giving rise to the TRK-T3 oncogene, which is associated with thyroid papillary carcinoma (Greco et al. 1995). TFG is part of signal-transduction pathways, and interacts with the SH2 domain of SHP-1, a protein–tyrosine phosphatase, whose activity it modulates (Roccato et al. 2005). It also interacts with TANK and NEMO, two proteins involved in the NF-κB pathway (Miranda et al. 2006). To our knowledge, there was no evidence that TFG can bind RNA or regulate RNA metabolism.
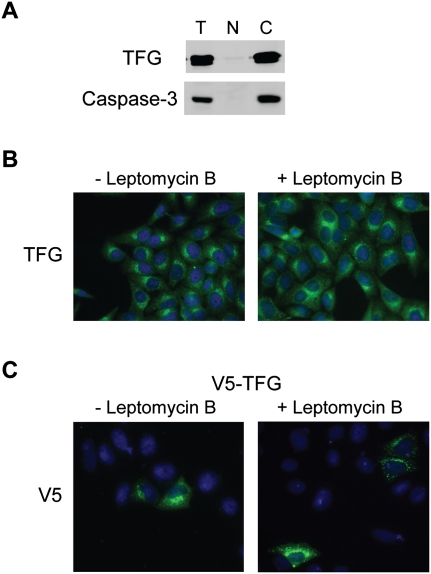
To investigate possible mechanisms through which TFG regulates alternative splicing induced by RBFOX, we first determined the subcellular localization of the protein. Previous reports focused on TRK-T3, which comprises only an N-terminal fragment of TFG, so the localization of full-length TFG was not known. We prepared nuclear and cytoplasmic fractions of HeLa cells, and analyzed them by Western blotting. We probed for Caspase-3 as a cytoplasmic marker. Surprisingly, TFG was mainly localized in the cytoplasm, with only a very weak signal in the nucleus (Fig. 6A). We also carried out immunofluorescence with antibodies against TFG or the V5 tag to examine the localization of both endogenous and transfected V5-tagged TFG. We observed that TFG protein localized in the cytoplasm, possibly in the endoplasmic reticulum (Fig. 6B,C). We could not detect a signal in the nucleus. To test whether TFG can shuttle between the nucleus and the cytoplasm, we treated HeLa cells with leptomycin B, a nuclear-export inhibitor. However, this treatment did not cause the TFG protein to accumulate in the nucleus (Fig. 6B,C). But as leptomycin B specifically inhibits the nuclear export receptor CRM1 (Yoshida and Horinouchi 1999), we cannot exclude the possibility that TFG shuttles through other pathways. Thus, it is possible that TFG transiently localizes in the nucleus, where it modulates RBFOX activity; alternatively, RBFOX1/2 might shuttle to the cytoplasm, where it would interact with TFG, and this would somehow be important for its nuclear function in alternative splicing.
FIGURE 6.
TFG is localized in the cytoplasm. (A) Western blot analysis of HeLa cell subcellular fractionations using TFG and caspase-3 antibody. (T) Total; (N) nucleus; (C) cytoplasm. (B) Indirect immunofluorescence of HeLa cells with or without Leptomycin B treatment using TFG antibody. (C) Indirect immunofluorescence of HeLa cells transfected with V5-TFG was performed using V5 antibody.
DISCUSSION
We characterized the function of RBFOX1 using an MS2-tethering assay. We found that the C-terminal domain—especially the last 100 amino acids—is important for both RBFOX1 nuclear localization and exon inclusion activity. It was recently reported that the C-terminal sequence comprises a putative hPY-NLS, a nuclear localization signal recognized by karyopherin β2 (Boillee et al. 2006; Kuroyanagi 2009). This sequence is conserved in both RBFOX1 and RBFOX2, from C. elegans to human, and it is also present in some other splicing factors, such as hnRNP A1, hnRNP D, hnRNP F, etc. This is consistent with our localization results. It has also been noted that some RBFOX1 isoforms lack the second half of the C-terminal domain because of frame-shifting, and therefore, they are not functional in splicing regulation (Nakahata and Kawamoto 2005; Lee et al. 2009).
The RBFOX1 C-terminal domain alone is sufficient to promote alternative exon inclusion when tethered to the downstream intron via MS2. This result suggests that like modular transcription factors and some splicing factors, such as SR proteins and hnRNP proteins, RBFOX1 also has modular nucleic acid-binding and functional domains. However, both the RRM and the C-terminal domain are required for alternative exon repression when tethered to the upstream intron. This observation implies that different mechanisms are involved in RBFOX1 activation versus repression. It is possible that the RRM is also involved in protein–protein interactions (Ding et al. 1999), which could be critical for exon repression, but not for exon activation. An alternative scenario is that RBFOX1 also needs to interact with other RNA sequences—for example, elsewhere on the pre-mRNA to loop out the exon, or with an snRNA in the spliceosome—in addition to the engineered MS2-binding site for its repressive activity, especially considering that the amino acid substitutions in the canonical RNA-binding surface of the RRM completely abolished the repression activity.
There are precedents for different domains of a splicing factor being required in the context of its different activities. For example, all SR proteins have one or two RRMs, and one RS domain that is involved primarily in protein–protein interactions (Long and Caceres 2009; Zhong et al. 2009). SR proteins function in two distinct aspects of splicing: on one hand, they are required for constitutive splicing and spliceosome assembly; on the other hand, they also regulate alternative splicing, e.g., upon binding to exonic splicing-enhancer elements (Long and Caceres 2009). The RS domain alone is sufficient for enhancer-dependent splicing, but not for constitutive splicing in MS2-tethering in vitro splicing assays; the latter reaction requires a full-length SR protein (Graveley and Maniatis 1998). But, under some conditions the RS domain is dispensable for general splicing in vitro (Zhu and Krainer 2000; Shaw et al. 2007).
Using co-IP and mass spectrometry, we identified several RBFOX1-interacting proteins. Previously, the only known interacting protein of RBFOX1 was Ataxin-2 (Shibata et al. 2000), which is how RBFOX1 was first discovered. We did not find this protein in our analysis, probably because of its tissue-restricted expression. We selected three prominent candidates for further analysis: hnRNP H1, RALY, and TFG. IP-Western validation showed that both RBFOX1 and RBFOX2's C-terminal domains bind to all of the three proteins specifically, both before and after nuclease treatment. The C-terminal domains of the RBFOX1 and RBFOX2 isoforms we studied are 58% identical, rich in Ala, Tyr, and Gly residues, and have an overall positive charge. This is consistent with the two paralogs having similar interacting partners and functions. Using functional assays in combination with RNAi, we showed that hnRNP H1 and TFG modulate the activity of RBFOX1/2 in alternative splicing, both in activation and repression. In contrast, RALY had no effect on any of the targets we tested.
hnRNP H1 belongs to the superfamily of heterogeneous nuclear ribonucleoproteins (hnRNPs). It binds to intronic oligo-(G) sequences and regulates alternative exons negatively or positively, depending upon the context (Chen et al. 1999; Chou et al. 1999; Jacquenet et al. 2001; Caputi and Zahler 2002; Garneau et al. 2005; Crawford and Patton 2006). It was recently shown that hnRNP F and hnRNP H form a complex with RBFOX2, and this interaction enhances hnRNP H/F's ability to antagonize SRSF1 binding to FGFR2 exon IIIc (Mauger et al. 2008). Our experiments showed that RBFOX1 can also interact with hnRNP H1, and that hnRNPH1 enhances the activity of RBFOX1/2 in both exon activation and exon repression. Thus, our data confirm and extend the previous study, and suggest that RBFOX1/2 and hnRNP H1 modulate each other's activities in alternative splicing regulation.
RALY was originally identified as an autoantigen that cross-reacts with EBNA-1 of the Epstein-Barr virus in infectious mononucleosis (Rhodes et al. 1997). It too is a member of the hnRNP superfamily, and is also known as hnRNP C-like 2. It is associated with the spliceosomal C complex (Jurica et al. 2002), and is therefore suspected to be involved in pre-mRNA splicing. However, to date there is no direct functional evidence of such involvement, and no extensive characterization of this protein. Our data showed that RALY specifically binds to RBFOX1/2. However, the functional significance of this interaction remains unknown, as knockdown of this protein did not affect alternative splicing of RBFOX1/2 endogenous targets.
TFG is a signaling protein (Roccato et al. 2005; Miranda et al. 2006). We included it in our functional study because it strongly and specifically interacted with the C-terminal domain of RBFOX1 and RBFOX2. Knockdown of TFG markedly affected alternative splicing of several RBFOX1/2 endogenous targets, suggesting that TFG is indeed involved in modulating the activity of RBFOX1/2 splicing factors. However, these two proteins do not appear to colocalize at steady state when analyzed by immunofluorescence. It is possible that they interact transiently while shuttling, and that this interaction is important for the nuclear activity of RBFOX. Alternatively, TFG may modulate RBFOX1/2's functions indirectly, such as by controlling post-translational modifications through signaling pathways. Our data indicate that TFG and RBFOX1 are part of the same protein complex, at least in vitro, and it is possible that they do not contact each other directly, but that each can physically interact with a common binding partner. More extensive studies will be required to uncover the precise mechanisms underlying the effect of TFG on splicing regulation by RBFOX proteins.
MATERIALS AND METHODS
Plasmids
We inserted the MS2-binding site (5′-GCGTACACCATCAGGGTACGC-3′) into SMN2 (Hua et al. 2008) and β-globin PB1 (Zhang et al. 2008) minigenes by site-directed mutagenesis (Supplemental Methods). Three RBFOX binding sites in the upstream intron and two in the exon were mutated in PB1 (Supplemental Methods). We used cDNAs corresponding to the best-characterized variants, known as RBFOX1 isoform 4 (397 amino acids) and RBFOX2 isoform 1 (380 amino acids); the accession numbers in NCBI are NM_018723.3 and NM_001031695.2, respectively. The sequences flanking the RRM in pcDNA-Flag-Fox1 (Zhang et al. 2008) were mutated to introduce NcoRI and BamHI sites, and then the RRM (aa 118–189) was replaced by T7-tagged MS2 coding sequences. We subcloned MS2-Fox1C (aa 190–397), MS2-Fox1N (aa 1–117), and MS2-Fox1Ca (aa 190–297) into pcDNA-Flag via BamHI/BglII and XhoI sites. We deleted the first 108 amino acids of the C-terminal domain in MS2-Fox1C by site-directed mutagenesis to create MS2-Fox1Cb (aa 298–397). We inserted a DNA fragment coding for an SV40 NLS (5′-CCTAAGAAGAAACGTAAGGTC-3′) before the stop codon by mutagenesis to construct MS2-NLS, MS2-Fox1N-NLS, and MS2-Fox1Ca-NLS. We cleaved MS2-Fox1C with BamHI and XhoI to release the C-terminal domain of Fox1, and subcloned the full-length Fox-1 via BglII and XhoI sites to create MS2-Fox1; or subcloned both the RRM and C-terminal domain to construct MS2-Fox1ΔN; or subcloned the RRM only to construct MS2-Fox1RRM; or subcloned the C-terminal domain of RBFOX2 (aa 184–380) to create MS2-Fox2C. We mutated the two phenylalanines (F158 and F160) to aspartic acids by site-directed mutagenesis.
Cell culture and transfection
We cultured HeLa cells in DMEM supplemented with 10% (v/v) FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin. We used Fugene 6 (Roche) or Lipofectamine 2000 (Invitrogen) to transfect plasmids, and Oligofectamine (Invitrogen) to transfect siRNAs. For knockdown experiments, we transfected siRNAs 72 h prior to sample collection. The siRNA sequences were as follows: hnRNP H1 5′-CAAACAACGUUGAAAUGGA-3′; hnRNP F 5′-CGACCGAGAACGACAUUUA-3′; RALY 5′-UAACGUACCUGUCAAGCUC-3′; TFG: 5′-GAGGAAAACUUCUGAGUAA-3′; luciferase: 5′-GCCAUUCUAUCCUCUAGAGGA-3′). For the nuclear-export inhibition experiment, cells were treated with 10 ng/mL Leptomycin B (Calbiochem) for 12 h.
Cell fractionation
We lysed cells in gentle lysis buffer (10 mM HEPES at pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.5% [v/v] NP-40). We pelleted the nuclei at 2300g for 5 min at 4°C, and transferred the supernatant (cytoplasm) to another tube. We washed the nuclei once with the same buffer, and repelleted them.
Western blotting
We harvested the cells and lysed them in Laemmli buffer. The primary antibodies included β-catenin (Sigma), Flag tag (Sigma), hnRNP H1 (Abcam), hnRNP F (Santa Cruz), RALY (Abcam), and TFG (Abcam). The secondary antibodies were goat anti-mouse or anti-rabbit IgG (H+L) HRP-conjugated (Pierce), or labeled with IRDye 800CW (LI-COR). For detection, we used an ECL kit (Roche), or an Odyssey Infrared Imaging System (LI-COR).
RNA isolation and RT–PCR
To isolate total RNA from cells, we used Trizol (Invitrogen) and treatment with RQ1 DNase I (Promega). For first-strand cDNA synthesis, we used random hexamers and ImProm-II reverse transcriptase (Promega). For radioactive PCR, we used AmpliTaq (Roche), added [γ-32P]dCTP, and amplified for 23–26 cycles. We separated the PCR products on 6% nondenaturing polyacrylamide gels, and detected them with an Image Reader FLA-5100 (Fuji). PCR primer sequences are shown in the Supplemental Methods.
Immunofluorescence
We fixed HeLa cells with 4% (v/v) paraformaldehyde in PBS for 30 min, 48 h after transfection. We permeabilized cells in 0.2% (v/v) Triton X-100 for 5 min. We then incubated the cells with Flag antibody (5 μg/mL; Sigma) for 1 h, washed with PBS, and then incubated with Alexa Fluor 594 or 488-conjugated goat anti-rabbit IgG antibody (1:1000; Invitrogen) for 1 h. We imaged the cells with a fluorescence microscope (Axioskop, Carl Zeiss).
Immunoprecipitation
Dynabeads Protein G was washed twice with Citrate-Phosphate Buffer (CPB; 25 mM citric acid, 50 mM Na2HPO4 at pH 5.0), and incubated with T7-tag monoclonal antibody for 1 h at room temperature (2 mL of culture supernatant/10 μL of beads). We washed the beads twice in 500 μL of CPB, and twice in 0.2 M triethanolamine (pH 8.2) (Sigma). We then resuspended the beads in 1 mL of 20 mM dimethyl pimelimidate (Sigma) in 0.2 M triethanolamine (pH 8.2), and incubated at 20°C for 30 min with gentle mixing to cross-link the antibody to the beads. To stop the reaction, we resuspended the beads in 1 mL of 50 mM Tris-HCl (pH 7.5) and mixed for 15 min at room temperature. Finally, we washed the beads three times in PBS. We lysed four 15-cm plates of HeLa cells in 4 mL of lysis buffer (0.3% [v/v] NP-40, 200 mM NaCl, 50 mM Tris at pH 7.4, 1 mM DTT, 0.1 mM EDTA, 0.1 mM EGTA, with freshly added 1 mM sodium vanadate, 50 mM sodium fluoride and protease-inhibitor cocktail), and sheared the DNA by passing sequentially through a syringe with 20, 22, and 26G needles, three times each. We added nuclease cocktail (1 U/mL RNase cocktail [Ambion], 500 U/mL Benzonase [Novagen], and 2 mM MgCl2), and incubated on ice for 30 min. We then spun the lysates at 13,000g for 20 min at 4°C. We passed the supernatant through a 0.45-μm syringe filter (with HT Tuffryn membrane, Pall Corporation). We then added 80 μL beads (1:1 suspension) to the cleared lysates and incubated for 1 h at 4°C. After washing five times in lysis buffer, we resuspended the beads in Laemmli buffer and loaded the supernatant on an SDS–polyacrylamide gel.
Mass spectrometry
We ran the samples on a NuPAGE Novex Bis-tris Mini Gel (Invitrogen) and stained with Gelcode Blue Stain Reagent (Pierce). We cut out specific bands from the gel and washed in 50% (v/v) ethanol/5% (v/v) acetic acid prior to reduction with DTT and alkylation with iodoacetamide. We in-gel digested the proteins with sequencing-grade trypsin overnight at room temperature. We extracted peptides from the gel and analyzed them by LC-MS/MS. We used a Proxeon nano-flow HPLC pump. The buffer solutions were 5% (v/v) acetonitrile/0.1% (v/v) formic acid (buffer A), and 80% (v/v) acetonitrile/0.1% (v/v) formic acid (buffer B). Samples were loaded with the autosampler onto a 180-μm i.d. fused silica capillary precolumn containing 3 cm of 5-μm Aqua C 18 material (Phenomenex). The precolumn was then connected to an analytical column of a 100-μm i.d. capillary with a 5-μm pulled tip and packed with 12–13 cm of 3-μm Aqua C 18 material. Peptides were separated using a linear gradient from 5% (v/v) acetonitrile, increasing at 0.5% per min for 90 min. The flow rate was set at 300 nL/min (splitless). Peptides eluted from the microcapillary fritless column were directly electrosprayed into a linear ion-trap (LTQ) mass spectrometer (Thermo Finnigan) with the application of a distal 2.4 kV spray voltage. A cycle of one full-scan mass spectrum (400–1700 m/z), followed by five data-dependent MS/MS spectra at a 35% normalized collision energy, was repeated continuously throughout the chromatographic separation.
Obtained spectra were processed with MASCOT Distiller (Matrix Science) using the default parameters for ion-trap data analysis. LTQ MSMS spectra were searched with MASCOT version 2.2.04 against the human IPI nonredundant database (version 3.35) for tryptic peptides, allowing for one missed cleavage. The peptide mass tolerance was set for 30 ppm, and the fragment ion tolerance was set for 0.6 Da. The modifications included in the search were carbamidomethylation of cysteines as a fixed modification, oxidation of methionine as a variable modification, and deamidation of asparagines and glutamines as additional variable modifications. Peptide-spectrum matches were accepted for expectation value <0.5, resulting in a false discovery rate <1% when compared with decoy database hits.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Cristian Ruse for mass spectrometry analysis and Zhaozhu Qiu and Stephen Hearn for help with microscopy imaging. This work was supported by grant GM42699 from the National Institute of General Medical Sciences.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.030486.111.
REFERENCES
- Auweter SD, Fasan R, Reymond L, Underwood JG, Black DL, Pitsch S, Allain FH 2006. Molecular basis of RNA recognition by the human alternative splicing factor Fox-1. EMBO J 25: 163–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baraniak AP, Chen JR, Garcia-Blanco MA 2006. Fox-2 mediates epithelial cell-specific fibroblast growth factor receptor 2 exon choice. Mol Cell Biol 26: 1209–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boillee S, Yamanaka K, Lobsiger CS, Copeland NG, Jenkins NA, Kassiotis G, Kollias G, Cleveland DW 2006. Onset and progression in inherited ALS determined by motor neurons and microglia. Science 312: 1389–1392 [DOI] [PubMed] [Google Scholar]
- Caputi M, Zahler AM 2001. Determination of the RNA binding specificity of the heterogeneous nuclear ribonucleoprotein (hnRNP) H/H′/F/2H9 family. J Biol Chem 276: 43850–43859 [DOI] [PubMed] [Google Scholar]
- Caputi M, Zahler AM 2002. SR proteins and hnRNP H regulate the splicing of the HIV-1 tev-specific exon 6D. EMBO J 21: 845–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey J, Lowary PT, Uhlenbeck OC 1983. Interaction of R17 coat protein with synthetic variants of its ribonucleic acid binding site. Biochemistry 22: 4723–4730 [DOI] [PubMed] [Google Scholar]
- Chen CD, Kobayashi R, Helfman DM 1999. Binding of hnRNP H to an exonic splicing silencer is involved in the regulation of alternative splicing of the rat β-tropomyosin gene. Genes Dev 13: 593–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou MY, Rooke N, Turck CW, Black DL 1999. hnRNP H is a component of a splicing enhancer complex that activates a c-src alternative exon in neuronal cells. Mol Cell Biol 19: 69–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford JB, Patton JG 2006. Activation of α-tropomyosin exon 2 is regulated by the SR protein 9G8 and heterogeneous nuclear ribonucleoproteins H and F. Mol Cell Biol 26: 8791–8802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damianov A, Black DL 2010. Autoregulation of Fox protein expression to produce dominant negative splicing factors. RNA 16: 405–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, Hayashi MK, Zhang Y, Manche L, Krainer AR, Xu RM 1999. Crystal structure of the two-RRM domain of hnRNP A1 (UP1) complexed with single-stranded telomeric DNA. Genes Dev 13: 1102–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumura K, Kato A, Jin Y, Ideue T, Hirose T, Kataoka N, Fujiwara T, Sakamoto H, Inoue K 2007. Tissue-specific splicing regulator Fox-1 induces exon skipping by interfering E complex formation on the downstream intron of human F1γ gene. Nucleic Acids Res 35: 5303–5311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumura K, Taniguchi I, Sakamoto H, Ohno M, Inoue K 2009. U1-independent pre-mRNA splicing contributes to the regulation of alternative splicing. Nucleic Acids Res 37: 1907–1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau D, Revil T, Fisette JF, Chabot B 2005. Heterogeneous nuclear ribonucleoprotein F/H proteins modulate the alternative splicing of the apoptotic mediator Bcl-x. J Biol Chem 280: 22641–22650 [DOI] [PubMed] [Google Scholar]
- Graveley BR, Maniatis T 1998. Arginine/serine-rich domains of SR proteins can function as activators of pre-mRNA splicing. Mol Cell 1: 765–771 [DOI] [PubMed] [Google Scholar]
- Greco A, Mariani C, Miranda C, Lupas A, Pagliardini S, Pomati M, Pierotti MA 1995. The DNA rearrangement that generates the TRK-T3 oncogene involves a novel gene on chromosome 3 whose product has a potential coiled-coil domain. Mol Cell Biol 15: 6118–6127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y, Vickers TA, Okunola HL, Bennett CF, Krainer AR 2008. Antisense masking of an hnRNP A1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice. Am J Hum Genet 82: 834–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquenet S, Mereau A, Bilodeau PS, Damier L, Stoltzfus CM, Branlant C 2001. A second exon splicing silencer within human immunodeficiency virus type 1 tat exon 2 represses splicing of Tat mRNA and binds protein hnRNP H. J Biol Chem 276: 40464–40475 [DOI] [PubMed] [Google Scholar]
- Jin Y, Suzuki H, Maegawa S, Endo H, Sugano S, Hashimoto K, Yasuda K, Inoue K 2003. A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. EMBO J 22: 905–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurica MS, Licklider LJ, Gygi SR, Grigorieff N, Moore MJ 2002. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA 8: 426–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KK, Adelstein RS, Kawamoto S 2009. Identification of neuronal nuclei (NeuN) as Fox-3, a new member of the Fox-1 gene family of splicing factors. J Biol Chem 284: 31052–31061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroyanagi H 2009. Fox-1 family of RNA-binding proteins. Cell Mol Life Sci 66: 3895–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JA, Tang ZZ, Black DL 2009. An inducible change in Fox-1/A2BP1 splicing modulates the alternative splicing of downstream neuronal target exons. Genes Dev 23: 2284–2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorian M, Schwartz S, Clark TA, Hollander D, Tan LY, Spellman R, Gordon A, Schweitzer AC, de la Grange P, Ast G, et al. 2010. Position-dependent alternative splicing activity revealed by global profiling of alternative splicing events regulated by PTB. Nat Struct Mol Biol 17: 1114–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JC, Caceres JF 2009. The SR protein family of splicing factors: master regulators of gene expression. Biochem J 417: 15–27 [DOI] [PubMed] [Google Scholar]
- Mauger DM, Lin C, Garcia-Blanco MA 2008. hnRNP H and hnRNP F complex with Fox2 to silence fibroblast growth factor receptor 2 exon IIIc. Mol Cell Biol 28: 5403–5419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeda A, Munroe SH, Cáceres JF, Krainer AR 1994. Function of conserved domains of hnRNP A1 and other hnRNP A/B proteins. EMBO J 13: 5483–5495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda C, Roccato E, Raho G, Pagliardini S, Pierotti MA, Greco A 2006. The TFG protein, involved in oncogenic rearrangements, interacts with TANK and NEMO, two proteins involved in the NF-κB pathway. J Cell Physiol 208: 154–160 [DOI] [PubMed] [Google Scholar]
- Nakahata S, Kawamoto S 2005. Tissue-dependent isoforms of mammalian Fox-1 homologs are associated with tissue-specific splicing activities. Nucleic Acids Res 33: 2078–2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkura N, Takahashi M, Yaguchi H, Nagamura Y, Tsukada T 2005. Coactivator-associated arginine methyltransferase 1, CARM1, affects pre-mRNA splicing in an isoform-specific manner. J Biol Chem 280: 28927–28935 [DOI] [PubMed] [Google Scholar]
- Ponthier JL, Schluepen C, Chen W, Lersch RA, Gee SL, Hou VC, Lo AJ, Short SA, Chasis JA, Winkelmann JC, et al. 2006. Fox-2 splicing factor binds to a conserved intron motif to promote inclusion of protein 4.1R alternative exon 16. J Biol Chem 281: 12468–12474 [DOI] [PubMed] [Google Scholar]
- Rhodes GH, Valbracht JR, Nguyen MD, Vaughan JH 1997. The p542 gene encodes an autoantigen that cross-reacts with EBNA-1 of the Epstein Barr virus and which may be a heterogeneous nuclear ribonucleoprotein. J Autoimmun 10: 447–454 [DOI] [PubMed] [Google Scholar]
- Roccato E, Miranda C, Raho G, Pagliardini S, Pierotti MA, Greco A 2005. Analysis of SHP-1-mediated down-regulation of the TRK-T3 oncoprotein identifies Trk-fused gene (TFG) as a novel SHP-1-interacting protein. J Biol Chem 280: 3382–3389 [DOI] [PubMed] [Google Scholar]
- Shaw SD, Chakrabarti S, Ghosh G, Krainer AR 2007. Deletion of the N-terminus of SF2/ASF permits RS-domain-independent pre-mRNA splicing. PLoS ONE 2: e854 doi: 10.1371/journal.pone.0000854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata H, Huynh DP, Pulst SM 2000. A novel protein with RNA-binding motifs interacts with ataxin-2. Hum Mol Genet 9: 1303–1313 [DOI] [PubMed] [Google Scholar]
- Tang ZZ, Zheng S, Nikolic J, Black DL 2009. Developmental control of CaV1.2 L-type calcium channel splicing by Fox proteins. Mol Cell Biol 29: 4757–4765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J, Stefani G, Mele A, Ruggiu M, Wang X, Taneri B, Gaasterland T, Blencowe BJ, Darnell RB 2006. An RNA map predicting Nova-dependent splicing regulation. Nature 444: 580–586 [DOI] [PubMed] [Google Scholar]
- Underwood JG, Boutz PL, Dougherty JD, Stoilov P, Black DL 2005. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol Cell Biol 25: 10005–10016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables JP, Klinck R, Koh C, Gervais-Bird J, Bramard A, Inkel L, Durand M, Couture S, Froehlich U, Lapointe E, et al. 2009. Cancer-associated regulation of alternative splicing. Nat Struct Mol Biol 16: 670–676 [DOI] [PubMed] [Google Scholar]
- Yeo GW, Coufal NG, Liang TY, Peng GE, Fu XD, Gage FH 2009. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat Struct Mol Biol 16: 130–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M, Horinouchi S 1999. Trichostatin and leptomycin. Inhibition of histone deacetylation and signal-dependent nuclear export. Ann N Y Acad Sci 886: 23–36 [DOI] [PubMed] [Google Scholar]
- Zhang C, Zhang Z, Castle J, Sun S, Johnson J, Krainer AR, Zhang MQ 2008. Defining the regulatory network of the tissue-specific splicing factors Fox-1 and Fox-2. Genes Dev 22: 2550–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong XY, Ding JH, Adams JA, Ghosh G, Fu XD 2009. Regulation of SR protein phosphorylation and alternative splicing by modulating kinetic interactions of SRPK1 with molecular chaperones. Genes Dev 23: 482–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou HL, Lou H 2008. Repression of prespliceosome complex formation at two distinct steps by Fox-1/Fox-2 proteins. Mol Cell Biol 28: 5507–5516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou HL, Baraniak AP, Lou H 2007. Role for Fox-1/Fox-2 in mediating the neuronal pathway of calcitonin/calcitonin gene-related peptide alternative RNA processing. Mol Cell Biol 27: 830–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Krainer AR 2000. Pre-mRNA splicing in the absence of an SR protein RS domain. Genes Dev 14: 3166–3178 [DOI] [PMC free article] [PubMed] [Google Scholar]