Ribotoxins cleave essential RNAs involved in protein synthesis as a strategy for cell killing. RNA repair systems exist in nature to counteract the lethal actions of ribotoxins, as first demonstrated by the RNA repair system from bacteriophage T4 25 yr ago. Recently, the authors found that two bacterial proteins, named Pnkp and Hen1, form a stable complex and are able to repair ribotoxin-cleaved tRNAs in vitro. However, unlike the well-studied T4 RNA repair system, the natural RNA substrates of the bacterial Pnkp/Hen1 RNA repair system are unknown. Here they present comprehensive RNA repair assays with the recombinant Pnkp/Hen1 proteins from Anabaena variabilis using a total of 33 different RNAs as substrates that might mimic various damaged forms of RNAs present in living cells and find that, unlike the RNA repair system from bacteriophage T4, the bacterial Pnkp/Hen1 RNA repair system exhibits broad substrate specificity.
Keywords: RNA repair, Pnkp/Hen1, substrate specificity, RNA damage, ribotoxin
Abstract
Ribotoxins cleave essential RNAs involved in protein synthesis as a strategy for cell killing. RNA repair systems exist in nature to counteract the lethal actions of ribotoxins, as first demonstrated by the RNA repair system from bacteriophage T4 25 yr ago. Recently, we found that two bacterial proteins, named Pnkp and Hen1, form a stable complex and are able to repair ribotoxin-cleaved tRNAs in vitro. However, unlike the well-studied T4 RNA repair system, the natural RNA substrates of the bacterial Pnkp/Hen1 RNA repair system are unknown. Here we present comprehensive RNA repair assays with the recombinant Pnkp/Hen1 proteins from Anabaena variabilis using a total of 33 different RNAs as substrates that might mimic various damaged forms of RNAs present in living cells. We found that unlike the RNA repair system from bacteriophage T4, the bacterial Pnkp/Hen1 RNA repair system exhibits broad substrate specificity. Based on the experimental data presented here, a model of preferred RNA substrates of the Pnkp/Hen1 repair system is proposed.
INTRODUCTION
RNA in living cells is highly susceptible to damage, primarily due to breakage of the phosphodiester backbone. Phosphodiester bond cleavages are usually triggered by ribonucleases, assisted by the adjacent 2′ OH group in RNA at the site of breakage, generating 2′,3′-cyclic phosphate at the 3′ end and 5′ OH at the 5′ end in most cases. Most RNA breakdown is part of normal metabolism, carried out by endogenous ribonucleases. However, cleavage of essential RNAs is exploited by some organisms as a strategy for cell killing. The agents responsible for the RNA cleavage are proteins called ribotoxins, which are usually site-specific ribonucleases. The ribotoxins identified to date kill cells by targeting rRNAs and tRNAs involved in protein translation. Known ribotoxins targeting rRNAs are sarcin (Endo and Wool 1982), restrictocin (Fando et al. 1985), colicin E3 (Bowman et al. 1971), and Biota orientalis ribonuclease (Xu et al. 2004). Several ribotoxins such as colicin E5 (Ogawa et al. 1999), colicin D (Tomita et al. 2000), and Kluyveromyces lactis γ-toxin (Lu et al. 2005) are tRNA-specific ribotoxins, which make a single cut in the anticodon loop of a specific tRNA. In addition to the external ribotoxins that invade cells and damage RNA, a cell sometimes produces its own site-specific ribonucleases to regulate gene expression and cell fate. For example, when infected by bacteriophage T4, some strains of Escherichia coli activate ribonuclease PrrC, which cleaves tRNALys at the wobble position in the anticodon loop (Amitsur et al. 1987). RelE is activated to cleave mRNAs at the ribosome A site to restrict global protein production when a cell is starved (Pedersen et al. 2003; Neubauer et al. 2009). More recently, VapC was shown to have the same effect as RelE but by cleaving the initiator tRNAfMet in the anticodon loop (Winther and Gerdes 2011). In eukaryotic organisms, stress causes significant RNA damage, which may involve ribonucleases (Lee and Collins 2005; Thompson et al. 2008; Yamasaki et al. 2009).
Two proteins from bacteriophage T4, Pnkp (polynucleotide kinase-phosphatase) and Rnl1 (RNA ligase 1), counteract the suicidal action of PrrC in infected E. coli by RNA repair (Amitsur et al. 1987). Specifically, bifunctional T4Pnkp hydrolyzes the 2′,3′-cyclic phosphate at the 3′ end and adds a phosphate group at the 5′ end of the cleaved tRNALys. Subsequently, T4Rnl1 ligates the two processed ends to restore the tRNALys to its original form. Thus, the RNA repair carried out by bacteriophage T4 precludes disruption of T4 late translation and thus rescues the infection.
Recently, we found that two bacterial proteins repair ribotoxin-cleaved tRNAs in vitro (Chan et al. 2009b). The two proteins, bacterial Pnkp and Hen1, physically interact with each other, forming a heterotetramer in vitro. The bacterial Pnkp from Clostridium thermocellum was first characterized by Martins and Shuman (2005), who showed 5′ kinase, 2′,3′ phosphatase, and adenylyltransferase activities. Interestingly, Pnkp alone could not repair a cleaved RNA (Keppetipola et al. 2007). We demonstrated that RNA repair requires both Pnkp and Hen1 (Chan et al. 2009b). The Pnkp/Hen1 RNA repair system is distinct from the T4Pnkp/Rnl1 system due to the involvement of a fourth enzymatic activity, 2′-O-methyltransferase, which is located at the C terminus of bacterial Hen1. During RNA repair, bacterial Hen1 carries out 2′-O-methylation at the 3′ cleavage end after dephosphorylation of the 2′,3′-cyclic phosphate but before ligation of the two cleavage ends (Chan et al. 2009b). Due to the 2′-O-methylation at the junction of repair, the same RNA damage can no longer occur in the repaired RNA.
In contrast to the extensively characterized T4 system, we know much less about the more recently discovered bacterial Pnkp/Hen1 RNA repair system. Prior studies from the Shuman laboratory and our laboratory have focused on in vitro characterization, either of the Pnkp/Hen1 complex (Chan et al. 2009b) or of components of the complex (Martins and Shuman 2005; Keppetipola et al. 2007; Chan et al. 2009a; Jain and Shuman 2010, 2011). Perhaps the biggest gap is the lack of knowledge about the in vivo RNA substrate(s) of Pnkp/Hen1 and the biological effect of RNA repair in strains containing the Pnkp/Hen1 RNA repair system. If RNA damage is caused internally in bacterial species possessing the Pnkp/Hen1 RNA repair system, it is possible that an in vivo study on one of these bacterial species would provide insight into RNA damage and repair. On the other hand, it is possible that RNA damage in these bacteria could be caused by external agents such as ribotoxins released from other surrounding organisms. If that were the case, then RNA repair would only occur when these bacteria live under conditions that induce such damage, which might not be readily reproduced in a laboratory setting.
Here we present a comprehensive in vitro RNA repair study using Pnkp/Hen1 from Anabaena variabilis (AvaPnkp/Hen1) and a variety of RNA substrates, with the aim of providing insight into the likely in vivo RNA substrates of the Pnkp/Hen1 RNA repair system. We found that PnkP/Hen1 may exhibit broader substrate specificity than T4PnkP/Rnl1. Based on the studies presented here, the likely in vivo RNA substrates of Pnkp/Hen1 and the possible biological roles of Pnkp/Hen1 are discussed.
RESULTS
AvaPnkp/Hen1 efficiently repairs tRNA deletion mutants
Prior to finding the RNA repair capabilities of bacterial Pnkp/Hen1, several RNA ligase enzymes, with implications for RNA editing or repair, have been biochemically characterized (Amitsur et al. 1987; Ho and Shuman 2002; Simpson et al. 2003; Martins and Shuman 2004a,b). Among them, the T4Pnkp/Rnl1 is the most extensively characterized as an RNA repair system. The in vivo RNA repair target of the T4 system was well defined: to repair E. coli tRNALys cleaved by the endogenous ribonuclease PrrC. Furthermore, the T4Pnkp/Rnl1 repair system appears to have evolved to optimize RNA repair of the cleaved tRNA; this has been suggested by respective T4Pnkp/tRNA and T4Rnl1/tRNA docking experiments (Galburt et al. 2002; El Omari et al. 2006), as well as a subsequent study by Shuman and coworkers (Nandakumar et al. 2008). Using both the cleaved full-length and tRNA deletion mutants, Shuman and coworkers were able to demonstrate that the efficiency of repair for the cleaved full-length tRNA is significantly higher than the ones for the cleaved tRNA deletion mutants (Nandakumar et al. 2008).
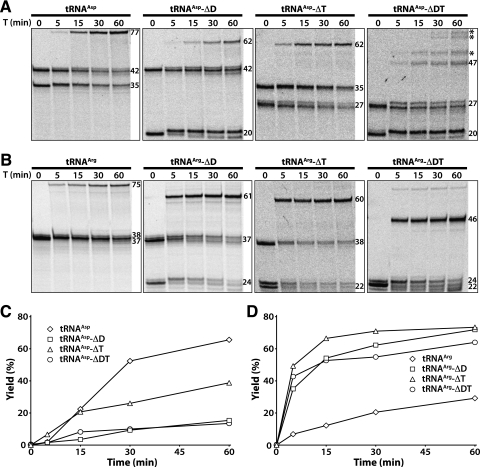
Considering the possibility that like the T4 RNA repair system, the cleaved tRNAs could also be the natural repair targets of the bacterial Pnkp/Hen1 RNA repair system, we performed similar experiments with the AvaPnkp/Hen1 proteins. The cleaved tRNAAsp and its deletion derivatives were our first choice for the experiments because, like the E. coli tRNALys cleaved by PrrC, these tRNAs are cleaved by colicin E5 at a position near the wobble nucleotide (Fig. 1). The cleaved full-length tRNAAsp was efficiently repaired by AvaPnkp/Hen1, with a repair yield as high as 65% after 60 min of repair reaction (Fig. 2A,C, labeled with tRNAAsp). Deletion of the D stem–loop in tRNAAsp resulted in a significantly worse RNA substrate, with a repair yield of 15% after 60 min of repair reaction (Fig. 2A,C, labeled with tRNAAsp-ΔD). Deletion of the TΨC stem–loop was not as bad, with a final repair yield of 40% (Fig. 2A,C, labeled with tRNAAsp-ΔT). Also, AvaPnkp/Hen1 was still able to repair a tRNAAsp deletion mutant with both D and TΨC stem–loops deleted, with a repair yield comparable to the deletion of D stem–loop alone (Fig. 2A,C, labeled with tRNAAsp-ΔDT). In addition to the repaired tRNAAsp-ΔDT, some ligation products of higher molecular weight (Fig. 2A, marked with *) were also produced with the cleaved tRNAAsp-ΔDT as the substrate. We noticed that tRNAAsp-ΔDT-3′ has five continuous Gs (Table 1). Therefore, it is possible that repair of abnormal annealed RNA substrates resulted in production of additional ligation products.
FIGURE 1.
Cloverleaf view of full-length and tRNA deletion mutants employed in this study. Nucleotides are depicted as cycles, and they are linked with each other by short thin lines. Base pairs are represented with thick lines. The cleavage sites of tRNAAsp by colicin E5 (ColE5) and tRNAArg by colicin D (ColD) are marked with arrows. Three nucleotides constituting the anticodon of tRNA are highlighted in gray.
FIGURE 2.
Repair of cleaved full-length and tRNA deletion mutants depicted in Figure 1 by AvaPnkp/Hen1. (A,C) Time course of RNA repair of tRNAAsp and its deletion derivatives. (B,D) Time course of RNA repair of tRNAArg and its deletion derivatives. The positions of RNA substrates and repair products are marked with numbers representing their sizes in nucleotides. T indicates time in minutes.
TABLE 1.
RNAs used in this study
Next, we carried out RNA repair of the cleaved tRNAArg and its deletion derivatives (Fig. 2B,D). tRNAArg and its deletion derivatives were cleaved by colicin D at a position that is significantly different from the one by colicin E5 (Fig. 1). Compared with the repair of the cleaved tRNAAsp, cleaved full-length tRNAArg was repaired less efficiently, with a repair yield of 30% after 60 min of repair reaction (Fig. 2B,D, labeled with tRNAArg). Surprisingly, unlike tRNAAsp deletion derivatives, deletion of the D stem–loop, TΨC stem–loop, or both in tRNAArg resulted in RNA substrates that were more efficiently repaired (Fig. 2B,D). The efficiency of repair of these cleaved tRNAArg deletion derivatives by AvaPnkp/Hen1 is quite remarkable. For example, after only 15 min of repair reaction, the repair yields of the cleaved tRNAArg-ΔD, tRNAArg-ΔT, and tRNAArg-ΔDT were 54%, 67%, and 53%, respectively (Fig. 2D). At that time point, the repair yields of the cleaved tRNAAsp and tRNAArg were 22% and 12%, respectively.
Efficient repair by AvaPnkp/Hen1 requires an RNA substrate of more than a short stem–loop RNA
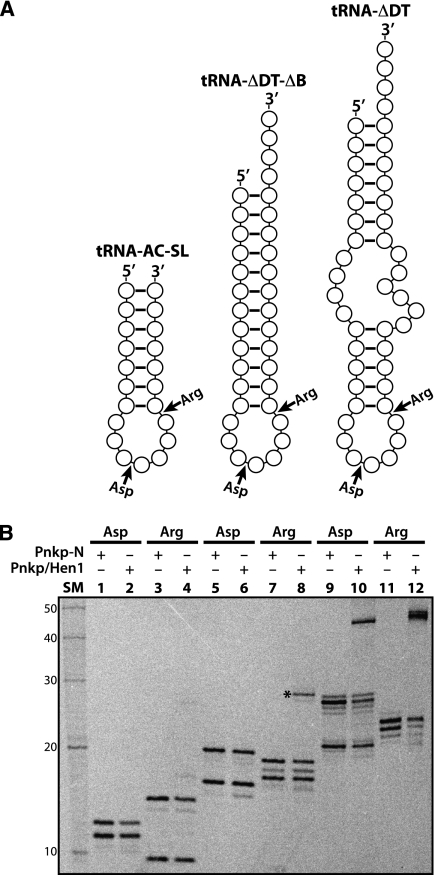
Having demonstrated that the drastically deleted tRNAs (e.g., tRNA-ΔDT) could still be effectively repaired by AvaPnkp/Hen1, we next tested if further truncated RNA substrates would be repaired, aiming to define a minimum RNA repair substrate. A total of four additional RNA substrates were prepared for the RNA repair assays. The first two are further deletion of tRNA-ΔDT, with the bulge region between the acceptor stem and anticodon stem deleted (named tRNA-ΔDT-ΔB). The deletion of the bulge region produces a stem–loop RNA with a long and continuous stem (Fig. 3A, middle). The other two RNAs correspond to the anticodon stem–loops of tRNAAsp and tRNAArg (named tRNA-AC-SL). To increase the stability of these two RNAs and to make the in vitro repair assay feasible, two additional base pairs were added on the stem (Fig. 3A, left).
FIGURE 3.
Repair of broken stem–loop RNAs by AvaPnkp/Hen1. (A) Schematic view of three sets of broken stem–loop RNAs used for the assay. Since each substrate is reconstituted by mixing and annealing separately prepared 5′-half and 3′-half RNAs, its breaking point is marked with an arrow and labeled as Asp (for nucleotide sequence matching tRNAAsp) or Arg (for nucleotide sequence matching tRNAArg). (B) Analysis of phosphorylation and repair reactions by DPAGE. Pnkp-N indicates the N-terminal half of Pnkp consisting of the kinase and phosphatase domains. SM indicates RNA size marker.
Because colicin D did not cleave tRNAArg-ΔDT-ΔB and tRNAArg-AC-SL, we had to reconstitute these four RNA substrates differently. Specifically, we first prepared the 5′ half and the 3′ half of each RNA substrate individually via chemical synthesis (tRNA-AC-SL) or in vitro transcription (tRNA-ΔDT-ΔB), followed by mixing and annealing the two halves. Thus, these RNA substrates do not have the 2′,3′-cyclic phosphate at the 3′ terminus of the 5′ half. To ensure that the missing 2′,3′-cyclic phosphate does not significantly affect the outcome of RNA repair, we used the same approach to prepare tRNA-ΔDT substrates as positive controls (Fig. 3A, right).
Each of the six RNA substrates was incubated with either Pnkp-N (containing only the kinase and phosphatase domains of Pnkp) or Pnkp/Hen1 in the presence of 33P-γ-ATP, and the resulting products were analyzed by a denaturing gel (Fig. 3B). The broken tRNAAsp-ΔDT and tRNAArg-ΔDT were efficiently repaired (Fig. 3B, lanes 10,12, cf. with lanes 9,11, respectively), and their repair yields were comparable to the ones observed previously (Fig. 2), indicating that the lack of the 2′,3′-cyclic phosphate does not significantly affect the outcome of RNA repair. Deleting the bulge in tRNA-ΔDT has a significant effect on RNA repair. Although a repaired product was produced with the broken tRNAArg-ΔDT-ΔB as the substrate (Fig. 3B, lane 8 marked with an *), its size (∼29 nt) did not match the expected size of 35 nt that would have resulted from the ligation of the 5′ half to the 3′ half. Therefore, the identity of the repaired product is unknown. It is possible that, as in the case of tRNAAsp-ΔDT (Fig. 2A), continuous 4Gs in tRNAArg-ΔDT-ΔB-3′ (Table 1) could be the culprit of this repaired product. This uncertainty, however, does not affect our general conclusion that the deletion of the bulge region results in an RNA substrate with significantly reduced repair efficiency by AvaPnkp/Hen1 (Fig. 3B, cf. lanes 8 and 12). Furthermore, deletion of the bulge region in tRNAAsp-ΔDT resulted in an RNA substrate that was not repaired at all by AvaPnkp/Hen1 (Fig. 3B, lane 6). Further shortening the stem of tRNAArg-ΔDT-ΔB produced an unrepaired RNA substrate (Fig. 3B, lane 4), and like tRNAAsp-ΔDT-ΔB, tRNAAsp-AC-SL was not a substrate of AvaPnkp/Hen1 (Fig. 3B, lane 2).
AvaPnkp/Hen1 does not efficiently repair nicked and bulged double-stranded RNAs
The single-stranded regions of RNAs are the most susceptible to RNA damage in vivo. Indeed, all ribotoxins discovered to date target single-stranded RNAs (ssRNAs) for cleavage. In addition to pure ssRNAs and loops of the stem–loop RNAs, bulges that interrupt continuation of double-stranded RNAs are also frequently present as ssRNAs in living cells. Therefore, we decided to carry out an investigation to see if AvaPnkp/Hen1 was able to repair RNAs damaged in the bulge region. Furthermore, we were also interested in knowing if AvaPnkp/Hen1 could repair a nicked double-stranded RNA.
Our strategy for substrate preparation of the study described in this section was to utilize two ssRNAs available in our laboratory (named Lead and Tail RNAs, respectively) as the targets of repair. These two ssRNAs were brought together by base-pairing with a complementary strand (named Bridge). By varying the nucleotide sequence of Bridge RNA, four RNA repair substrates differing in their secondary structures were produced (Fig. 4A–D). To simplify interpretation of repair results, only Tail RNA was radiolabeled (Fig. 4, marked with a *). These four RNA substrates, together with other various combinations of Tail, Lead, and Bridge RNAs (used as negative controls), were incubated with AvaPnkp/Hen1 for RNA repair and the results were surprising (Fig. 4E).
FIGURE 4.
Repair of broken double-stranded RNAs by AvaPnkp/Hen1. (A–D) Schematic depiction of different secondary structures of RNA repair substrates with single RNA break in the regions of double-stranded RNA (Nick) and bulges (Bulge, Double Bulge1, and Double Bulge2). Only Tail RNA, which is depicted with white cycles, is radiolabeled (with *). Lead RNA, the potential repair partner of Tail, is colored black, and the complementary strand bridging Tail and Lead RNAs (Bridge1 to Bridge4) is in gray. (E) Analysis of RNA repair products by DPAGE. B1 to B4 indicate Bridge1 to Bridge4.
First, only a small amount of the repaired products was produced with the nicked and bulged RNA substrates (Fig. 4E, lanes 8–11), indicating that, unlike the cleaved stem–loop RNAs, RNAs damaged within the double-stranded region (nicked RNA) (Fig. 4A) or in the bulge region of the double-stranded RNA cannot be effectively repaired by AvaPnkp/Hen1.
Efficient RNA repair was observed with some RNA substrates originally designed as negative controls. Specifically, in the absence of Lead RNA, Tail RNA was efficiently ligated to Bridge3 and Bridge4 RNAs (Fig. 4E, lanes 6,7), but not Bridge1 and Bridge2 RNAs (Fig. 4E, lanes 4,5). Without Lead RNA, Tail RNA and Bridge3 or Bridge4 RNAs form a double-stranded RNA with a 4-nt single-stranded 5′ overhang of Tail RNA, and a 27-nt or 31-nt single-stranded 3′ overhang of Bridge3 or Bridge4 (Supplemental Fig. S1C,D). Efficient RNA repair of Tail/Bridge3 and Tail/Bridge4 combinations indicates ligation of the 5′ overhang of Tail RNA to the 3′ overhang of the Bridge3 or Bridge4 RNAs to form a 31-nt or 35-nt size loop. The inability of Tail RNA to be ligated to Bridge1 (Fig. 4E, lane 4) can be explained by the lack of the flexible single-stranded 5′ overhang of Tail RNA in the Tail/Bridge1 combination (Supplemental Fig. S1A). On the other hand, the negative result of RNA repair in the Tail/Bridge2 combination was initially puzzling, as it should have produced a result similar to the Tail/Bridge3 combination because they appeared to have similar single-stranded 5′ and 3′ overhangs (Fig. 4B,C). Further inspection of the nucleotide sequences of these two Bridge RNAs explains why. In the absence of Lead RNA, three of the four nucleotides in Bridge2 originally designed to base-pair with the 3′ end of Lead RNA are now able to base-pair with the single-stranded 5′ overhang of Tail RNA (Supplemental Fig. S1B). In essence, the Tail/Bridge2 combination has a secondary structure similar to the one of the Tail/Bridge1 combination (Supplemental Fig. S1B, cf. with Supplemental Fig. S1A). It is therefore only logical that RNA repair of these two combinations by AvaPnkp/Hen1 produced similar results (Fig. 4E, lanes 4,5).
Another observation was the efficient ligation of Tail RNA to Lead RNA in the absence of any Bridge RNAs (Fig. 4E, lane 3). Again, inspection of the nucleotide sequences of these two RNAs provides rationale as to why these two RNAs were efficiently ligated by AvaPnkp/Hen1. Within a region of 11 nt near the 3′ end of Lead RNA and the 5′ end of Tail RNA, these two RNAs are able to form a duplex of a total of 10 bp, and eight of them are continuous (Supplemental Fig. S2A). Base-pairing between Lead RNA and Tail RNA produces an RNA substrate with a 12-nt single-stranded 5′ overhang of Lead RNA and a 19-nt single-stranded 3′ overhang of Tail RNA. Presumably, these two ends were ligated by AvaPnkp/Hen1 during RNA repair, producing a stem–loop RNA with a 31-nt size loop.
In addition to the efficient RNA repair of the Tail/Lead, Tail/Bridge3, and Tail/Bridg4 combinations, a small amount of self-ligated product of Tail RNA was also observed in the absence of other RNAs (Fig. 4E, lane 2). A possible explanation for this inefficient self-ligation is the potential formation of four GC pairs via 5′ ends of two Tail RNAs.
AvaPnkp/Hen1 does not ligate two unrelated ssRNAs
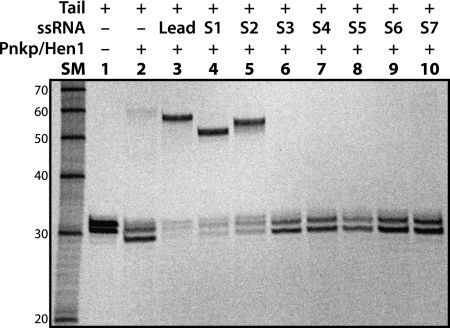
Due to the unexpected result of RNA repair of the Tail/Lead combination (Fig. 4E, lane 3), we decided to further investigate potential RNA ligation of Tail RNA with other ssRNAs available in our laboratory. We have chosen seven ssRNAs (Table 1, S1 to S7) for the study. Based on their nucleotide sequences, these seven ssRNAs can be divided into three groups. S1 and S2 RNAs belong to the first group, both of which are related to Lead RNA due to their high nucleotide sequence identities (Supplemental Fig. S2B). Indeed, like Lead RNA, S1 and S2 RNAs were efficiently ligated to Tail RNA by AvaPnkp/Hen1 (Fig. 5, lanes 4,5, cf. lane 3). The positions of the ligation products in the gel are consistent with the expected ligation sizes of Lead (27 nt), S1 (21 nt), and S2 (24 nt) RNAs to Tail RNA (30 nt).
FIGURE 5.
Repair of RNA substrates consisting of two single-stranded RNAs (ssRNAs) by AvaPnkp/Hen1. As in the case of Figure 4, only Tail RNA is radiolabeled.
S3 is the only RNA that belongs to the second group. S3 is also related to Lead RNA. But instead of sharing the same nucleotide sequences with Lead RNA as S1 and S2, S3 is complementary to Lead RNA (Supplemental Fig. S2C). RNA ligation of S3 to Tail RNA was not observed (Fig. 5, lane 6). Therefore, the positive results from S1 and S2, and the negative result from S3, provide further support for the explanation that efficient ligation of Tail RNA to Lead RNA is due to formation of a secondary structure as depicted in Supplemental Figure S2A.
RNAs in the third group are S4, S5, S6, and S7 (ranging in size from 17–35 nt) (Table 1). Based on their nucleotide sequences, these RNAs are not related to Lead RNA or to Tail RNA. None of them were able to ligate to Tail RNA (Fig. 5, lanes 7–10). Therefore, we conclude that if two ssRNAs do not physically interact with each other via base-pairing to bring them together, their ligation by AvaPnkp/Hen1 will not occur.
It is interesting to notice that whereas a small amount of self-ligated product was produced with Tail RNA alone (Fig. 5, lane 2), such a ligation product was not observed with the addition of S3–S7 (Fig. 5, lanes 6–10). Presumably, the addition of excess ssRNAs S3–S7 sequesters a significant amount of enzyme, making inefficient self-ligation of Tail RNA unlikely to occur.
DISCUSSION
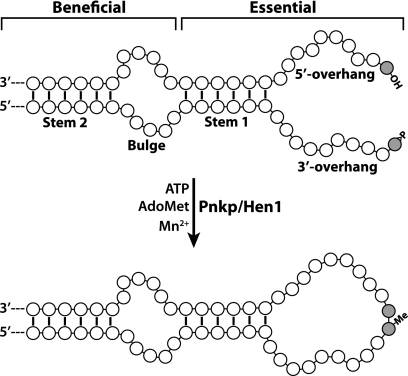
Based on results of RNA repair assays presented in this study, we summarize an effective RNA substrate for repair by AvaPnkp/Hen1 as schematically depicted in Figure 6.
FIGURE 6.
Schematic depiction of an effective RNA substrate of AvaPnkp/Hen1 and its repaired product based on experimental data presented in this study. The two terminal nucleotides to be ligated by AvaPnkp/Hen1 are highlighted in gray, with the depicted 5′-OH and 2′,3′-cyclic phosphate groups that are directly involved in repair chemistry. ATP is required for phosphorylation of the 5′-OH group, and AdoMet is the methyl donor for 2′-O-methylation on the 3′-terminal nucleotide during repair, which requires a manganese ion (Mn2+) to carry out the reaction. The numbers of nucleotides for the different regions of RNA (5′ overhang, 3′ overhang, Stem 1, Bulge, and Stem 2) were chosen arbitrarily. Secondary structures of the Bulge, the 5′ overhang, the 3′ overhang, and the resulting repaired loop were also arbitrary depicted.
To be an effective RNA repair substrate of AvaPnkp/Hen1, two elements are essential. The first element is the two single-stranded ends from the original RNA cleavage, which are the repair targets and are depicted here as 5′ and 3′ overhangs (Fig. 6). The second element is a means to bring these two ends together to facilitate repair by AvaPnkp/Hen1. Our data indicate that the lengths of both 5′ and 3′ overhangs are quite flexible, and a stable stem is the most effective method to bring these two overhangs together for efficient repair.
For the 5′ overhang, we have observed efficient repair of RNA substrates with its length from 4–12 nt (Fig. 4E, lanes 3,6,7; Supplemental Figs. S1C,D, S2A). It is likely that RNA substrates with the 5′ overhang >12 nt can also be efficiently repaired by AvaPnkp/Hen1, but it requires future experiments for confirmation. The shortest length of 5′ overhang allowed for effective repair is in fact zero, demonstrated by the effective repair of the cleaved tRNAArg and its truncated derivatives (Figs. 1, 2). However, tRNAArg and its deletion derivatives are likely to be an exception as similar configurations in the Tail/Bridge1 and Tail/Bridge2 combinations did not work well for repair (Fig. 4E, lanes 4,5; Supplemental Fig. S1A,B). It is possible that, after cleavage by colicin D, the 5′-terminal nucleotide (nucleotide 39) in tRNAArg and its deletion derivatives might no longer base-pair with its partner (nucleotide 31) due to limited stability of the anticodon stem (5 bp). This would provide the 5′ end some flexibility for the repair reactions (phosphorylation and ligation) to be carried out. This hypothesis is consistent with the observations that the cleaved tRNAArg deletion mutant is repaired more efficiently than the full-length tRNAArg (Fig. 2B,D). Deleting the D or/and TΨC stem–loop in tRNAArg would likely produce less stable RNAs, which might be better substrates of AvaPnkp/Hen1 due to more potential flexibility of the 5′-terminal nucleotide. Conversely, because both Tail/Bridge1 and Tail/Bridge2 have long stems (Supplemental Fig. S1A,B), the flexibility of the 5′-terminal nucleotide amenable for RNA repair seen in tRNAArg and its deletion derivatives might not be present in the Tail/Bridge1 and Tail/Bridge2 combinations, resulting in inefficient repair of Tail/Bridge1 and Tail/Bridge2 (Fig. 4E, lanes 4,5).
For the 3′ overhang, RNA substrates with its length ranging from 3 nt (tRNAAsp and its derivative) (Fig. 2A,C) to 31 nt (Tail/Bridge4 combination) (Fig. 4E, lane 7) were seen to be efficiently repaired by AvaPnkp/Hen1. It is possible that the 3′ overhang could even be <3 nt, but we have not tested this possibility.
We propose that a stable stem, as depicted Stem 1 in Figure 6, is the best means to bring two overhangs together for efficient repair by AvaPnkp/Hen1 based on the following observations. First, AvaPnkp/Hen1 is unable to repair an RNA substrate that does not form a secondary structure to bring the two ends together. This is supported by the negative results of ligating Tail RNA to other unrelated ssRNAs (Fig. 5, lanes 6–10). Second, employing two double-stranded RNAs to bring the two ends together for repair did not work well, as AvaPnkp/Hen1 was unable to repair the damaged bulged RNAs (Fig. 4E, lanes 8–11). Third, of the all the RNA substrates we have tested for repair so far, RNAs that were efficiently repaired by AvaPnkp/Hen1 are always having secondary structures of some variations of the cleaved stem–loop RNAs (tRNAAsp and its deletion derivatives, tRNAArg and its deletion derivatives, Tail/Lead combination, Tail/Bridge3 combination, and Tail/Bridge4 combination). The discrimination of the cleaved stem–loop RNAs vs. the cleaved bulged RNAs for repair by AvaPnkp/Hen1 might reflect the structure of RNA-binding groove/pocket in the Pnkp/Hen1 complex. It is possible that the RNA binding groove/pocket of Pnkp/Hen1 is not suitable for binding of a damaged bulged RNA, which requires accommodation of two double-stranded RNAs on both sides of the cleaved bulge RNA. A crystal structure of the Pnkp/Hen1 complex should confirm or refute this possibility.
In addition to the two essential elements described above, other RNA elements beyond the required stem might be beneficial, and in some cases are essential, for efficient RNA repair (Fig. 6). While tRNA-ΔDT was efficiently repaired, deletion of the bulge region in tRNA-ΔDT produced RNAs that were not good substrates of AvaPnkp/Hen1 (Fig. 3). We postulate that some single-stranded elements in RNA substrates are needed to provide sufficient interaction with Pnkp/Hen1 for efficient repair. Cleaved tRNA-ΔDT has the bulge region to provide such elements. Although Tail/Bridge3, Tail/Bridge4, and Tail/Lead combinations do not have bulges (Supplemental Figs. S1C,D, S2A), they have long single-stranded overhangs as the single-stranded elements. On the other hand, cleaved tRNA-ΔDT-ΔB and tRNA-AC-SL do not have bulges, and their 5′ and 3′ overhangs are too short to be the providers of the single-stranded elements. Again, a crystal structure of Pnkp/Hen1, which we are actively working on, should provide insight into whether interaction with the single-stranded region of an RNA substrate by Pnkp/Hen1 is required for efficient RNA repair.
As we have discussed previously, the biological roles of Pnkp/Hen1 in vivo are unknown. Nevertheless, we could speculate about the possible situations within living cells where RNA repair is required. If RNA damage in bacteria carrying Pnkp/Hen1 is induced by exogenous agents, we could then imagine two possible scenarios in which a bacterium requires RNA repair. The first is in an emergency, such as when a bacterium is invaded by a ribotoxin. A ribotoxin could cleave an essential RNA so rapidly that new rounds of transcription might not be fast enough to save the cell. This scenario requires only a short-term solution, and bacterial Pnkp/Hen1 could play such a role. Evidence supporting such a possibility is shown by more efficient repair of the cleaved full-length tRNAAsp compared with the cleaved tRNAAsp deletion derivatives (Fig. 2A,C). However, the difference in efficiency of repairing the cleaved full-length tRNAAsp vs. the deletion derivative appears to be much smaller in the bacterial Pnkp/Hen1 system than the T4Pnkp/Rnl1 system (Nandakumar et al. 2008). This observation, together with the more efficient RNA repair of the tRNAArg deletion mutant vs. the full-length one, is consistent with the Pnkp/Hen1 employed for RNA repair in a second scenario described below.
In a second scenario, repair of damaged RNAs might be employed as a long-term strategy to “improve the quality of life” of bacteria that suffer constant RNA damage. In such a case, the damaged RNAs need to be repaired in such a way that the RNAs after repair could no longer be cleaved again. Otherwise, the cycle of RNA damage followed by RNA repair would keep repeating. We have previously demonstrated that, because of the 2′-O methylation at the junction of RNA repair, repaired RNA can no longer be cleaved again at the site of repair (Chan et al. 2009b). The data presented in this study indicate that Pnkp/Hen1 is able to repair a variety of damaged RNAs with high efficiency. Therefore, the mechanism of RNA repair carried out by Pnkp/Hen1 and the broad RNA substrate specificity of AvaPnkp/Hen1 described in this study are consistent with the second scenario. From a logical point of view, how an RNA is damaged is not the concern of the Pnkp/Hen1 RNA repair system. The presence of a significant amount of a particular RNA cleaved at a specific site is sufficient to indicate the vulnerability of the site. One round of repair of the cleaved RNA by Pnkp/Hen1 with the 2′-O-methylation at the repair junction helps the cell solve the RNA damage problem at that particular site.
We believe that the general conclusions based on our in vitro RNA repair study of Pnkp/Hen1 from A. variabilis are applicable for Pnkp/Hen1 from other bacterial species. However, we cannot rule out the possibility that, during the long history of bacterial evolution, Pnkp/Hen1 in different bacterial species have maintained the same chemistry for RNA repair (5′-phosphorylation, 3′-dephosphorylation, 5′,3′-ligation, and 2′-O-methylation) but have evolved different RNA substrate specificity due to the requirement of repairing specific RNA damages caused by RNA-damaging agents present in their local living environments. Future studies similar to the one presented here, but using Pnkp/Hen1 from other organisms, should address this issue. In addition, we have recently found a second subfamily of the bacterial Pnkp/Hen1 RNA repair system that is significantly different from the first subfamily in the amino acid sequences of both Pnkp and Hen1 (data not shown). Therefore, it is possible that Pnkp/Hen1 from the second subfamily might have a different RNA substrate specificity from those in the first subfamily, of which AvaPnkp/Hen1 is a member.
MATERIALS AND METHODS
Recombinant proteins
AvaPnkp/Hen1 and AvaPnkp-N were overexpressed in E. coli BL21(DE3) and purified from soluble bacterial lysates as described previously (Chan et al. 2009b).
Preparation of full-length and cleaved RNAs
All the RNAs listed in Table 1 were in vitro transcribed using T7 polymerase with the exception of tRNAAsp-AC-SL-5′, tRNAAsp-AC-SL-3′, tRNAArg-AC-SL-5′, tRNAArg-AC-SL-3′, Lead, S1, S2, S3, S4, and S5 RNAs, which were purchased from IDT. Both in vitro transcribed and purchased RNAs were purified by DPAGE. In vitro transcription and cleavage of tRNAs by colicin E5 and colicin D were carried out as described previously (Chan et al. 2009b).
RNA repair assays with tRNAs and its deletion derivatives RNAs
The RNA repair reaction was carried out in a reaction mixture containing 25 mM Tris-HCl (pH 8.0), 50 mM KCl, 2.5 mM MgCl2, 0.05 mM EDTA, 2.5% glycerol, 5 mM DTT, 0.2 mM ATP, 0.5 mM MnCl2, 0.05 mM S-AdoMet, 1.0 μM internally 33P-radiolabeled cleaved tRNA or its deletion derivatives, and 0.5 μM of AvaPnkp/Hen1 heterotetramer. The reaction mixture was incubated for 0, 5, 15, 30, and 60 min at 37°C. The reactions were quenched by adding DPAGE loading buffer. The samples were heated for 3 min at 95°C and analyzed by 15% DPAGE. The radioactivity of the repaired and unrepaired tRNAs was quantified using a PhosphorImager system (Molecular Dynamics).
RNA repair assays with stem–loop RNAs
The RNA repair reaction was carried out in a reaction mixture containing 25 mM Tris-HCl (pH 8.0), 50 mM KCl, 2.5 mM MgCl2, 0.05 mM EDTA, 2.5% glycerol, 5 mM DTT, 0.5 mM MnCl2, 0.05 mM S-AdoMet, 1 μM of an RNA substrate (tRNA-AC-SL, tRNA-ΔDT-ΔB, or tRNA-ΔDT), and 0.5 μM of AvaPnkp/Hen1 or AvaPnkp-N in the presence of 0.2 mM ATP/33P-γ-ATP mixture. The reaction mixture was incubated for 40 min at 37°C. The samples were heated for 3 min at 95°C and analyzed by 20% DPAGE. The radioactivity of the repaired and unrepaired tRNAs was quantified using a PhosphorImager system (Molecular Dynamics).
RNA repair assays with the nicked and broken bulged double-stranded RNAs and with ssRNAs
The RNA repair reaction was carried out in a reaction mixture containing 25 mM Tris-HCl (pH 8.0), 50 mM KCl, 2.5 mM MgCl2, 0.05 mM EDTA, 2.5% glycerol, 5 mM DTT, 0.2 mM ATP, 0.5 mM MnCl2, 0.05 mM S-AdoMet, 1 μM of an RNA substrate with only Tail RNA internally radiolabeled with 33P-α-ATP, and 0.5 μM of AvaPnkp/Hen1 heterotetramer. The reaction mixture was incubated for 40 min at 37°C. The reactions were quenched by adding DPAGE loading buffer. The samples were heated for 3 min at 95°C and analyzed by 15% DPAGE. The radioactivity of the repaired and unrepaired tRNAs was quantified using a PhosphorImager system (Molecular Dynamics).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank K. Selvadurai for critical reading of the manuscript. This research was supported by NSF grant MCB-0920966.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.030502.111.
REFERENCES
- Amitsur M, Levitz R, Kaufmann G 1987. Bacteriophage T4 anticodon nuclease, polynucleotide kinase and RNA ligase reprocess the host lysine tRNA. EMBO J 6: 2499–2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman CM, Dahlberg JE, Ikemura T, Konisky J, Nomura M 1971. Specific inactivation of 16S ribosomal RNA induced by colicin E3 in vivo. Proc Natl Acad Sci 68: 964–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C, Zhou C, Brunzelle JS, Huang RH 2009a. Structural and biochemical insights into 2′-O-methylation at the 3′-terminal nucleotide of RNA by Hen1. Proc Natl Acad Sci 106: 17699–17704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CM, Zhou C, Huang RH 2009b. Reconstituting bacterial RNA repair and modification in vitro. Science 326: 247. [DOI] [PubMed] [Google Scholar]
- El Omari K, Ren J, Bird LE, Bona MK, Klarmann G, LeGrice SF, Stammers DK 2006. Molecular architecture and ligand recognition determinants for T4 RNA ligase. J Biol Chem 281: 1573–1579 [DOI] [PubMed] [Google Scholar]
- Endo Y, Wool IG 1982. The site of action of α-sarcin on eukaryotic ribosomes. The sequence at the α-sarcin cleavage site in 28 S ribosomal ribonucleic acid. J Biol Chem 257: 9054–9060 [PubMed] [Google Scholar]
- Fando JL, Alaba I, Escarmis C, Fernandez-Luna JL, Mendez E, Salinas M 1985. The mode of action of restrictocin and mitogillin on eukaryotic ribosomes. Inhibition of brain protein synthesis, cleavage and sequence of the ribosomal RNA fragment. Eur J Biochem 149: 29–34 [DOI] [PubMed] [Google Scholar]
- Galburt EA, Pelletier J, Wilson G, Stoddard BL 2002. Structure of a tRNA repair enzyme and molecular biology workhorse: T4 polynucleotide kinase. Structure 10: 1249–1260 [DOI] [PubMed] [Google Scholar]
- Ho CK, Shuman S 2002. Bacteriophage T4 RNA ligase 2 (gp24.1) exemplifies a family of RNA ligases found in all phylogenetic domains. Proc Natl Acad Sci 99: 12709–12714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R, Shuman S 2010. Bacterial Hen1 is a 3′ terminal RNA ribose 2′-O-methyltransferase component of a bacterial RNA repair cassette. RNA 16: 316–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R, Shuman S 2011. Active site mapping and substrate specificity of bacterial Hen1, a manganese-dependent 3′ terminal RNA ribose 2′O-methyltransferase. RNA 17: 429–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keppetipola N, Nandakumar J, Shuman S 2007. Reprogramming the tRNA-splicing activity of a bacterial RNA repair enzyme. Nucleic Acids Res 35: 3624–3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SR, Collins K 2005. Starvation-induced cleavage of the tRNA anticodon loop in Tetrahymena thermophila. J Biol Chem 280: 42744–42749 [DOI] [PubMed] [Google Scholar]
- Lu J, Huang B, Esberg A, Johansson MJ, Bystrom AS 2005. The Kluyveromyces lactis γ-toxin targets tRNA anticodons. RNA 11: 1648–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins A, Shuman S 2004a. An RNA ligase from Deinococcus radiodurans. J Biol Chem 279: 50654–50661 [DOI] [PubMed] [Google Scholar]
- Martins A, Shuman S 2004b. Characterization of a baculovirus enzyme with RNA ligase, polynucleotide 5′-kinase, and polynucleotide 3′-phosphatase activities. J Biol Chem 279: 18220–18231 [DOI] [PubMed] [Google Scholar]
- Martins A, Shuman S 2005. An end-healing enzyme from Clostridium thermocellum with 5′ kinase, 2′,3′ phosphatase, and adenylyltransferase activities. RNA 11: 1271–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandakumar J, Schwer B, Schaffrath R, Shuman S 2008. RNA repair: an antidote to cytotoxic eukaryal RNA damage. Mol Cell 31: 278–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubauer C, Gao YG, Andersen KR, Dunham CM, Kelley AC, Hentschel J, Gerdes K, Ramakrishnan V, Brodersen DE 2009. The structural basis for mRNA recognition and cleavage by the ribosome-dependent endonuclease RelE. Cell 139: 1084–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T, Tomita K, Ueda T, Watanabe K, Uozumi T, Masaki H 1999. A cytotoxic ribonuclease targeting specific transfer RNA anticodons. Science 283: 2097–2100 [DOI] [PubMed] [Google Scholar]
- Pedersen K, Zavialov AV, Pavlov MY, Elf J, Gerdes K, Ehrenberg M 2003. The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell 112: 131–140 [DOI] [PubMed] [Google Scholar]
- Simpson L, Sbicego S, Aphasizhev R 2003. Uridine insertion/deletion RNA editing in trypanosome mitochondria: a complex business. RNA 9: 265–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DM, Lu C, Green PJ, Parker R 2008. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA 14: 2095–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita K, Ogawa T, Uozumi T, Watanabe K, Masaki H 2000. A cytotoxic ribonuclease which specifically cleaves four isoaccepting arginine tRNAs at their anticodon loops. Proc Natl Acad Sci 97: 8278–8283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winther KS, Gerdes K 2011. Enteric virulence associated protein VapC inhibits translation by cleavage of initiator tRNA. Proc Natl Acad Sci 108: 7403–7407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, He WJ, Liu WY 2004. A novel ribotoxin with ribonuclease activity that specifically cleaves a single phosphodiester bond in rat 28S ribosomal RNA and inactivates ribosome. Arch Biochem Biophys 427: 30–40 [DOI] [PubMed] [Google Scholar]
- Yamasaki S, Ivanov P, Hu GF, Anderson P 2009. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol 185: 35–42 [DOI] [PMC free article] [PubMed] [Google Scholar]