Abstract
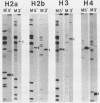
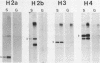
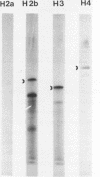
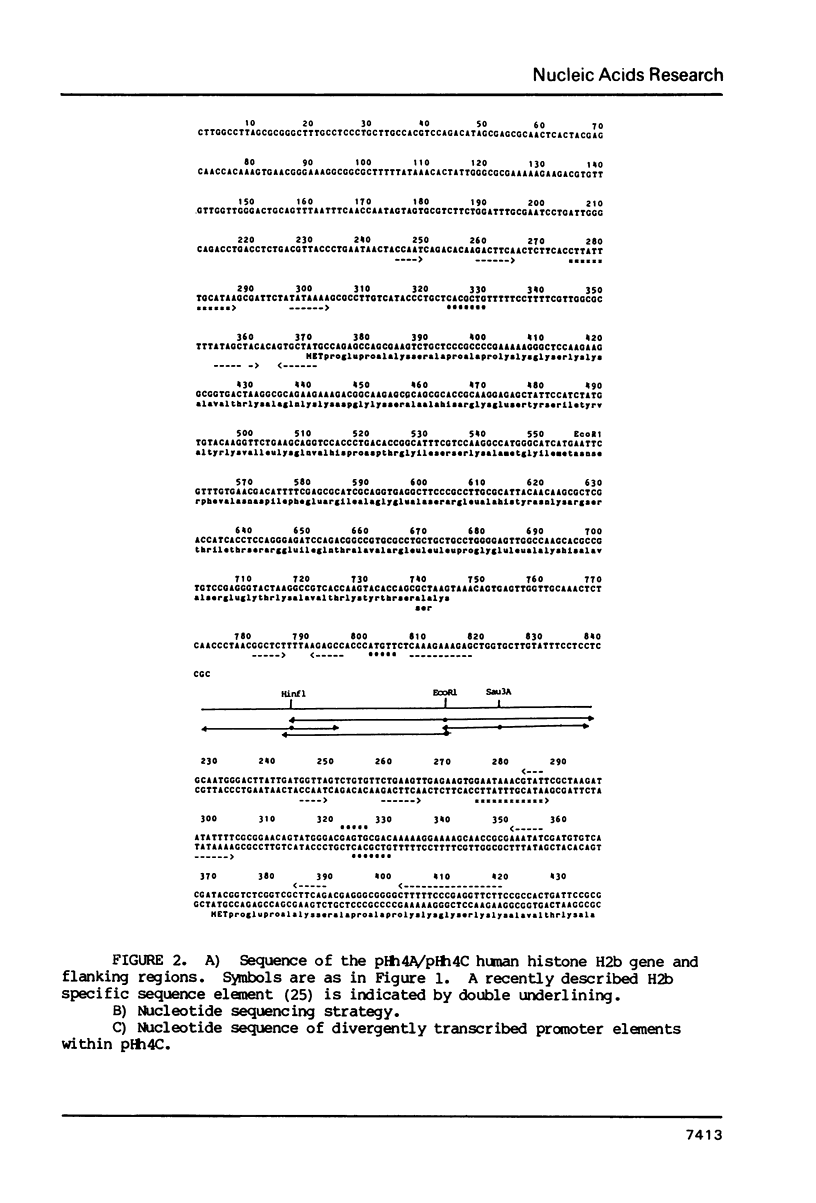
The complete nucleotide sequence of four human histone genes has been determined. Each gene codes for a core histone protein which is very homologous with the corresponding calf thymus of rat histones. The 5' and 3' flanking regions of the human histone genes contain previously identified concensus sequences: the TATA box, the GACTTC element; the CCAAT sequence; the 3' terminal dyad symmetry element thought to be involved in transcription termination; and a recently identified H2b specific upstream sequence. A putative H2a specific upstream sequence 5'-TTCTTGGACTCCTCTTTTC-3' is present approximately 40 base pairs upstream from the TATA box in the human H2a gene promoter. Nuclease S1 analysis of the human histone mRNAs encoded within each of these clones demonstrates that the mRNA terminii map to the expected positions relative to the known concensus sequences, and that the abundance of each mRNA is regulated during the HeLa cell cycle. Finally, in contrast to the H2b, H3 and H4 mRNAs encoded within clones pHh 4A/pHh4C, pHh5B and pHu4A, respectively, the H2a mRNA encoded by Hh5G is not present in human placental RNA.
Full text
PDF















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bendig M. M., Hentschel C. C. Transcription of sea urchin histone genes in HeLa cells. Nucleic Acids Res. 1983 Apr 25;11(8):2337–2346. doi: 10.1093/nar/11.8.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Spliced early mRNAs of simian virus 40. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1274–1278. doi: 10.1073/pnas.75.3.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boime I., Boothby M., Hoshina M., Daniels-McQueen S., Darnell R. Expression and structures of human placental hormone genes as a function of placental development. Biol Reprod. 1982 Feb;26(1):73–91. doi: 10.1095/biolreprod26.1.73. [DOI] [PubMed] [Google Scholar]
- Corden J., Wasylyk B., Buchwalder A., Sassone-Corsi P., Kedinger C., Chambon P. Promoter sequences of eukaryotic protein-coding genes. Science. 1980 Sep 19;209(4463):1406–1414. doi: 10.1126/science.6251548. [DOI] [PubMed] [Google Scholar]
- Efstratiadis A., Posakony J. W., Maniatis T., Lawn R. M., O'Connell C., Spritz R. A., DeRiel J. K., Forget B. G., Weissman S. M., Slightom J. L. The structure and evolution of the human beta-globin gene family. Cell. 1980 Oct;21(3):653–668. doi: 10.1016/0092-8674(80)90429-8. [DOI] [PubMed] [Google Scholar]
- Engel J. D., Dodgson J. B. Histone genes are clustered but not tandemly repeated in the chicken genome. Proc Natl Acad Sci U S A. 1981 May;78(5):2856–2860. doi: 10.1073/pnas.78.5.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin S. G., Zweidler A. Non-allelic variants of histones 2a, 2b and 3 in mammals. Nature. 1977 Mar 17;266(5599):273–275. doi: 10.1038/266273a0. [DOI] [PubMed] [Google Scholar]
- Harvey R. P., Robins A. J., Wells J. R. Independently evolving chicken histone H2B genes: identification of a ubiquitous H2B-specific 5' element. Nucleic Acids Res. 1982 Dec 11;10(23):7851–7863. doi: 10.1093/nar/10.23.7851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintz N., Zernik M., Roeder R. G. The structure of the human histone genes: clustered but not tandemly repeated. Cell. 1981 Jun;24(3):661–668. doi: 10.1016/0092-8674(81)90092-1. [DOI] [PubMed] [Google Scholar]
- Hentschel C. C., Birnstiel M. L. The organization and expression of histone gene families. Cell. 1981 Aug;25(2):301–313. doi: 10.1016/0092-8674(81)90048-9. [DOI] [PubMed] [Google Scholar]
- Hentschel C., Irminger J. C., Bucher P., Birnstiel M. L. Sea urchin histone mRNA termini are located in gene regions downstream from putative regulatory sequences. Nature. 1980 May 15;285(5761):147–151. doi: 10.1038/285147a0. [DOI] [PubMed] [Google Scholar]
- Isenberg I. Histones. Annu Rev Biochem. 1979;48:159–191. doi: 10.1146/annurev.bi.48.070179.001111. [DOI] [PubMed] [Google Scholar]
- Kozak M. Mechanism of mRNA recognition by eukaryotic ribosomes during initiation of protein synthesis. Curr Top Microbiol Immunol. 1981;93:81–123. doi: 10.1007/978-3-642-68123-3_5. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Osley M. A., Hereford L. M. Yeast histone genes show dosage compensation. Cell. 1981 May;24(2):377–384. doi: 10.1016/0092-8674(81)90327-5. [DOI] [PubMed] [Google Scholar]
- Seiler-Tuyns A., Birnstiel M. L. Structure and expression in L-cells of a cloned H4 histone gene of the mouse. J Mol Biol. 1981 Oct 5;151(4):607–625. doi: 10.1016/0022-2836(81)90426-5. [DOI] [PubMed] [Google Scholar]
- Sierra F., Leza A., Marashi F., Plumb M., Rickles R., Van Dyke T., Clark S., Wells J., Stein G. S., Stein J. L. Human histone genes are interspersed with members of the Alu family and with other transcribed sequences. Biochem Biophys Res Commun. 1982 Jan 29;104(2):785–792. doi: 10.1016/0006-291x(82)90706-9. [DOI] [PubMed] [Google Scholar]
- Sittman D. B., Chiu I. M., Pan C. J., Cohn R. H., Kedes L. H., Marzluff W. F. Isolation of two clusters of mouse histone genes. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4078–4082. doi: 10.1073/pnas.78.7.4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sures I., Lowry J., Kedes L. H. The DNA sequence of sea urchin (S. purpuratus) H2A, H2B and H3 histone coding and spacer regions. Cell. 1978 Nov;15(3):1033–1044. doi: 10.1016/0092-8674(78)90287-8. [DOI] [PubMed] [Google Scholar]
- Weaver R. F., Weissmann C. Mapping of RNA by a modification of the Berk-Sharp procedure: the 5' termini of 15 S beta-globin mRNA precursor and mature 10 s beta-globin mRNA have identical map coordinates. Nucleic Acids Res. 1979 Nov 10;7(5):1175–1193. doi: 10.1093/nar/7.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R. S., Tsai S., Bonner W. M. Patterns of histone variant synthesis can distinguish G0 from G1 cells. Cell. 1982 Dec;31(2 Pt 1):367–374. doi: 10.1016/0092-8674(82)90130-1. [DOI] [PubMed] [Google Scholar]
- Zernik M., Heintz N., Boime I., Roeder R. G. Xenopus laevis histone genes: variant H1 genes are present in different clusters. Cell. 1980 Dec;22(3):807–815. doi: 10.1016/0092-8674(80)90557-7. [DOI] [PubMed] [Google Scholar]
- van Dongen W., de Laaf L., Zaal R., Moorman A., Destrée O. The organization of the histone genes in the genome of Xenopus laevis. Nucleic Acids Res. 1981 May 25;9(10):2297–2311. doi: 10.1093/nar/9.10.2297. [DOI] [PMC free article] [PubMed] [Google Scholar]