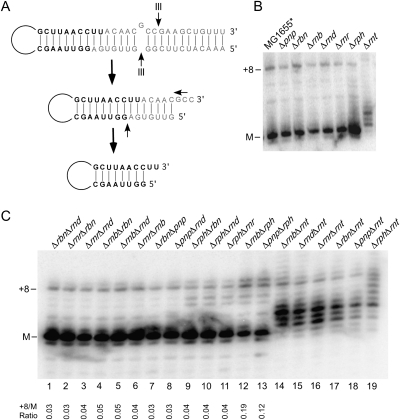
FIGURE 1.
Effect of 3′ to 5′ exonucleases on 23S rRNA 3′ end processing. (A) Schematic description of 23S rRNA processing. The precursor sequences at the 5′ and 3′ ends of 23S rRNA are complementary and form long-range base-pairs. This 23S rRNA precursor is initially cleaved by the double strand–specific RNase, RNase III. The resulting product is subsequently digested by 3′ to 5′ exonucleases to generate a mature 3′ end and by an unknown endonuclease to generate a mature 5′ end. Mature residues are indicated in black lettering; precursor residues, in gray. (B) Effect of deletions in genes encoding 3′ to 5′ exonucleases on 23S rRNA processing. RNA was isolated from a wild-type strain (MGG1655*) or from derivative strains containing Δpnp (PNPase), Δrbn (RNase BN), Δrnb (RNase II), Δrnd (RNase D), Δrnr (RNase R), Δrph (RNase PH), or Δrnt (RNase T) deletions; the encoded enzymes are indicated in brackets. The RNAs were cleaved 50 nt from the mature 3′ end of 23S rRNA using oligonucleotide-directed RNase H cleavage of 23S rRNA, followed by fractionation of the resulting product on a denaturing gel and Northern blotting using a 3′ end probe for 23S rRNA. The migration of mature RNA (M) and a prominent precursor containing eight unprocessed residues (+8) is indicated. (C) RNA was extracted from strains containing 19 different combinations of dual exonuclease gene deletions, as indicated, and analyzed by Northern blotting as described above. The +8 precursor to the mature RNA ratio is indicated below for all strains except those containing Δrnt deletions, which contain negligible levels of fully processed 23S rRNA.