Abstract
The emergence of an RNA entity capable of synthesizing peptides was a key prebiotic development. It is hypothesized that a precursor of the modern ribosomal exit tunnel was associated with this RNA entity (e.g., “protoribosome” or “bonding entity”) from the earliest time and played an essential role. Various compounds that can bind and activate amino acids, including extremely short RNA chains carrying amino acids, and possibly di- or tripeptides, would have associated with the internal cavity of the protoribosome. This cavity hosts the site for peptide bond formation and adjacent to it a relatively elongated feature that could have evolved to the modern ribosomal exit tunnel, as it is wide enough to allow passage of an oligopeptide. When two of the compounds carrying amino acids or di- or tripeptides (to which we refer, for simplicity, as small aminoacylated RNAs) were in proximity within the heart of the protoribosome, a peptide bond could form spontaneously. The growing peptide would enter the nearby cavity and would not disrupt the attachment of the substrates to the protoribosome or interfere with the subsequent attachment of additional small aminoacylated RNAs. Additionally, the presence of the peptide in the cavity would increase the lifetime of the oligopeptide in the protoribosome. Thus, subsequent addition of another amino acid would be more likely than detachment from the protoribosome, and synthesis could continue. The early ability to synthesize peptides may have resulted in an abbreviated RNA World. Key Words: Ribosome—RNA World—Prebiotic synthesis—Chirality—Ribosomal RNA. Astrobiology 12, 57–60.
It is widely accepted that the modern ribosome originated in an RNA World that predates the last universal common ancestor of life. This reflects the fact that the peptidyl transferase reaction has been shown to be catalyzed by the large ribosomal subunit RNA at a location known as the peptidyl transferase center (PTC), which is devoid of protein. A region of a semi-2-fold symmetry that has a pocket-like structure and includes the PTC and its environs has been described in detail (Bashan et al., 2003; Agmon et al., 2005). The three-dimensional structure of the RNA backbone fold of this region, and almost all of its primary sequence, are virtually unchanged in the large subunit structures seen in the archaeon Haloarcula marismortui, the eubacteria Escherichia coli, Thermus thermophilus, and Deinococcus radiodurans, as well as the mitochondria and the eukaryote Saccharomyces cerevisiae (Krupkin et al., 2011). The suggestion based on this finding is that an RNA comprising just the pocket-like symmetrical region may be capable of catalyzing peptide bond formation if provided with activated amino acids, which would make it the RNA World progenitor of the modern translation machinery.
Although missing most of the features that enhance and characterize the modern ribosome, the pocket-like symmetrical region is not devoid of all features other than the catalytic site. This early bonding entity, which may have become the symmetrical region of the modern ribosome, includes the entrance to what would later be the exit tunnel, Fig. 1. This, coupled with a recent breakthrough discovery that a four-base RNA can be readily aminoacylated by a five-residue ribozyme (Turk et al., 2011), has led us to hypothesize that an exit cavity was a key feature of the prebiotic bonding machine that led to the eventual development of the modern ribosome.
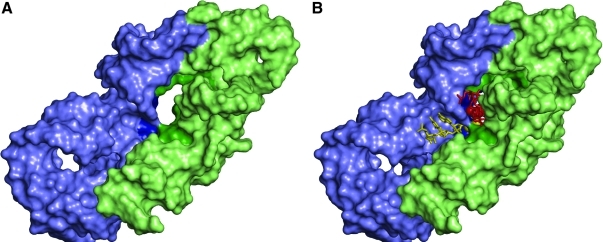
FIG. 1.
Pocket-like symmetrical region as defined by Agmon et al. (2005) showing the entry to the exit tunnel with and without aminoacylated RNAs present. Nucleotides 2058–2079, 2241–2258, 2430–2463, and 2487–2501 comprise the portion of the ribosome that accommodates the 3′ end of the P-site tRNA, while nucleotides 2502–2522 and 2543–2610 comprise the portion of the ribosome that accommodates the 3′ end of the A-site tRNA, with the former colored lime and the latter slate. The entrance of the tunnel is the darker version of the colors. The entrance nucleotides are 2061–2064, 2439, 2441–2442, and 2450–2452 from the P site and nucleotides 2505–2506 and 2585–2587 from the A site. All numbers follow the E. coli numbering convention. The coordinates were taken from structures 2DWK and 2DWL for the Thermus thermophilus ribosome and rendered using PyMOL. The apparent smaller hole near the bottom left does not exist in the modern ribosome. (A) The protoribosome is shown without aminoacylated RNAs. (B) The protoribosome with hypothetical aminoacylated RNAs attached. Aminoacylated RNAs representing the last three residues of a modern tRNA are shown in association with what are the A and P sites of the modern ribosome. The RNA portion of each substrate is rendered as sticks, while the amino acid moiety is shown as spheres. The A site substrate is colored in yellow and the P site substrate is red.
Our hypothesis is as follows. Activated amino acids (e.g., possibly bound to compounds that existed in the RNA World, such as extremely small RNAs) approach the ancient RNA bonding entity (called here the protoribosome) and associate with it. If two such substrates are present at the same time and properly juxtaposed, a peptide bond could be formed and stay on the protoribosome as an activated dipeptide. If the dipeptide dissociates from the protoribosome before the now naked RNA leaves and another activated amino acid arrives, a successive reaction will not occur. However, if the dipeptide remains for an extended period of time, then a new aminoacylated RNA may join with the result that a second reaction may occur and produce a tripeptide. The presence of the exit cavity would be the deciding factor in that the newly formed dipeptide would immediately enter the cavity adjacent to its active site. This would occur due to the favorable architecture that accommodates the components of the amide bond in an orientation that dictates entry into the cavity. Being partly in this cavity, the dipeptide may stay longer within the pocket possibly due to space limitations and stabilizing interactions with the cavity edges, which might be dependent on the type of amino acid. Alternatively, and perhaps most importantly, by placing the extended peptide deep within the core of the protoribosome, its interactions would be maintained; hence the probability of the dipeptide remaining associated within the pocket would be enhanced. As the oligopeptide chain grows, it would be positioned deeper into the cavity; hence the lifetime of its association with the protoribosome may be increased further. Although we expect that the presumed protoribosome will ultimately be shown to have catalytic ability, the argument described above is not changed if a larger portion of the modern RNA is required for its catalytic activity as long as a portion of the exit cavity is present. Likewise, no change is necessary for cases when the carriers/activators are short RNAs if one of them is carrying a di- or tripeptide (Turk et al., 2011), as the preexisting small peptide could enter the tunnel cavity prior to the first synthesis step.
Although small RNAs seem to be an appropriate carrier and activator, the protoribosome and tunnel hypothesis presented above does not actually require that the amino acid be brought to the bonding entity by RNA. This might actually be accomplished by an alternative carrier such as ribose or phosphoribose, though the presence of the RNA bases might facilitate attachment to the protoribosome by noncovalent interactions, including Watson-Crick base pairs between the nucleoside components of activated amino acids and the protoribosome, in a fashion similar to that which occurs in the PTC of the contemporary ribosome between the C and CC bases of the tRNA 3′ ends. In addition, it should be appreciated that it is the growing peptide that is actually the entity of interest. It has to remain associated with the tunnel, regardless of whether it is still bound or already detached from its carrier/activator, which may or may not be RNA. Finally, it should be noted that the hypothesis is not based on some ultimate biological advantage to RNA-catalyzed peptide synthesis. Rather, what is proposed is simply a competition between two states where the one that is longer-lived is more likely to dominate (Lukes et al., 2011). Entities containing such a primitive protein synthesizing systems would be properly regarded as progenotes (Woese and Fox, 1977).
The importance of the exit tunnel in modern ribosomes has not gone unnoticed. It is involved in protein biosynthesis stalling that occurs in association with cellular events, and antibiotic sensitivity is caused by blockage of the tunnel (Nakatogawa and Ito, 2002; Lu and Deutsch, 2005; Yonath, 2005; Ito et al., 2010; Ramu et al., 2009, 2011; Chiba et al., 2011). In addition, the modern ribosome provides an optimized center for its polymerase function (Yonath, 2009). In particular, in the modern ribosome, the 3′ end of the A-site tRNA rotates into the P-site while the peptide bond is being formed (Gindulyte et al., 2006). The PTC is configured in such a manner that this rotary motion places the tRNA that is now carrying the nascent peptide into a configuration that is appropriate for the next biosynthetic step. In conjunction with this rotation, the nascent peptide is directed into the exit tunnel (Bashan et al., 2003; Yonath, 2003, 2009; Zarivach et al., 2004; Agmon et al., 2005). In the absence of the tunnel, for example, when it is blocked, the modern machine stalls, as the nascent peptide does not have anywhere to go.
The exit cavity hypothesis proposed here has significant implications for the origin of life. In particular, the proposed mechanism offers the first detailed hypothesis for the emergence of the modern protein-synthesizing machinery from a predecessor RNA World. It also provides an alternative model for the synthesis of peptides in the late prebiotic world. In this regard, it is noteworthy that despite the considerable effort that has been undertaken (Rode, 1999; Leman et al., 2004; Brack, 2007; Fitz et al., 2008), no clear prebiotic mechanism for peptide synthesis has yet been demonstrated.
The emergence of the protoribosome would have greatly enhanced the level and possibly the size of peptides produced in the prebiotic world. How long would they have been? Structural examination of the portion of the exit tunnel within the proposed pocket-like protoribosome indicates it could initially protect 3–5 residues. Moreover, because it is relatively wide, the tunnel could accommodate a large variety of short peptides composed of any of the standard amino acids with little likelihood of stalling in the tunnel. Although only a small portion of the emerging peptide would be protected, in a prebiotic world without modern enzymes the product peptides might have remained stable even as they exited the primitive tunnel. If so, they could have grown to an extended length and potentially contributed to the maintenance and function of entities in the RNA World.
Being random, noncoded, and likely lacking complex structures, what function might these peptides perform? The obvious possibility is stabilization of the structure of any RNA that might have been present in the RNA World, including that of the protoribosome itself (Fox and Naik, 2004; Belousoff et al., 2010), which would have resulted in further increased lifetime and hence greater survival potential. In the modern ribosome, key proteins have extended regions that lack the common protein secondary structure features and enter deep into the ribosome, where they apparently stabilize the rRNA (Ramakrishnan and Moore, 2001; Hsiao et al., 2009).
Although it is conceivable that short mixed chirality proteins may be able to stabilize RNAs, it may be necessary to have largely chiral peptides to achieve significant enzymatic activity. The protoribosome active site and the tunnel entrance adjacent to it would have provided the stereochemistry that would have allowed for peptide bond formation and led to the formation of dipeptides. The direction of these dipeptides into the tunnel entrance may be assisted by the chirality of the system. However, deviations may occur so that the chirality of the product will not be kept and/or its position within the protoribosome will dictate other directionality. Rather short products would be preferred, owing to the limited size of the surface of the tunnel entrance that is available for stabilizing the products within the protoribosome. If chiral preference exists in the attachment of the amino acid to its carrier, then the combined effect may produce peptides that are sufficiently chiral to act as catalysts (Fox, 2010).
Finally, another key implication is that the emergence of the protoribosome may have provided for an abbreviated RNA World. The recently described aminoacylated RNAs and the extremely small ribozymes that produce them (Turk et al., 2011) are highly consistent with the RNA World. The pocket-like protoribosome proposed previously (Davidovich et al., 2009; Krupkin et al., 2011) is as small as 120–180 nucleotides and might actually be constructed by hybridization of several smaller pieces as well as by ligation or duplication of RNA chains. It is also possible that an even smaller RNA entity could provide the same core functionality as the modern version, which is clearly an optimized ancient entity (Yonath, 2009). Importantly, efforts to demonstrate the existence of an RNA-based RNA replicase have been difficult, with the best working models (Lincoln and Joyce, 2009; Wochner et al., 2011) no less complex than the pocket-like protoribosome considered here. In addition, these engineered ribozymes have yet to synthesize RNAs comparable to their own size (Yarus, 2011), as a true replicase must. Moreover, there is no clear evidence of the prior existence of such a prebiotic entity in extant organisms. An alternative possibility is that the peptides produced by the protoribosome may have acquired the ability to replicate RNA before RNA-based RNA replicases could evolve in the RNA World. Alternatively, an RNA-peptide complex may have been the first to accomplish the task. In either case, the transition from the RNA World to an RNA-Protein World may have occurred much more rapidly and easily than is traditionally thought.
Acknowledgments
This work was funded in part by grants to G.E.F. from the Georgia Institute of Technology Center for Ribosomal Evolution and Adaptation, the Center for Bionanotechnology and Environmental Research at Texas Southern University (NASA Cooperative Agreement NNX08B4A47A), and the Institute of Space Systems Operations at the University of Houston, as well as a NASA Earth and Space Science Fellowship for Mr. Quyen Tran. A.Y.'s efforts were supported by National Institutes of Health grant GM34360 and by the Kimmelman Center for Macromolecular Assemblies. A.Y. holds the Martin and Helen Kimmel Professorial Chair.
Author Disclosure Statement
No competing financial interests exist.
Abbreviation
PTC, peptidyl transferase center.
References
- Agmon I. Bashan A. Zarivach R. Yonath A. Symmetry at the active site of the ribosome: structural and functional implication. Biol Chem. 2005;386:833–844. doi: 10.1515/BC.2005.098. [DOI] [PubMed] [Google Scholar]
- Bashan A. Zarivach R. Schluenzen F. Agmon I. Harms J. Auerbach T. Baram D. Berisio R. Bartels H. Hansen H.A. Fucini P. Wilson D. Peretz M. Kessler M. Yonath A. Ribosomal crystallography: peptide bond formation and its inhibition. Biopolymers. 2003;70:19–41. doi: 10.1002/bip.10412. [DOI] [PubMed] [Google Scholar]
- Belousoff M.J. Davidovich C. Zimmerman E. Caspi Y. Wekselman I. Rozenszajn L. Shapira T. Sade-Falk O. Taha R. Bashan A. Weiss M.S. Yonath A. Ancient machinery embedded in the contemporary ribosome. Biochem Soc Trans. 2010;38:422–427. doi: 10.1042/BST0380422. [DOI] [PubMed] [Google Scholar]
- Brack A. From interstellar amino acids to prebiotic catalytic peptides: a review. Chem Biodivers. 2007;4:665–679. doi: 10.1002/cbdv.200790057. [DOI] [PubMed] [Google Scholar]
- Chiba S. Kanamori T. Ueda T. Akiyama Y. Pogliano K. Ito K. Recruitment of a species-specific translational arrest module to monitor different cellular processes. Proc Natl Acad Sci USA. 2011;108:6073–6078. doi: 10.1073/pnas.1018343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidovich C. Belousoff M. Bashan A. Yonath A. The evolving ribosome: from non-coded peptide bond formation to sophisticated translation machinery. Res Microbiol. 2009;160:487–492. doi: 10.1016/j.resmic.2009.07.004. [DOI] [PubMed] [Google Scholar]
- Fitz D. Jakschitz T. Rode B.M. The catalytic effect of L- and D-histidine on alanine and lysine peptide formation. J Inorg Biochem. 2008;102:2097–2102. doi: 10.1016/j.jinorgbio.2008.07.010. [DOI] [PubMed] [Google Scholar]
- Fox G.E. Origin and evolution of the ribosome. Cold Spring Harb Perspect Biol. 2010;2:a003483. doi: 10.1101/cshperspect.a003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox G.E. Naik A.K. The evolutionary history of the translation machinery, In The Genetic Code and the Origin of Life. In: Ribas de Pouplana L., editor. Eurekah.com/Landes Bioscience; Georgetown, TX: 2004. pp. 92–105. [Google Scholar]
- Gindulyte A. Bashan A. Agmon I. Massa L. Yonath A. Karle J. The transition state for formation of the peptide bond in the ribosome. Proc Natl Acad Sci USA. 2006;103:13327–13332. doi: 10.1073/pnas.0606027103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao C. Mohan S. Kalahar B.K. Williams L.D. Peeling the onion: ribosomes are ancient molecular fossils. Mol Biol Evol. 2009;26:2415–2425. doi: 10.1093/molbev/msp163. [DOI] [PubMed] [Google Scholar]
- Ito K. Chiba S. Pogliano K. Divergent stalling sequences sense and control cellular physiology. Biochem Biophys Res Commun. 2010;393:1–5. doi: 10.1016/j.bbrc.2010.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupkin M. Matzov D. Tang H. Metz M. Kalaora R. Belousoff M.J. Zimmerman E. Bashan A. Yonath A. A vestige of a prebiotic bonding machine is functioning within the contemporary ribosome. Philos Trans R Soc Lond B Biol Sci. 2011;366:2972–2978. doi: 10.1098/rstb.2011.0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leman L. Orgel L. Ghadiri M.R. Carbonyl-sulfide mediated prebiotic formation of peptides. Science. 2004;306:283–286. doi: 10.1126/science.1102722. [DOI] [PubMed] [Google Scholar]
- Lincoln T.A. Joyce G.F. Self-sustained replication of an RNA enzyme. Science. 2009;323:1229–1232. doi: 10.1126/science.1167856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J. Deutsch C. Folding zones inside the ribosomal exit tunnel. Nat Struct Mol Biol. 2005;12:1123–1129. doi: 10.1038/nsmb1021. [DOI] [PubMed] [Google Scholar]
- Lukes J. Archibald J. Keeling P. Doolittle F. Gray M. How a neutral evolutionary ratchet can build cellular complexity. IUBMB Life. 2011;63:528–537. doi: 10.1002/iub.489. [DOI] [PubMed] [Google Scholar]
- Nakatogawa H. Ito K. The ribosomal exit tunnel functions as a discriminating gate. Cell. 2002;108:629–636. doi: 10.1016/s0092-8674(02)00649-9. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan V. Moore P.B. Atomic structures at last: the ribosome in 2000. Curr Opin Struct Biol. 2001;11:144–154. doi: 10.1016/s0959-440x(00)00184-6. [DOI] [PubMed] [Google Scholar]
- Ramu H. Mankin A. Vazquez-Laslop N. Programmed drug-dependent ribosome stalling. Mol Microbiol. 2009;71:811–824. doi: 10.1111/j.1365-2958.2008.06576.x. [DOI] [PubMed] [Google Scholar]
- Ramu H. Vazquez-Laslop N. Klepacki D. Dai Q. Piccrilli J. Micura R. Mankin A.S. Nascent peptide in the ribosome exit tunnel affects functional properties of the A-site of the peptidyl transferase center. Mol Cell. 2011;41:321–330. doi: 10.1016/j.molcel.2010.12.031. [DOI] [PubMed] [Google Scholar]
- Rode B.M. Peptides and the origin of life. Peptides. 1999;20:773–786. doi: 10.1016/s0196-9781(99)00062-5. [DOI] [PubMed] [Google Scholar]
- Turk R.M. Illangasekare M. Yarus M. Catalyzed and spontaneous reactions on ribozyme ribose. J Am Chem Soc. 2011;133:6044–6050. doi: 10.1021/ja200275h. [DOI] [PubMed] [Google Scholar]
- Wochner A. Attwater J. Coulson A. Holliger P. Ribozyme-catalyzed transcription of an active ribozyme. Science. 2011;332:209–212. doi: 10.1126/science.1200752. [DOI] [PubMed] [Google Scholar]
- Woese C.R. Fox G.E. The concept of cellular evolution. J Mol Evol. 1977;10:1–6. doi: 10.1007/BF01796132. [DOI] [PubMed] [Google Scholar]
- Yarus M. Molecular biology. Climbing in 190 dimensions. Science. 2011;332:181–182. doi: 10.1126/science.1205379. [DOI] [PubMed] [Google Scholar]
- Yonath A. Ribosomal tolerance and peptide bond formation. Biol Chem. 2003;384:1411–1419. doi: 10.1515/BC.2003.156. [DOI] [PubMed] [Google Scholar]
- Yonath A. Antibiotics targeting ribosomes: resistance, selectivity, synergism and cellular regulation. Annu Rev Biochem. 2005;74:649–679. doi: 10.1146/annurev.biochem.74.082803.133130. [DOI] [PubMed] [Google Scholar]
- Yonath A. Large facilities and the evolving ribosome, the cellular machine for genetic-code translation. J R Soc Interface. 2009;6:S575–S585. doi: 10.1098/rsif.2009.0167.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarivach R. Bashan A. Berisio R. Harms J. Auerbach T. Schluenzen F. Bartels H. Baram D. Pyetan E. Sittner A. Amit M. Hansen H.A.S. Kessler M. Liebe C. Wolff A. Agmon I. Yonath A. Functional aspects of ribosomal architecture: symmetry, chirality and regulation. J Phys Org Chem. 2004;17:901–912. [Google Scholar]