Abstract
Comparative studies of bulk samples of hydrolytically-degradable poly(lactic acid) (PLA) vs. core-shell block copolymer micelles having PLA cores revealed remarkable acceleration in the proteinase K enzymatic hydrolysis of the nanoparticulate forms, and demonstrated that even with amidation-based shell crosslinking, the core domain remained accessible. Kinetic analyses by 1H NMR spectroscopy showed less than 20% lactic acid released from enzymatically- catalyzed hydrolysis of poly(L-lactic acid) in bulk, whereas ca. 70 % of the core degraded within 48 h for block copolymer micelles of poly(N-(acryloyloxy)succinimide-copolymer-N-acryloylmorpholine)-block-poly(L-lactic acid) (P(NAS-co-NAM)-b-PLLA), with only a slight reduction to ca. 50 % for the shell crosslinked derivatives. Rigorous characterization measurements by NMR spectroscopy, fluorescence spectroscopy, dynamic light scattering, atomic force microscopy, and transmission electron microscopy were employed to confirm core excavation. These studies provide important fundamental understanding of the effects of nanoscopic dimensions on protein-polymer interactions and polymer degradability, which will guide development of these degradable nanoconstructs to reach their potential for controlled release of therapeutics and biological clearance.
Keywords: Poly(lactic acid), degradable, nanoparticles, micelles, enzymatic, hydrolysis
INTRODUCTION
There has been a growing interest in employing degradable materials for applications in biomedical settings, due to their reduced toxicity and ability to clear through biological systems. Biodegradable polymers were first developed for sutures in the 1960s1 and this technology was later adopted for controlled drug delivery purposes, followed by clinical translations in the 1980s.2 After one of the earliest works on drug-loaded degradable microparticles based on PLA-peptide drug formulations, reported by DuPont in 1973,3 the utilization of biodegradable materials for sustained release has emerged immensely.
Therapeutics can be loaded into polymeric nanoconstructs and the release of these encapsulated guest molecules can then be triggered by external stimuli such as changes in pH,4,5 light6 and temperature.7 However, for rapid and effective clinical translation, it is imperative that these nanostructures are comprised of biocompatible materials,8 which can package therapeutics, gate their release and provide efficient delivery. Complex polymeric nanostructures with core-shell morphologies that constitute degradable polyesters in the core domain have shown great potential as vehicles for delivery of active therapeutics.9–20
Our group has a long-standing interest in the design and development of shell crosslinked nanomaterials with tunable size,4 shape,21,22 core flexibility,23,24 and the ability to perform chemical modifications selectively within the nanoscopic framework.25,26 These materials are derived from block copolymer micellar assemblies, for which crosslinking performed between reactive units within the shell domain produces shell crosslinked knedel-like (SCK) nanoparticles, providing structural stability to the resulting nanostructures,27 and also gating the trafficking of guest molecules to and from the core.4,28 Significant efforts have been made to integrate degradability into polymeric nanoparticles by the use of various types of hydrolytically-cleavable polymers, such as poly(ε-caprolactone) (PCL), as the hydrophobic core domain of crosslinked13,15,16 and worm-like29 micellar assemblies, enzymatically- and hydrolytically-degradable PLA in PEG-based nanoparticles,9–12,14 crystallization-driven cylindrical assemblies,30 UV-induced thiolene crosslinked nanocapsules,31 as well as in nanosized sugar balls.32 The selective excavation of the degradable core or cleavage of a single shell-core connection site of some of these materials via environmental triggers has generated hollow nanocages.15,32–35 Additionally, chromophore- linked degradable crosslinkers have been incorporated into the shell of SCKs for programmed disassembly of nanostructures and controlled release of reporter molecules,36 with inspiration from Fréchet’s explodable micelles.37
As an extension to the previous studies focused on incorporating chemical and hydrolytic degradability into nanostructures, this current research integrates degradable PLA, and specifically investigates the hydrolytic degradation behaviors of PLA within the core regions of mi- celles and SCKs, under acid and enzyme catalysis. As the enzymatic cleavage of the PLA core requires the enzyme to be accessible to the core domain, this work also provides information on the permeability of the crosslinked shell.
RESULTS AND DISCUSSION
Core-degradable SCK nanoparticles were constructed by the supramolecular assembly of a novel amphiphilic diblock copolymer P(NAS0.24-co-NAM0.76)210-b-PLLA43, followed by crosslinking between the N-hydroxysuccinimide (NHS)-activated acrylic acid (NAS) functionalities presented within the shell of the nanoparticles by the addition of diamino crosslinkers. Sequential ring opening polymerization (ROP) and reversible addition-fragmentation chain transfer (RAFT) polymerization were employed to obtain the initial PLLA43 homopolymer and subsequent P(NAS0.24-co-NAM0.76)210-b-PLLA43 diblock copolymer, each having a narrow molecular weight distribution (Scheme 1). Our design of the amphiphilic diblock copolymer P(NAS0.24-co-NAM0.76)210-b-PLLA43 precursor to the micelles and SCKs incorporates a hydrophobic, degradable PLA segment, and a P(NAS-co-NAM) copolymer segment that provides built-in functionality and hydrophilicity. The choice of P(NAS-co-NAM) copolymer as the hydrophilic block was inspired by previous demonstrations of the utility of NAS as a convenient functional handle,38,39 that also allows in-situ synthesis of SCKs by direct crosslinking with elimination of the need for coupling agents.40 Block copolymer micelles and SCKs were prepared from P(NAS0.24-co-NAM0.76)210-b-PLLA43 by treating the samples to the same conditions throughout the process, with the exception of the addition of a crosslinker. Self assembly of the diblock copolymer into micelles was afforded by its dissolution in tetrahydrofuran (THF, 1.77 mg/mL) followed by dropwise addition of an equal volume of nanopure water using a syringe pump (7.5 mL/h). The solution was dialyzed against nanopure water for 4 d to afford micellar assemblies (0.5 mg/mL). The solution of micelles was divided into two equal volumes and one of the portions was subjected to a nominal crosslinking density of 20% throughout the shell region by the addition of a diamine crosslinker (2,2′-(ethylenedioxy)bis(ethylamine)) to a stirring solution of micelles to yield SCKs. The solution of SCKs was dialyzed against nanopure water for 2 d to remove the NHS by-products and unreacted crosslinkers. The crosslinking density of 20% refers to the stoichiometry of the amines of the crosslinkers relative to the initial NAS residues. Additionally, the micellar solution was allowed to undergo dialysis for the same period of time to compensate for any hydrolytic degradation of PNAS and/or PLLA units. The pH values of the two solutions were adjusted to 7–8 by dialysis against 1 mM PBS buffer containing 0.05% w/v NaN3 for 3 d. Scheme 2 illustrates the preparation of SCK nanoparticles from the polymer precursor and the enzymatic degradation process of the SCKs.
Scheme 1.
(a) Synthesis of PLLA43 homopolymer and P(NAS0.24-co-NAM0.76)210-b-PLLA43 diblock co-polymer, (b) size exclusion chromatography traces of the homopolymer and diblock copolymer samples, showing narrow molecular weight distributions.
Scheme 2.
Preparation of SCK nanoparticles by self assembly of amphiphilic diblock copolymer P(NAS0.24-co-NAM0.76)210-b-PLLA43 followed by crosslinking, and production of a nanocage-like structure from selective hydrolysis of the PLA core of the SCK template.
Enzymatic degradation of the PLA core was accomplished by the addition of proteinase K into the two solutions of micelles and SCKs at 37 °C.41–43 Quantitative analyses of PLA degradation from the nanoparticles were performed by solvent suppression 1H NMR spectroscopy. Micelles and SCKs were prepared in deuterated PBS buffer at pH 7–8, and 0.6 mL of each of the solutions was transferred into NMR tubes for analysis. Assembly of the amphiphilic diblock copolymers into micellar structures in aqueous solution was confirmed by reduction and/or broadening of the NMR peaks corresponding to the hydrophobic PLA segment and observable for the diblock copolymer when solvated in an organic solvent (-CH, 5.22-5.10 ppm and –CH3, 1.64-1.52 ppm), due to the inability of the aqueous media to solvate the hydrophobic core region of the nanoparticles (Figure 1-a and 1-b). The degradation experiments were conducted by addition of proteinase K (20 μL in tris-HCl buffer, 800 U/mL) into the NMR tubes and collection of spectra at 30 min time intervals for 48 h, maintaining the instrument temperature at 37 °C. To compare the rate of PLA degradation from the cores of the nanostructures vs. bulk PLA, the same kinetics studies were also performed on the PLA homopolymer, as a thin film within an NMR tube.
Figure 1.
1H NMR spectra: (a) diblock copolymer P(NAS0.24-co-NAM0.76)210-b-PLLA43 in CDCl3; (b) SCKs in deuterated PBS buffer (solvent suppressed) confirming self assembly of the diblock copolymer by the reduction and/or broadening of the hydrophobic PLA peak intensities; (c) SCKs after 15 min of adding the enzyme (solvent suppressed) confirming degradation of PLA by the appearance of two sets of new OLA/LA peaks.
Real-time monitoring of the enzymatic degradation was accomplished by observing the methyl protons of oligo(lactic acid) (OLA) and lactic acid (LA) ranging from 1.64 ppm to 1.44 ppm in deuterated buffer, in comparison to an external chloroform reference. Interestingly, within 15 min of adding the enzyme, two new sets of peaks were apparent at 1.64-1.56 ppm and 1.48-1.44 ppm (Figure 1-c), in which the intensity of the peaks corresponding to the methyl protons from LA (1.48-1.44 ppm) continued to increase at the expense of the peaks corresponding to OLA (1.64-1.56 ppm). Figure 2-a represents the real-time 1H NMR spectra of the OLA/LA methyl protons for micelles monitored for 3 d following addition of the enzyme.
Figure 2.
(a) Real-time 1H NMR spectra of the OLA (1.64-1.56 ppm) and LA (1.48-1.44 ppm) methyl protons for micelles monitored over 3 d following addition of the enzyme, (b) percent degradation products vs. degradation time plot from integration of OLA or LA peaks from micelles, SCKs and bulk PLA monitored at 30 min intervals over 48 h of enzyme exposure.
To confirm that the degradation of PLA was facilitated by the enzyme and not by non-catalyzed hydrolysis, control experiments were performed. In the absence of the enzyme, the NMR spectra did not show any OLA or LA proton signals within 48 h of monitoring (data not shown). Also, as an identification and confirmation of the production of LA from PLA, at the end of the degradation experiment, one of the NMR tubes containing degraded PLA was spiked with LA externally, and an increase in the peak intensity at 1.48-1.44 ppm was observed.
Having demonstrated that PLA degradation occurred in the presence of the enzyme, our primary goal of the NMR studies was to quantify and compare the degradation behavior of the micelles as opposed to the crosslinked material, in order to determine if the crosslinking provided a barrier to the enzyme to permeate through the shell. We were also interested in the rates of hydrolysis for the nanomaterials vs. bulk PLA. The hydrolysis rates of PLA were measured by comparing the relative amounts of LA and OLA generated from micelles and SCKs to the 100% PLA determined by lyophilization of an equal volume of micelles followed by analysis in d6-DMSO, a solvent for the entire diblock copolymer. For the bulk sample, the total amount of PLA was known, due to the use of a stock solution for the sample preparation. During the quantification process, the same external reference of chloroform, in the form of a sealed capillary tube, was used in all the NMR tubes for accurate integration purposes. As illustrated in Figure 2-b, which summarizes the kinetic analysis of the degradation processes determined from NMR spectroscopy, ca. 80% of the total OLA was observable for the micelles and SCKs within 15 min, whereas continued degradative conversion to LA occurred more rapidly for the micelles than for the SCKs. LA production from SCKs began to plateau after ca. 15 h of exposure to enzyme, reaching a maximum percent degradation of approximately 50% of the PLA core. However, the LA generation from micelles continued to increase to about 70% through 48 h of exposure to the enzyme. In comparison, the bulk thin film PLA underwent only <20% LA generation over 48 h incubation with the enzyme solution, and gave an OLA signal that reached only ca. 5%. These results confirm the ability of the enzyme to gain access to the PLA, when packaged within a micelle or SCK core and when fully exposed. Given the dimensions of the enzyme (2–3 nm), it is expected that migration through the SCK shell requires some degree of cleavage of the amide crosslinkers, as migration of macromolecules through SCK shells has been shown to be limited by the crosslinks.44 It is intriguing that neither the OLA nor the LA proton signals were observed until the addition of the enzyme was performed, yet it remains uncertain whether the OLA signal is due to free oligomers in solution or to mobile lactic acid repeat units that remained connected to the PLA of the nanostructures or thin film. The differences observed for continued breakdown into LA for the micelles vs. SCKs suggest that the OLA remained as parts of the nanostructures. The crosslinks within the shells of the SCKs were shown to behave as gates, limiting migration of the enzyme through the shell to gain access to the PLA/OLA and/or mobility of the PLA/OLA to the SCK surface to expose some portion of the PLA chain segments, initially and upon potential reorganization events as the degradation proceeded. The similar kinetics for the initial OLA production for both the micelles and SCKs suggest that the crosslinked shell posed no barrier to the enzyme, and merely reduced the rate of conversion of OLA to LA.
The relatively rapid enzymatic degradation kinetics of the PLA core, especially within 15 min of adding the enzyme may be attributed to the possible interactions between the negatively-charged surface of the nanoparticles and positively-charged enzyme at the physiological pH conditions applied. Partial hydrolysis of the NAS units in the shell of the nanoparticles gave rise to an anionic shell (zeta potential: -35 ± 2 mV and -48 ± 3 mV for freshly-prepared micelles and SCKs, respectively, at pH 7–8) that could readily attract the positively-charged proteinase K (isoelectric point 8.9)45 to the nanoconstructs, following rapid degradation of the core material. A control study using lipase (isoelectric point 4.9), which should be electrostatically repelled from the SCKs observed no PLA degradation.
Because proteinase K is capable of cleaving amide-based crosslinks within the SCK shells, in addition to the observed rapid degradation of the PLA core, we investigated the integrity of the nanoparticle upon core excavation. In order to thoroughly evaluate and compare the fate of the nanoparticles, especially the stability of micelles compared to SCKs upon hydrolysis of PLA, a series of analytical tools including dynamic light scattering (DLS), transmission electron microscopy (TEM) and atomic force microscopy (AFM) was utilized.
DLS studies provided insight into the dimensional changes in the hydrodynamic diameters of the nanoparticles upon core excavation. It was observed that upon hydrolysis of the PLA segment, the resulting hydrophilic nanocage-like structures underwent swelling, due to the diffusion of water into the core region, increasing progressively as the hydrophobic components were eliminated. The swelling process was monitored for both micelles and SCKs upon hydrolytic as well as enzymatic degradation of PLA. As shown in Figure 3, the initial number-average hydrodynamic diameters (Dh(n)) of micelles and SCKs were 34 ± 10 nm and 30 ± 9 nm, respectively. For micelles, while hydrolytic degradation for 3 months in nanopure water at pH 5–6 expanded the hydrodynamic diameter to 93 ± 25 nm, after 12 h of enzymatic degradation micelles were no longer detectable from DLS, indicating complete disassembly of the particles. The significant increase in Dh under hydrolytic conditions is expected to be the result of micellar reorganization events, including polymer chain exchange and micelle aggregation, as the hydrophobic chain segment lengths decrease over time and hydrophilic surface chains are released from the micelle assemblies. Although the crosslinked analog showed a similar trend for expansion upon hydrolytic degradation over the same period of 3 months, their Dh increased only to 48 ± 22 nm, suggesting shell expansion in the absence of additional micellar reorganization and aggregation. Also in contrast to the micelles, 12 h of enzymatic degradation of SCKs did not result in complete disassembly, but rather showed a dual distribution in the hydrodynamic diameter to 18 ± 4 nm and 72 ± 21 nm, and further expansion to 21 ± 4 nm and 118 ± 32 nm after 48 h of enzymatic incubation.
Figure 3.
Examination of changes in hydrodynamic diameters by dynamic light scattering for micelles and SCKs as a function of acidic and enzymatic hydrolysis times.
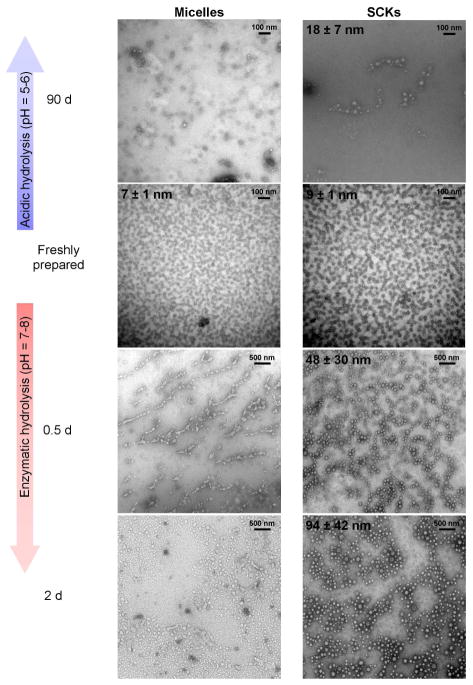
To further assess the dimensional changes observed from DLS, the nanoparticles were observed under TEM. Freshly-prepared nanoparticles were well-defined and uniform in size as shown in Figure 4, with average core diameters of 7 ± 1 nm and 9 ± 1 nm for micelles and SCKs, respectively. After 90 days of hydrolysis in water, micelles showed ill-defined non-spherical particles in the dry state while SCKs showed approximately 50% increases in the core diameters, expanding to 18 ± 7 nm. These results were consistent with the increases in the hydrodynamic diameters upon partial hydrolysis of the PLA core observed by DLS measurements. Interestingly, enzymatically-hydrolyzed micelles, which showed from NMR ca. 55% of the PLA cores degraded within 12 h, appeared as ill-defined polymer aggregates under TEM imaging. In contrast to micelles, SCKs remained robust and appeared as hollowed-out nanocage-like structures with a broad distribution of core diameters (48 ± 30 nm) in agreement with DLS measurements. After 48 h of enzyme treatment, completely degraded micellar structures looked similar to the 12 h time point under TEM, while SCKs showed further expansion to 94 ± 42 nm.
Figure 4.
Examination of changes in average core diameters in the dry state by transmission electron microscopy for micelles and SCKs as a function of acidic and enzymatic hydrolysis times.
In our previous work, it has been observed from AFM that SCK nanoparticles composed of soft core material undergo a flattening process upon adsorption on to the mica substrate, resulting in a decreased particle height and increased diameter compared to the hydrodynamic diameters observed from DLS.46–48 As illustrated in Figure 5, AFM images revealed theconversion of SCK nanoparticles to hollowed nanocages (which appear as donut-like structures after collapse onto the mica substrate) upon hydrolytic degradation of the PLA core for 3 months, with a reduction of the particle heights (from 3 ± 1 nm to <1 nm, before and after hydrolytic core degradation, respectively) and a substantial increase in the average particle width (from 38 ± 10 nm to 88 ± 12 nm, before and after hydrolytic core degradation, respectively). Even though the DLS data indicated further expansion of the SCKs upon enzymatic deg radation, particles were not detectable from AFM after 2 d excavation of the core.
Figure 5.
Tapping-mode AFM images of freshly-prepared SCKs and hydrolytically-degraded SCKs, showing collapse of the nanocage-like structures on the mica substrate upon core excavation.
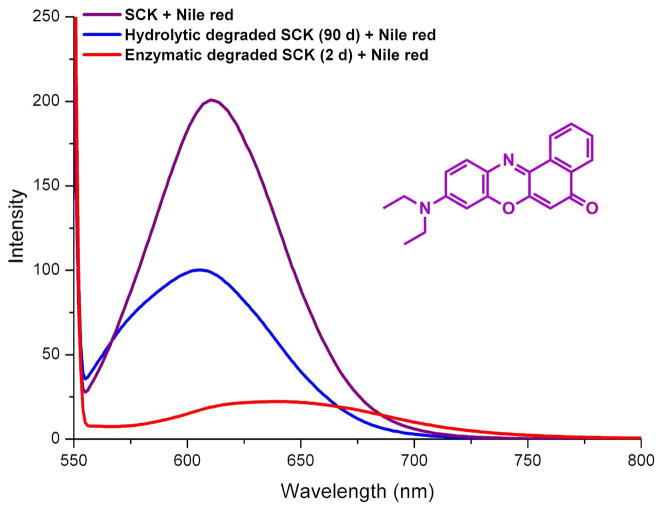
As a measure of the reduction of hydrophobicity within the SCKs upon removal of the PLA region, a reporter molecule Nile red was encapsulated into the nanoparticles. Nile red is a solvatochromic dye that fluoresces intensely under hydrophobicenvironments but displays weak emission in aqueous media.49 Equal volumes of freshly prepared SCKs, enzymatically-degraded SCKs (2 d) and hydrolytically-degraded SCKs (90 d) were incubated with an excess amount of Nile red. The dye was dispersed into aqueous solutions of the nanostructures from THF and allowed to stir overnight. The solutions were sparged with nitrogen to remove THF and residual dye was removed by filtration through a 0.45 μm Teflon® membrane to afford a bright magenta solution of freshly-prepared SCKs, a light magenta solution of partially hydrolyzed SCKs, and a pale magenta-to-colorless solution of enzymatically-degraded SCKs. Fluorescence measurements were conducted by collecting emission spectra (λex = 535 nm) of the three solutions (Figure 6). The profound loss in fluorescence intensity of the Nile red loaded into the solutions that had undergone degradation confirmed the loss of hydrophobicity upon (enzymatic) hydrolysis of the PLA core of the SCKs.
Figure 6.
Fluorescence intensity when reporter molecule Nile red was encapsulated into SCKs vs. hydrolyzed SCKs.
CONCLUSIONS
In summary, we have reported fundamental advances in the synthetic methodologies for the preparation of hydrolytically- degradable, functionalizable nanoparticles, together with rigorous characterization of their degradation properties. Specifically, we have demonstrated enzyme –triggered selective excavation of the polyesterbased core of block copolymer micelle assemblies and their shell crosslinked nanoparticle analogs. The rates of hydrolysis of the PLA from the cores of the block copolymer micelles and SCKs were significantly greater than for bulk PLA. Although it is not surprising that having the PLA dispersed in nanoscopic form throughout the solutions resulted in greater ability of the enzymes to access the PLA, rapid migration of the enzyme through the crosslinked shell layer is curious and worth further investigation, to determine whether the mechanism is by diffusion only or also facilitated by cleavage of some portion of the crosslinks. In contrast to their non-crosslinked analogs, which undergo complete disassembly upon loss of their hydrophobic moieties, SCK nanoparticles maintain their nanoparticulate integrity even in the absence of its amphiphilicity. Therefore, any cleavage of the amide-based crosslinks is not detrimental to the resulting nanocage framework. Studies are being continued to explore the effect of varying shell crosslinking densities on the core degradation properties of the SCKs. Additionally, loading of active therapeutics into the PLA based SCKs and the release kinetics of the guest molecules as a function of core degradation are being evaluated, and will be reported in the near future.
Supplementary Material
Acknowledgments
We gratefully acknowledge financial support from the National Heart Lung and Blood Institute of the National Institutes of Health as a Program of Excellence in Nanotechnology (HHSN268201000046C) and Covidien, Inc. The Welch Foundation is gratefully acknowledged for support through the W. T. Doherty-Welch Chair in Chemistry, Grant No. A-0001. Dr. A. d’Avignon and Dr. K. P. Sarathy of the NMR facilities at Washington University in St. Louis and Texas A&M University, respectively, are gratefully acknowledged for assistance with NMR experiments. The authors would also like to thank Dr. Krishnamurthy Shanmuganandamurthy for initiating this research work, Shiyi Zhang and Dr. Jiong Zou for discussions and support, and Hasitha Samarajeewa for creating the Autodesk 3ds Max images.
Footnotes
Supporting Information. Detailed experimental procedures including polymer synthesis, nanoparticle preparation, degradation conditions and characterization. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Schmitt EE, Polistina RA. 3297033. US Patent. 1967
- 2.Hoffman AS. J Controlled Release. 2008;132:153–163. doi: 10.1016/j.jconrel.2008.08.012. [DOI] [PubMed] [Google Scholar]
- 3.Boswell G, Scribner R. 3773919. US Patent. 1973
- 4.Lin LY, Lee NS, Zhu JH, Nystrom AM, Pochan DJ, Dorshow RB, Wooley KL. J Controlled Release. 2011;152:37–48. doi: 10.1016/j.jconrel.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Broaders KE, Pastine SJ, Grandhe S, Frechet JMJ. Chem Commun. 2011;47:665–667. doi: 10.1039/c0cc04190d. [DOI] [PubMed] [Google Scholar]
- 6.Lendlein A, Jiang HY, Junger O, Langer R. Nature. 2005;434:879–882. doi: 10.1038/nature03496. [DOI] [PubMed] [Google Scholar]
- 7.Lendlein A, Langer R. Science. 2002;296:1673–1676. doi: 10.1126/science.1066102. [DOI] [PubMed] [Google Scholar]
- 8.Peer D, Karp JM, Hong S, FaroKhzad OC, Margalit R, Langer R. Nature Nanotechnology. 2007;2:751–760. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- 9.Yamamoto Y, Yasugi K, Harada A, Nagasaki Y, Kataoka K. J Controlled Release. 2002;82:359–371. doi: 10.1016/s0168-3659(02)00147-5. [DOI] [PubMed] [Google Scholar]
- 10.Iijima M, Nagasaki Y, Okada T, Kato M, Kataoka K. Macromolecules. 1999;32:1140–1146. [Google Scholar]
- 11.Rijcken CJ, Snel CJ, Schiffelers RM, van Nostrum CF, Hennink WE. Biomaterials. 2007;28:5581–5593. doi: 10.1016/j.biomaterials.2007.08.047. [DOI] [PubMed] [Google Scholar]
- 12.Sun J, Chen XS, Lu TC, Liu S, Tian HY, Guo ZP, Jing XB. Langmuir. 2008;24:10099–100106. doi: 10.1021/la8013877. [DOI] [PubMed] [Google Scholar]
- 13.Van Horn BA, Wooley KL. Macromolecules. 2007;40:1480–1488. [Google Scholar]
- 14.Kang N, Perron ME, Prud’homme RE, Zhang YB, Gaucher G, Leroux JC. Nano Lett. 2005;5:315–319. doi: 10.1021/nl048037v. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Q, Remsen EE, Wooley KL. J Am Chem Soc. 2000;122:3642–3651. [Google Scholar]
- 16.Li YL, Zhu L, Liu ZZ, Cheng R, Meng FH, Cui JH, Ji SJ, Zhong ZY. Angew Chem-Int Ed. 2009;48:9914–9918. doi: 10.1002/anie.200904260. [DOI] [PubMed] [Google Scholar]
- 17.Tan JPK, Kim SH, Nederberg F, Appel EA, Waymouth RM, Zhang Y, Hedrick JL, Yang YY. Small. 2009;5:1504–1507. doi: 10.1002/smll.200801756. [DOI] [PubMed] [Google Scholar]
- 18.Kim SH, Tan JPK, Nederberg F, Fukushima K, Yang YY, Waymouth RM, Hedrick JL. Macromolecules. 2009;42:25–29. [Google Scholar]
- 19.Gref R, Minamitake Y, Peracchia MT, Trubetskoy V, Torchilin V, Langer R. Science. 1994;263:1600–1603. doi: 10.1126/science.8128245. [DOI] [PubMed] [Google Scholar]
- 20.Karnik R, Valencia PM, Hanewich-Hollatz MH, Gao WW, Karim F, Langer R, Farokhzad OC. Biomaterials. 2011;32:6226–6233. doi: 10.1016/j.biomaterials.2011.04.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pochan DJ, Chen ZY, Cui HG, Hales K, Qi K, Wooley KL. Science. 2004;306:94–97. doi: 10.1126/science.1102866. [DOI] [PubMed] [Google Scholar]
- 22.Lee NS, Sun G, Lin LY, Neumann WL, Freskos JN, Karwa A, Shieh JJ, Dorshow RB, Wooley KLJ. Mater Chem. 2011;21:14193–14202. doi: 10.1039/C1JM11854D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Q, Wang M, Wooley KL. Curr Org Chem. 2005;9:1053–1066. [Google Scholar]
- 24.Nystrom AM, Wooley KL. Soft Matter. 2008;4:849–858. doi: 10.1039/b719314a. [DOI] [PubMed] [Google Scholar]
- 25.Sun G, Hagooly A, Xu J, Nystrom AM, Li ZC, Rossin R, Moore DA, Welch MJ, Wooley KL. Biomacromolecules. 2008;9:1997–2006. doi: 10.1021/bm800246x. [DOI] [PubMed] [Google Scholar]
- 26.Zhang S, Li Z, Samarajeewa S, Sun GR, Yang C, Wooley KLJ. Am Chem Soc. 2011;133:11046–11049. doi: 10.1021/ja203133h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sorrells JL, Shrestha R, Neumann WL, Wooley KL. J Mater Chem. 2011;21:8983–8986. [Google Scholar]
- 28.Lee NS, Lin LY, Neumann WL, Freskos JN, Karwa A, Shieh JJ, Dorshow RB, Wooley KL. Small. 2011;7:1998–2003. doi: 10.1002/smll.201100567. [DOI] [PubMed] [Google Scholar]
- 29.Geng Y, Discher DE. J Am Chem Soc. 2005;127:12780–12781. doi: 10.1021/ja053902e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petzetakis N, Dove AP, O’Reilly RK. Chemical Science. 2011;2:955–960. [Google Scholar]
- 31.Zou J, Hew CC, Themistou E, Li Y, Chen CK, Alexandridis P, Cheng C. Adv Mater. 2011;23:4274–4277. doi: 10.1002/adma.201101646. [DOI] [PubMed] [Google Scholar]
- 32.Ting SRS, Gregory AM, Stenzel MH. Biomacromolecules. 2009;10:342–352. doi: 10.1021/bm801123b. [DOI] [PubMed] [Google Scholar]
- 33.Ievins AD, Moughton AO, O’Reilly RK. Macromolecules. 2008;41:3571–3578. [Google Scholar]
- 34.Moughton AO, Stubenrauch K, O’Reilly RK. Soft Matter. 2009;5:2361–2370. [Google Scholar]
- 35.Zhang Y, Jiang M, Zhao J, Wang Z, Dou H, Chen D. Langmuir. 2005;21:1531–1538. doi: 10.1021/la047912p. [DOI] [PubMed] [Google Scholar]
- 36.Li YL, Du WJ, Sun GR, Wooley KL. Macromolecules. 2008;41:6605–6607. [Google Scholar]
- 37.Gillies ER, Frechet JMJ. Chem Commun. 2003:1640–1641. doi: 10.1039/b304251k. [DOI] [PubMed] [Google Scholar]
- 38.Li YT, Lokitz BS, McCormick CL. Macromolecules. 2006;39:81–89. [Google Scholar]
- 39.Handke N, Trimaille T, Luciani E, Rollet M, Delair T, Verrier B, Bertin D, Gigmes D. Journal of Polymer Science Part a-Polymer Chemistry. 2011;49:1341–1350. [Google Scholar]
- 40.Li YL, Akiba I, Harrisson S, Wooley KL. Adv Funct Mater. 2008;18:551–559. [Google Scholar]
- 41.Williams DF. Eng Med. 1981;10:5. [Google Scholar]
- 42.Xiong XY, Tam KC, Gan LH. J Controlled Release. 2005;108:263. doi: 10.1016/j.jconrel.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 43.MacDonald RT, McCarthy SP, Gross RA. Macromolecules. 1996;29:7356. [Google Scholar]
- 44.Murthy KS, Ma Q, Clark CG, Jr, Remsen EE, Wooley KL. Chem Commun. 2001:773–774. [Google Scholar]
- 45.Ebeling W, Hennrich N, Klockow M, Metz H, Orth HD, Lang H. Eur J Biochem. 1974;47:91–97. doi: 10.1111/j.1432-1033.1974.tb03671.x. [DOI] [PubMed] [Google Scholar]
- 46.Turner JL, Wooley KL. Nano Lett. 2004;4:683–688. [Google Scholar]
- 47.Huang HY, Remsen EE, Kowalewski T, Wooley KL. J Am Chem Soc. 1999;121:3805–3806. [Google Scholar]
- 48.Ma QG, Remsen EE, Kowalewski T, Schaefer J, Wooley KL. Nano Lett. 2001;1:651–655. [Google Scholar]
- 49.Greenspan P, Mayer EP, Fowler SD. J Cell Biol. 1985;100:965–973. doi: 10.1083/jcb.100.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.