Abstract
The fusion of myoblasts into multinucleate syncytia plays a fundamental role in muscle function, as it supports the formation of extended sarcomeric arrays, or myofibrils, within a large volume of cytoplasm. Principles learned from the study of myoblast fusion not only enhance our understanding of myogenesis, but also contribute to our perspectives on membrane fusion and cell-cell fusion in a wide array of model organisms and experimental systems. Recent studies have advanced our views of the cell biological processes and crucial proteins that drive myoblast fusion. Here, we provide an overview of myoblast fusion in three model systems that have contributed much to our understanding of these events: the Drosophila embryo; developing and regenerating mouse muscle; and cultured rodent muscle cells.
Keywords: Founder cell, Fusion-competent myoblast, Myoblast, Myocyte, Myotube, Myofiber, Satellite cell, Regeneration, Migration, Adhesion, Myogenesis
Introduction
The process of fusing two adjacent membranes accomplishes many goals that are crucial to the development and maintenance of living organisms. Broadly, membrane fusion can occur intracellularly, as with synaptic vesicles, or between cells, as for sperm-egg fusion. Cell fusion occurs in a broad range of organisms, including Saccharomyces cerevisiae and Caenorhabditis elegans, and between several cell types, such as macrophages, placental trophoblasts and myoblasts. Experimental analysis of fusion in these systems has revealed the involvement of a diverse array of specialized molecules.
Myoblast fusion, a fundamental step in the differentiation of muscle in most organisms, can involve tens of thousands of myoblasts, Given the complexity of the musculature, fusion must be a regulated process in which the appropriate number of cells fuse at the appropriate time and place. Often these cells migrate long distances prior to fusion, and fusion can involve multiple cell types, necessitating cell recognition and adhesion, which are crucial to accurate and efficient fusion. In addition to the early fusion events that occur during embryogenesis, vertebrate muscle tissue is able to regenerate in response to damage and disease. This regeneration involves proliferation of muscle satellite cells (see Glossary, Box 1) and their subsequent fusion to repair the damaged muscles. The focus of this review is the process of myoblast fusion, which involves cell migration, adhesion and signaling transduction pathways leading up to the actual fusion event. We review the crucial role of the actin cytoskeleton and actin polymerizing proteins, and recently revealed membrane protrusions that may drive fusion itself. We describe three powerful systems for this analysis: Drosophila embryogenesis; mouse embryogenesis and regeneration; and rodent tissue culture. We highlight the genes and morphological events involved in myoblast fusion, as revealed by each system. With the emergence of Danio rerio as another model for myoblast fusion, we also list relevant homologs in this system (Tables 1, 2).
Box. 1 Glossary
Embryonic myoblast. A proliferative muscle progenitor cell found in early embryonic mouse development that differentiates and fuses to form primary myofibers. In Drosophila it also refers to either the founder cells or fusion-competent myoblasts prior to fusion.
Fetal myoblast. A proliferative muscle progenitor cell found in later embryonic mouse development that differentiates and fuses to form secondary myofibers.
Founder cell. A mono-nucleate cell in Drosophila that determines the properties of a myofiber (size, shape and attachment to the epidermis). It arises by asymmetric division of a muscle progenitor and does not proliferate further. There is one founder cell for each embryonic/larval somatic muscle.
Fusion-competent myoblast. A mono-nucleate cell in Drosophila that is committed to a muscle-specific program of differentiation but not yet fused. It undergoes very limited additional proliferation. Many fusion-competent myoblasts will fuse with a single founder cell, taking on the identity of that founder cell.
Myofiber (also referred to as a muscle fiber). A single syncytia in Drosophila embryos that results from the fusion of one founder cell and multiple fusion-competent myoblasts. In mice, it refers to a multinucleated muscle cell in vivo formed by the fusion of multiple myocytes.
Mature myotube. A large mouse muscle cell in vitro that contains many nuclei. These cells are not as developed in size, myonuclear number or internal structure as myofibers in vivo.
Myocyte. A differentiated mononucleated muscle progenitor cell in mice.
Nascent myotube. A newly formed multinucleated mouse muscle cell in vitro that results from the fusion of a few myocytes.
Primary myofiber. A multinucleated muscle cell present early in embryonic mouse development.
Satellite cell. A muscle stem cell in mice that lies in close apposition to a myofiber underneath the basal lamina surrounding the myofiber. These cells are normally quiescent in uninjured muscle but begin to proliferate upon muscle injury to give rise to myoblasts and also self-renew to form new satellite cells.
Secondary myofiber. A multinucleated muscle cell present later in embryonic mouse development that develops in close apposition to primary myofibers.
Table 1.
Proteins that function in migration, adhesion and recognition events prior to myoblast fusion
Table 2.
Proteins that function in signal transduction, actin remodeling and membrane fusion
Experimental systems for the analysis of myoblast fusion
The ability to isolate and propagate mammalian myoblasts that could differentiate and fuse in vitro launched the analysis of myoblast fusion several decades ago. This experimental system was the first to implicate specific molecules in myoblast fusion. Drosophila then emerged as a model organism for the study of embryonic myoblast (see Glossary, Box 1) fusion owing to its easily manipulated genetics and the ability to screen for mutations that impact fusion (Fig. 1). One can readily examine the developing musculature in both fixed and live embryos (see Box 2). Indeed, the identification of crucial genes in Drosophila has provided a valuable entry into the study of myoblast fusion in higher organisms. Nevertheless, insect muscles differ in fundamental ways (see Box 2) from vertebrate muscles. With the emergence of gene knockout and siRNA strategies, and the ability to manipulate primary myoblasts and myoblast cell lines, rodent systems have become powerful models for the analysis of myoblast fusion that more accurately recapitulate that occurring in humans (see Box 2). The details of each of these systems are described below.
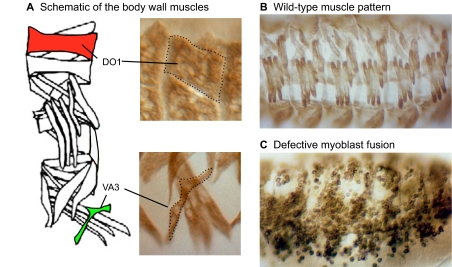
Fig. 1.
Muscle pattern and myoblast fusion in Drosophila. (A). Left: Schematic of the 30 distinct muscles per abdominal hemisegment in the Drosophila embryo. Highlighted are a small muscle (ventral acute 3; VA3 in green) and a large muscle (dorsal oblique 1; DO1 in red) illustrating the differences in size and shape. Right: High magnification view of VA3 and DO1, as visualized in a wild-type embryo by an antibody directed against muscle myosin. (B,C) The pattern of muscles and myoblasts in wild-type and fusion-defective embryos, as visualized by an antibody against muscle myosin. Multinucleate syncytia are apparent in the wild-type embryo shown in B. Defects in myoblast fusion are easily visible in the mutant embryo (C), highlighting the value of this model system. (B) Reproduced, with permission, from Bour et al. (Bour et al., 1995).
Box 2. Experimental systems for studying myoblast fusion: strengths and limitations
Drosophila
Strengths
Genetically tractable system with a rapid generation time.
Myogenesis in the embryo occurs within 12 hours.
Many processes are controlled by single-copy genes.
Amenable to live imaging with tagged proteins in wild-type and mutant embryos.
Limitations to the genetic analysis of the adult musculature are being overcome by RNAi technology.
Amenable to ultrastructural analysis of wild-type and mutant embryos.
Limitations
Limited availability of muscle-derived cultured cell lines.
Extent of gene redundancy unknown.
Primary cultures highly enriched for labeled myoblasts can be obtained, but are not pure and are available only in limited amounts.
Muscle development and structure are somewhat different from vertebrates, with muscles represented by single myotubes patterned by founder cells.
Mouse
Strengths
Long lifespan.
Can study fusion in various contexts, such as regeneration, aging, exercise and disease.
Muscle development and structure more closely resemble human.
Presence of slow and fast myofibers.
Cell culture models and approaches are well developed: can systemically control both the media and matrix of the cells; can easily manipulate cellular components with drugs, siRNA or DNA constructs; can analyze myoblast fusion in the absence of other cell types.
Limitations
Documented gene redundancy that appears to be more extensive than simpler genetic model organisms.
Limited ability to perform live cell imaging in vivo.
Length of time to generate mutants.
Limited number of offspring.
The Drosophila embryo
The muscles used by the Drosophila larva develop in the embryo over a period of 10-12 hours. These 30 segmentally repeated abdominal muscles differ from each other in size, shape, location, pattern of innervation and site of attachment to the epidermis (Bate, 1990) (Fig. 1A). In contrast to vertebrate muscles, which are composed of large bundles of myofibers (see Glossary, Box 1), each muscle is a single myofiber that arises by fusion of one founder cell (see Glossary, Box 1) with several fusion competent myoblasts (FCMs; see Glossary, Box 1). The founder cells and FCMs arise from the same population within the somatic mesoderm. Founder cells are initially selected in a complex process that involves overlapping signal transduction pathways, lateral inhibition mediated by Notch and asymmetric cell division (Tixier et al., 2010). This heterogeneous population of cells controls the muscle pattern, with a single founder cell for each muscle fiber (Bate, 1993). The identity and subsequent behavior of each muscle founder cell is then controlled through combinatorial expression of one or more muscle identity genes (Frasch, 1999; Enriquez et al., 2010; Tixier et al., 2010) that dictate the specific differentiation program of each muscle fiber (Bataillé et al., 2010; Tixier et al., 2010).
Once specified, the founder cell seeds the fusion process by recruiting FCMs. These myoblasts initially arise from the same cell population as the founder cells, but appear to be specified as a homogenous population by the Gli-family transcription factor Lame duck (Lmd) (Duan et al., 2001; Ruiz-Gomez et al., 2002). Following the initial fusion event between a founder cell and an FCM, additional rounds of fusion continue between the developing myotube and FCMs until the final muscle size is achieved. Thus, diversification of the muscle fibers is accomplished in a cell-autonomous manner, within the muscle cells themselves, rather than through the influence of external cues. Overall, the smallest muscles of the embryo will be formed by fusion of as few as two or three cells, whereas larger muscles can include more than a dozen cells (Bate, 1990; Richardson et al., 2007) (Fig. 1). Neither of these cell populations is highly proliferative. Progenitors of the founder cells often undergo only a single cell division subsequent to fusion (Tixier et al., 2010). Some FCMs also exhibit very limited cell division prior to fusion (Beckett and Baylies, 2007).
Muscle development and regeneration in rodents
Mice and rats are used extensively to study muscle formation in vertebrates with similar processes occurring in both species. In contrast to flies, specific subsets of myoblasts that seed the formation of myofibers have not been identified. Furthermore, myoblast fusion during rodent embryogenesis occurs in two stages known as primary and secondary myogenesis (Kelly and Zacks, 1969; Ontell and Kozeka, 1984; Ross et al., 1987), which results in individual muscles that are composed of multiple myofibers.
Embryonic muscle development
During primary myogenesis, embryonic myoblasts fuse to form primary myofibers (see Glossary, Box 1). Several days later, fetal myoblasts (see Glossary, Box 1), which are cells that develop from distinct, but related progenitors (Hutcheson et al., 2009) and differ in proliferation and fusion capacities (Biressi et al., 2007a; Biressi et al., 2007b), become prevalent and fuse with each other to form secondary myofibers (see Glossary, Box 1) in close apposition to primary myofibers (Duxson et al., 1989). As secondary myogenesis progresses, fetal myoblasts preferentially fuse with the ends of both primary and secondary myofibers (Zhang and McLennan, 1995). During secondary myogenesis, myofibers begin to differ in their expression of myosin heavy chain isoforms that distinguish between future fast- and slow-contracting muscles of the adult (Lyons et al., 1990). Late in development, a third type of muscle precursor cell, the adult satellite cell, is present between the basal lamina and the myofiber plasma membrane (Lepper and Fan, 2010). Myoblasts derived from satellite cells are responsible for adult muscle growth and regeneration (Lepper et al., 2011; Murphy et al., 2011; Sambasivan et al., 2011). During muscle growth in the postnatal period, large numbers of myoblasts fuse to the ends of myofibers, resulting in increased myofiber length and girth (Kitityakara and Angevine, 1963; Williams and Goldspink, 1971). Adult myofibers contain one nucleus approximately every 20 μm, depending on age and muscle type (Bruusgaard et al., 2003; Bruusgaard et al., 2006); thus, a 5 mm muscle fiber contains several hundred nuclei.
Regeneration in adult muscle
Myoblast fusion during regeneration in adult muscle is the best-studied model in vertebrates (Fig. 2). Degeneration can be induced by local physical or chemical trauma, resulting in rapid focal necrosis of myofibers with new membrane formation demarcating the ends of the surviving stumps and probably limiting the extent of myofiber necrosis (Carpenter and Karpati, 1989; Papadimitriou et al., 1990). A localized cellular immune response ensues (Snow, 1977; McLennan, 1996; Pimorady-Esfahani et al., 1997) that is crucial for normal regeneration (Grounds, 1987; Arnold et al., 2007). In response to factors found in the injured area, satellite cells begin to proliferate, differentiate and fuse with one another or with existing myofibers (Robertson et al., 1990; Robertson et al., 1993b). As in embryogenesis, no evidence exists for specialized subsets of myoblasts that seed the formation of regenerated myofibers. Multiple small myotubes form within the basal lamina sheath of the original myofiber (Schmalbruch, 1976; Snow, 1977), and fuse with one another (Robertson et al., 1990) and to the stumps of the parent myofiber (Papadimitriou et al., 1990; Robertson et al., 1993b) to regenerate the parent myofiber. Current technologies do not exist for directly visualizing myoblast fusion in vivo in mice, thus, morphological and biochemical measurements are used as an indirect readout.
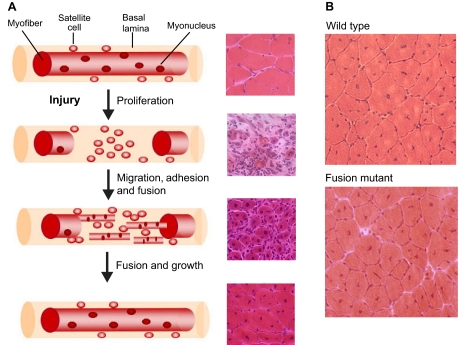
Fig. 2.
Muscle regeneration in adult mouse muscle. (A) Each myofiber in adult muscle is surrounded by a basal lamina sheath underneath which lie satellite cells in close apposition to the fiber. In response to injury, segmental necrosis of the myofiber occurs and satellite cells begin to proliferate and form myoblasts. These myoblasts differentiate and then migrate, adhere and fuse with one another to form multiple myotubes within the basal lamina sheath. Myoblasts/myotubes fuse with the stumps of the surviving myofiber and myotubes also fuse with each other to repair the injured myofiber. Regenerated myofibers are easily identified by the presence of centrally located nuclei. For each stage of regeneration in the schematic, a representative mouse muscle section is shown in cross-section and stained with Hematoxylin and Eosin to illustrate the morphological features of the tissue. (B) Cross-sections of regenerated myofibers 14 days after injury from wild-type and mutant mice stained with Hematoxylin and Eosin. Smaller myofibers are observed in the fusion mutant. Note the presence of centrally nucleated myofibers, a hallmark of muscle regeneration.
Cell culture models of myoblast fusion in mice
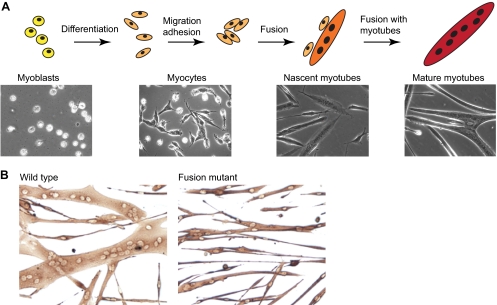
Much of the work pertaining to myoblast fusion in mammals derives from in vitro studies using either primary muscle cells isolated from mouse, rat or human muscles, or established mouse muscle cell lines, such as C2C12 (Fig. 3). Cell culture studies demonstrate that multinucleated myotubes form in a series of ordered steps. Initially, myoblasts differentiate into elongated myocytes (see Glossary, Box 1) that migrate, adhere and fuse to one another to form small nascent myotubes (see Glossary, Box 1) that contain few myonuclei. Nascent myotubes further fuse with additional myocytes or with other myotubes to generate mature myotubes (see Glossary, Box 1) that contain many myonuclei. Such in vitro experiments are often used to complement in vivo studies of adult regenerative myogenesis in mice to examine more fully the mechanisms that regulate myoblast fusion.
Fig. 3.
Myoblast fusion in cultured muscle cells. (A) Myoblasts proliferate in vitro in medium containing growth factors. To induce myotube formation, growth factors are removed and the majority of myoblasts will terminally differentiate into myocytes, which migrate, adhere and fuse with one another to form small nascent myotubes with few nuclei. Subsequently, nascent myotubes fuse with myocytes and other myotubes to form large mature myotubes with many nuclei. Representative phase contrast photos of mouse muscle cells are shown for each stage. (B) Muscle cells from adult wild-type and mutant mice cultured for 40 hours in vitro and visualized by immunostaining for myosin heavy chain.
Finding a fusion partner: recognition, migration and adhesion
This section examines our current understanding of how fusing myoblasts find their fusion partner(s). Although genetic loss-of-function studies in the Drosophila embryo have revealed genes essential for the early cell recognition and adhesion steps, cultured cell lines and primary mammalian cells have also allowed us to identify molecules that specifically direct migration of myoblasts to their targets. These molecules are summarized in Table 1 and discussed below in detail.
Myoblast migration, recognition and adhesion in Drosophila
In the Drosophila embryo, the FCMs and founder cells/myotubes arise in close proximity to each other, and are often adjacent cells that are in direct contact (Fig. 4). Moreover, the myotube will probably come in direct contact with additional FCMs as fusion proceeds and as the myotube becomes larger. Given these small distances, and in the absence of direct visualization of cell movements by live imaging in the Drosophila system, it is often difficult to distinguish genes involved in myoblast migration from those required for adhesion. However, indirect support for FCM migration comes from studies showing that these cells adopt a teardrop shape (Fig. 4A) with membrane processes oriented towards developing muscle fibers, consistent with their migration towards founder cells in fixed (Doberstein et al., 1997) and live (Richardson et al., 2007) embryos. Studies by Baylies et al. also revealed dramatic redistribution of FCMs to more external positions near founder cells in fixed embryos of different developmental stages (Beckett and Baylies, 2007). More direct evidence suggests that the FCMs are capable of migrating, and are dependent on transmembrane proteins of the immunoglobulin superfamily (IgSF) that include Kin-of-IrreC/Dumfounded (Kirre/Duf) (Ruiz-Gomez et al., 2000), Roughest/Irregular-optic-chiasma-C (Rst/Irre-C) (Strunkelnberg et al., 2001) and Sticks-and-stones (Sns) (Bour et al., 2000) for this behavior (Table 2, Fig. 5). Kirre is expressed specifically in the founder cells (Ruiz-Gomez et al., 2000), where it functions redundantly with Rst (Strunkelnberg et al., 2001) to recognize and associate with FCMs. Sns is required on the surface of the FCMs (Bour et al., 2000) for their ability to recognize founder cells and myotubes. The FCMs can clearly migrate to sites of ectopically expressed Kirre or Rst (Ruiz-Gomez et al., 2000; Strunkelnberg et al., 2001), and Sns is essential for this behavior (Kocherlakota et al., 2008). These data suggest that Kirre and Rst can function as Sns-dependent attractants for the FCMs, and that signaling cascades downstream of Sns direct migration. This process could involve filopodia that extend from the FCM to probe the environment and search for founder cells, either randomly finding membrane-associated Kirre on the surface of a founder cell or moving along a gradient of secreted Kirre. The latter possibility is indirectly supported by the presence of a truncated form of Kirre protein in the media of Drosophila S2 cells transfected with full-length Kirre (Chen and Olson, 2001). However, the presence of secreted or cleaved Kirre has not been established in vivo, and the mechanism by which FCMs find non-adjacent founder cells or myotubes is not clear.
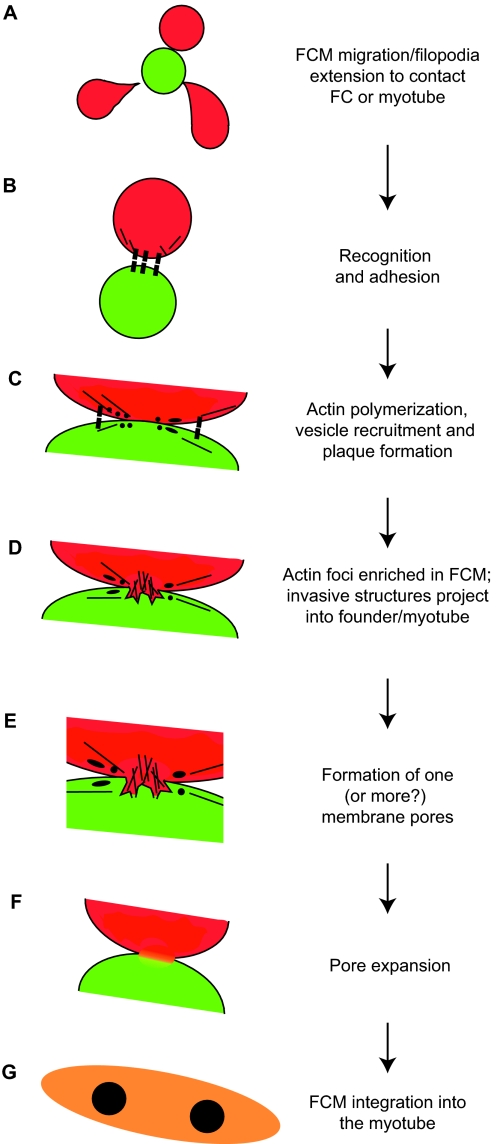
Fig. 4.
Hypothetical model of myoblast fusion in Drosophila embryos. (A) A fusion-competent myoblast (FCM; red) migrates or extends filopodia to contact a founder cell (FC; green) or, in subsequent rounds of fusion, a syncytial myotube (orange). (B) Cell-surface adhesion molecules (black boxes) mediate recognition and adhesion between cells. (C) Following cell-cell contact, and prior to fusion, electron-dense vesicles (black circles) are recruited to points of contact, possibly through vesicle trafficking mechanisms from the Golgi (not shown). This process may involve actin filaments (black lines). Such vesicles facilitate the fusion process, possibly by delivering fusion-associated components, such as lipids, fusogens or proteases, via targeted exocytosis near or at the sites of fusion. Vesicles may give rise to membrane plaques (black ellipses), which could reflect accumulation of adhesion proteins or other fusion machinery. (D) Actin accumulates in the FCM, forming a large F-actin-based protrusion that pushes into the founder cell. A thin sheath of actin is present in the founder cell (not shown). (E) One, or more, fusion pores form to allow mixing of cytoplasmic contents. (F) Expansion of the fusion pore(s) and elimination of membrane separating the cells. (G) The FCM is absorbed into the myotube, and the resulting syncytium continues additional rounds of fusion as needed.
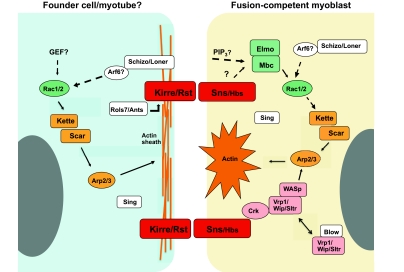
Fig. 5.
Genes and pathways associated with myoblast fusion in Drosophila. The indicated genes and pathways correspond to those, as discussed in the text, that appear to function in founder cells/myotubes and fusion-competent myoblasts of Drosophila embryos. The represented proteins include those for which a role in fusion has been shown experimentally. Generally, these comprise components of the Rac1, Scar and WASp pathways and their regulators, cell-adhesion molecules, and essential proteins such as Sing, for which a mechanistic role remains to be elucidated. In most instances, demonstration of an involvement in myoblast fusion has been established by direct loss-of-function studies experimentally. For some proteins, a role is inferred by biochemical interaction with known fusion proteins. Relationships indicated by broken lines are based on protein functions in other tissues, but have not yet been established in this system. Arp2/3, Actin-related protein 2/3; Ants, Antisocial; Arf6, ADP ribosylation factor 6; Blow, Blown Fuse; Crk, CT10 regulator of Kinase; Elmo, Engulfment and cell motility protein; GEF, Guanine nucleotide exchange factor; Hbs, Hibris; Kirre, Kin-of IrreC; Mbc, Myoblast city; PIP3, Phosphatidylinositol (3,4,5) triphosphate; Rac1, Ras-related C3 botulinum toxin substrate 1; Rst, Roughest; Sltr, Solitary; Rols7, Rolling pebbles isoform 7; Scar, Suppressor of cAMP receptor; Sing, Singles-bar; Sns, Sticks and stones; Vrp1, Verprolin 1; WASp, Wiscott-Aldrich syndrome protein; Wip, Drosophila WASp-interacting protein.
Candidate proteins with roles in migration of other cell types, that are essential for myoblast fusion, include the small GTPase Ras-related C3 botulinum toxin substrate 1 (Rac1) and its non-conventional guanine nucleotide exchange factor complex Myoblast City and Engulfment and Cell Motility Protein (Mbc/Elmo) (Erickson et al., 1997; Duchek et al., 2001; Geisbrecht et al., 2008). The Suppressor of cAMP Receptor/WASp family Verprolin-homologous (Scar/Wave) pathway, which is activated by Rac1 and plays a role in migration of other systems (see Box 3), is another candidate. Consistent with a role for Mbc and activated Rac1 in migration, the FCMs of embryos lacking Mbc or the combination of Rac1, Rac2 and Mig-2-like (Mtl) are more rounded and loosely associated with each other (Gildor et al., 2009). Scar is improperly localized in these mutant cells, suggesting an inability to activate Actin-related protein 2/3 (Arp2/3; Arp66B – FlyBase) and F-actin-dependent migration (Gildor et al., 2009). However, Scar is also improperly localized in embryos mutant for kette (Hem – FlyBase), a component of the Scar complex, but the FCMs of kette mutant embryos are still able to migrate to sites of ectopic Kirre expression (Rochlin et al., 2010) and electron-dense plaques characteristic of sites of fusion are present in these mutants (Schröter et al., 2004). Rac1 has also been implicated in stabilization of cell-cell contacts in vertebrates (Yamazaki et al., 2007), providing an alternative explanation for the loosely associated morphology of the FCMs in rac1 mutant embryos. Thus, the precise pathways regulating migration of myoblasts in flies remain to be clarified.
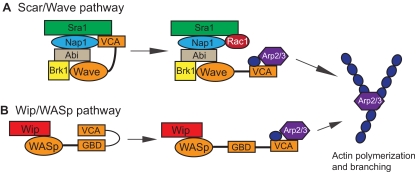
Box 3. The conserved Scar/Wave and Vrp1/WASp pathways for Arp2/3 activation
The Actin-related protein 2/3 (Arp2/3) complex is a conserved mediator of actin polymerization. It controls the formation of branched actin networks by binding to pre-existing filaments and promoting formation of new filaments by branching. This complex is activated by actin nucleation-promoting factors (NPFs) that include Suppressor of cAMP Receptor/WASp family Verprolin-homologous (Scar/Wave) (Scar in Drosophila; Wave in vertebrates) and Wiskott-Aldrich syndrome protein (WASp) (Machesky and Insall, 1998; Padrick et al., 2011). Scar/Wave exists in an inactive complex with Abi (Abl interactor protein), Kette/Nck-associated protein 1 (Nap1), Sra1 (Specifically Rac1-associated protein) and Brk1/Hspc300 (Breast tumor kinase/hematopoietic stem/progenitor cell protein 300) (Eden et al., 2002; Derivery and Gautreau, 2010; Kurisu and Takenawa, 2010) (see A). In this complex, the C-terminal verprolin central acidic (VCA) domain is blocked by interaction with Abi and Kette/Nap1 (Kim et al., 2000). Activation occurs upon binding of the complex to the small GTPase Rac1, releasing the VCA domain to bind to, and activate, the Arp2/3 complex (Kobayashi et al., 1998; Lebensohn and Kirschner, 2009). Some studies have suggested that Rac1 disrupts binding of the inhibitory Abi and Kette/Nap1 proteins (Ismail et al., 2009). However, more recent studies indicate that activated Rac1 alters the conformation of the Scar/Wave complex but does not physically disrupt it (Chen et al., 2010; Derivery and Gautreau, 2010). Arf GTPases may also activate Scar/Wave (Koronakis et al., 2011). The WASp NPF (see B) also activates the Arp2/3 complex and is present in the cell in an inactive state. Vrp1/Wip functions to stabilize WASp, protect it from degradation and contribute to its activation (Martinez-Quiles et al., 2001; Chou et al., 2006; Anton et al., 2007; de la Fuente et al., 2007). Similar to Scar/Wave, WASp is activated by protein binding to the autoinhibitory GTPase-binding domain (GBD), thereby releasing the VCA domain. Vrp1/Wip is also important for translocation of WASp to sites of actin polymerization.
The Kirre, Rst and Sns IgSF proteins mentioned above, with Kirre and Rst serving redundant functions, control cell-cell adhesion between founder cells and FCMs. Hibris (Hbs), a paralog of Sns (Artero et al., 2001; Dworak et al., 2001) can direct a small amount of fusion in the absence of Sns, but is quite inefficient by comparison (Shelton et al., 2009). Whereas flies lacking Hbs are viable, muscle fibers are absent in sns mutant embryos and these embryos do not hatch as a result (Bour et al., 2000). Non-muscle Drosophila S2 cells have been valuable in dissecting the specificity of adhesive interactions between these proteins (Dworak et al., 2001; Galletta et al., 2004). Sns can interact biochemically with Rst or Kirre, in trans to mediate cell adhesion (Galletta et al., 2004). Consistent with the need for FCMs to recognize founder cells and myotubes, but not other FCMs, neither Sns nor Hbs mediates homotypic interaction of S2 cells in trans. Although Kirre and Rst can interact, cells expressing these proteins have a strong preference for those expressing Sns, possibly accounting for the apparent absence of founder cell/founder cell interactions in vivo. No other cell-adhesion molecules have been implicated in the adhesion process. Studies examining the loss of integrins maternally and zygotically have revealed no defects in myoblast fusion (Zusman et al., 1993; Roote and Zusman, 1995; Prokop et al., 1998), and no studies have yet revealed a role for Ca2+-dependent adhesion molecules. It is particularly notable that muscles are normal in embryos lacking N-cadherin and N2-cadherin (Iwai et al., 1997; Prakash et al., 2005), the closest proteins to mammalian M-cadherin, for which a role in mammalian myoblast fusion is well established (see below). However, it remains to be determined whether maternally provided N-cadherin masks its role in embryonic myoblast fusion, or whether another cadherin functions redundantly with N-cadherin.
Myoblast migration, recognition and adhesion in mice
Most of our knowledge about migration of mouse myoblasts arises from in vitro studies, owing to the difficulty of studying this process in vivo. The basic processes that regulate myoblast migration can be carefully controlled and dissected in vitro, and probably reflect similar mechanisms occurring in vivo. Time-lapse analyses in vitro indicate that freshly isolated satellite cells migrate extensively on their associated myofibers, consistent with their expression of receptors for chemoregulatory molecules (Siegel et al., 2009). Furthermore, myoblasts cultured on various artificial substrates are also motile and migrate in response to a variety of factors, including chemokines (Corti et al., 2001; Odemis et al., 2007; Griffin et al., 2010), growth factors (Robertson et al., 1993a; Bischoff, 1997; Lee et al., 1999; Corti et al., 2001; Villena and Brandan, 2004) and other molecules (Nedachi et al., 2009; Tokura et al., 2011). Except for Cxcr4 [chemokine (C-X-C motif) receptor 4], the receptor for the chemokine Sdf1 (stromal-derived factor 1; Cxcl12 – Mouse Genome Informatics) (Griffin et al., 2010), the effect of depleting these molecules on cell fusion has not been tested. Cell motility changes greatly during differentiation, with myocytes exhibiting less motility than myoblasts (Powell, 1973; Griffin et al., 2010) and becoming unresponsive to several potent inducers of myoblast migration, such as hepatocyte growth factor (Hgf) and platelet-derived growth factor (Pdgf) (Griffin et al., 2010). This decrease in motility would increase the probability of cell-cell contact, thereby triggering differentiation (Krauss et al., 2005) and allowing myocytes to fuse with one another and with nascent myotubes (Nowak et al., 2009). As myotubes begin to form, muscle cells preferentially move into some fields and out of others (Chazaud et al., 1998), suggesting that migration is directed in response to chemotactic factors secreted by muscle cells (Bondesen et al., 2007; Griffin et al., 2010).
In vitro approaches have revealed details of how some regulatory factors influence muscle cell migration. Some factors modulate the velocity or direction of cell migration, whereas others regulate the clearance of the extracellular matrix at the leading edge of migrating cells, thereby enhancing cell motility (Horsley et al., 2003; Jansen and Pavlath, 2006; Lafreniere et al., 2006; Griffin et al., 2009). Myoblast fusion in vitro is enhanced by both positive regulators [e.g. CD164 (Bae et al., 2008), interleukin 4 (Il4) (Horsley et al., 2003; Lafreniere et al., 2006), mannose receptor (MR) (Jansen and Pavlath, 2006) and mouse odorant receptor 23 (Mor23; Olfr16 – Mouse Genome Informatics) (Griffin et al., 2009)] and negative regulators [e.g. prostacyclin (Bondesen et al., 2007)] of cell migration. Whereas positive migratory factors promote cell fusion by increasing the probability of myoblasts being close to one another, negative migratory factors may enhance cell fusion by acting as a ‘brake’ on migrating cells to facilitate cell-cell contact and adhesion (Bondesen et al., 2007). Thus, the net balance between these two classes of migratory regulators would be crucial for modulating myoblast fusion.
Myoblast migration in mice in vivo has mostly been studied during the course of development (Dietrich, 1999; Birchmeier and Brohmann, 2000; Christ and Brand-Saberi, 2002) where myogenic cells often migrate long distances to form muscles in the limb and tongue. Hepatocyte growth factor and the Cxcr4/Sdf1 axis are crucial regulators of myoblast migration in mouse embryos (Brand-Saberi et al., 1996; Bandow et al., 2004; Vasyutina et al., 2005). Until recently, the ability of myoblasts to migrate in adult muscle was inferred from studies using transplantation of either genetically marked myoblasts (Watt et al., 1987; Phillips et al., 1990; Watt et al., 1993; Jockusch and Voigt, 2003) or myofibers with their associated satellite cells (Hall et al., 2010), and histological examination of these genetic markers in fixed tissue sections. However, in vivo imaging of satellite cells in regenerating muscles provided the first direct evidence for myoblast migration in adult muscles (Ishido and Kasuga, 2011).
Cell-cell adhesion molecules and their role in muscle cell fusion in mice have been mostly studied using cell culture models with some experiments in regenerating adult muscles. Many more adhesion molecules have been identified to date in mice compared with flies. Nephrin, the vertebrate homolog of Sns, is the only adhesion molecule involved in fusion in both model systems (Sohn et al., 2009). Why adhesion in mouse muscle cells is controlled by a diversity of molecules is unknown, as is the interplay among these different molecules in preparing the cells for fusion. Such diversity may allow for activation of specific intracellular signaling pathways, such as Rac1 (Charrasse et al., 2007) or tyrosine kinases (Li et al., 2004), which are activated by cell-surface engagement of specific adhesion molecules and may be required for different molecular events required for cell fusion.
During differentiation in vitro, muscle cells extend lamellopodia and filopodia that contact neighboring muscle cells (Yoon et al., 2007; Mukai and Hashimoto, 2008; Mukai et al., 2009; Nowak et al., 2009; Stadler et al., 2010) and are sites for localized adhesion molecules (Abramovici and Gee, 2007; Mukai et al., 2009) and signaling molecules (Abramovici and Gee, 2007; Mukai and Hashimoto, 2008). These filopodia are reminiscent of axon growth cones and may be necessary for recognition of other muscle cells that are competent for fusion. In addition, filopodia may act as a zipper mechanism by which two cells are pulled in close contact to one another for eventual fusion (Abramovici and Gee, 2007).
Cell-contact sites are characterized by a number of molecular changes, both extracellular and intracellular, that facilitate recognition and adhesion. For example, molecules such as muscle-cadherin (M-cadherin), integrins and a disintegrin and metalloprotease 12 (Adam12) are commonly found localized at the contact sites in both contacting muscle cells (Cifuentes-Diaz et al., 1995; Schwander et al., 2003; Lafuste et al., 2005; Brzoska et al., 2006). M-cadherin function is required for myotube formation in vitro (Zeschnigk et al., 1995; Charrasse et al., 2006) but not in vivo (Hollnagel et al., 2002), suggesting that other cadherins or cell adhesion molecules may functionally compensate for the lack of M-cadherin. Eliminating the function of β1 integrin, α3 integrin and α9 integrin decreases myoblast fusion in vitro (Schwander et al., 2003; Lafuste et al., 2005; Brzoska et al., 2006) and in vivo (Schwander et al., 2003). Adam12 is a transmembrane protein that contains an integrin-binding site and can bind α9β1 integrin in muscle cells (Lafuste et al., 2005). Antisense oligonucleotides to Adam12 preferentially inhibit myoblast fusion with myotubes (Lafuste et al., 2005). Adhesion molecules at these cellular contact sites can be temporally regulated, as evidenced by N-cadherin localization at nascent intercellular contacts but not at established cell contacts (Abramovici and Gee, 2007). Cell-contact sites are further characterized by changes in the cell membranes. For example, phosphatidylserine (PS) is transiently exposed at these sites (van den Eijnde et al., 2001) and required for myotube formation, as inhibition of PS by annexin V inhibits fusion. Multiple roles for this transient exposure of PS have been proposed, including cell recognition and cell signaling. Additionally, lipid rafts containing cholesterol transiently accumulate at cell contact sites and are required for the accumulation of adhesion molecules (Mukai et al., 2009). Cholesterol is proposed to help maintain the proper rigidity of the lipid bilayers necessary for adhesion between the two myogenic cells. Finally, molecules that associate with the intracellular domains of specific cell-adhesion molecules, such as β-catenin (Vasyutina et al., 2009) and kindling 2 (Dowling et al., 2008), are also localized at cell contact sites. These intracellular molecules probably activate signal transduction pathways that ultimately lead to membrane fusion (Charrasse et al., 2007; Quach et al., 2009; Vasyutina et al., 2009). Molecules that regulate migration and adhesion in mouse muscle cells and for which a demonstrated role in contributing to myoblast fusion has been shown are summarized in Fig. 6 and Tables 1 and 2.
Fig. 6.
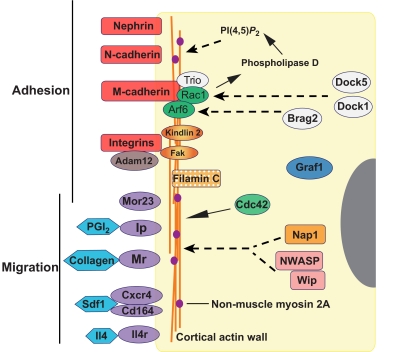
Genes and pathways associated with myoblast fusion in mice. The indicated proteins and pathways correspond to those, as discussed in the text, for which a role in fusion has been shown experimentally. Generally, these comprise cell-adhesion molecules and their associated adaptors, secreted molecules and their receptors that control cell migration, and molecules that signal to the actin cytoskeleton. For the most part, roles for these proteins have been identified through loss-of-function studies. Relationships indicated by broken lines are based on studies in other tissues, but have not yet been established in this system. Arf6, ADP ribosylation factor 6; Brag2, brefeldin A-resistant Arf GEF; CD164, cluster of differentiation 164; Cdc42, cell division cycle 42; Cxcr4, chemokine (C-X-C motif) receptor 4; Dock1 and Dock 5, dedicator of cytokinesis 1 and 5; Fak, focal adhesion kinase; Graf1, GTPase regulator associated with focal adhesion kinase; Il4, interleukin 4; Il4r, interleukin 4 receptor; Ip, prostacyclin receptor; Mr, mannose receptor; Mor23, mouse odorant receptor 23; Nap1, Nck-associated protein; NWASP, neural Wiskott-Aldrich syndrome protein; PGI2, prostacyclin; PI(4,5)P2, phosphatidylinositol 4,5-bisphosphate; Rac1, Ras-related C3 botulinum toxin substrate 1; SDF1, stromal-derived factor 1; Wip, WASp-interacting protein.
Membrane fusion and formation of the syncytium
Although the processes of migration, recognition and adhesion are the fundamental first steps in fusion, and perturbations in these processes lead to obvious defects in formation of myofibers, they are somewhat removed from the fusion process itself. Below, we discuss the genes, intracellular pathways and membrane events more intimately associated with the mechanics of myoblast fusion.
The process of myoblast fusion in the Drosophila embryo
A combination of approaches, including genetics, confocal microscopy, transmission electron microscopy (TEM) and biochemistry, has recently uncovered fundamental aspects of the fusion process itself and the molecules controlling this process. A hypothetical model describing these observations is provided in Fig. 4, with the known molecules shown in Fig. 5. Although several discrepancies remain in the morphological details, possibly as a consequence of the specific mutant alleles examined or the approach used, rapid progress is being made in deciphering the mechanism of myoblast fusion in this system.
From the membrane to the cytoplasm
Largely on the basis of genetic loss-of-function phenotypes associated with defects in myoblast fusion, many molecules have been identified that function downstream of the IgSF cell-surface receptors. The Sns cytodomain is essential for function, and redundant binding motifs probably link it to adaptor proteins that include the SH2-SH3 domain-containing protein CT10 regulator of kinase (Crk) (Kim et al., 2007). Crk, in turn, can recruit actin-polymerizing machinery to points of cell-cell contact [(Kim et al., 2007) see below]. The cytoplasmic domain of Kirre recruits both Rolling pebbles/Antisocial (Rols/Ants) (Chen and Olson, 2001; Menon and Chia, 2001; Rau et al., 2001), which contains numerous motifs for protein-protein interaction, and Schizo/Loner, a guanine nucleotide exchange factor (GEF) for the Arf6 GTPase (Arf51F – FlyBase) (Chen et al., 2003; Onel et al., 2004) to points of cell contact (Chen and Olson, 2001; Chen et al., 2003; Bulchand et al., 2010). Rols functions in a positive-feedback loop to stabilize Kirre at the myotube membrane, thereby ensuring its continued availability for interaction with FCMs (Menon et al., 2005). The finding that myoblasts separate in older embryos lacking rols is consistent with their inability to maintain adhesion (Rau et al., 2001). Though Rols can interact with Mbc biochemically, prompting the model that Rols functions to recruit Mbc to Kirre at sites of fusion (Chen and Olson, 2001), the relevance of this interaction is not clear, as Mbc does not appear to be required in the founder cells for fusion (Haralalka et al., 2011). Schizo/Loner might facilitate Arf-mediated recruitment of Rac1 to the membrane (Chen et al., 2003), probably resulting in activation of the Scar pathway by Rac1 (see Box 3). A Schizo-activated Arf GTPase might also, like Rac1, directly activate the Scar complex, as recently shown in mammalian cells (Koronakis et al., 2011). However, the cytoplasmic domain of Kirre is not absolutely essential for fusion (Bulchand et al., 2010), and so the mechanism by which Schizo/Loner is recruited to the membrane may be more complex. Moreover, Schizo is also present in the FCMs (Richardson et al., 2007), and may mediate the same processes in these cells independently of Kirre. Finally, as Arf GTPases are also associated with vesicle trafficking and membrane recycling (D’Souza-Schorey and Chavrier, 2006), the role of Schizo and its target Arf protein in myoblast fusion might also extend to other fusion-associated processes, as discussed below.
More than a decade ago, transmission electron microscopy (TEM) using conventional chemical fixation revealed distinct morphological features at sites of cell:cell contact, as depicted in Fig. 4. Electron-dense vesicles ∼40 nm in diameter are present at sites of myoblast contact in embryos. They appear to bud from the Golgi apparatus and become coated with actin (Kim et al., 2007). Though a causative role has not been established, these vesicles may give rise to membrane plaques along the apposed plasma membranes (Doberstein et al., 1997). Such vesicles and/or electron-dense membrane plaques have been observed in several subsequent studies and are perturbed in a variety of mutant backgrounds (Doberstein et al., 1997; Rau et al., 2001; Schröter et al., 2004; Estrada et al., 2007; Kim et al., 2007; Massarwa et al., 2007; Berger et al., 2008; Gildor et al., 2009) (S.A., unpublished). They are also reminiscent of the vesicles observed in cultured myoblasts (Shimada, 1971; Rash and Fambrough, 1973; Engel et al., 1986). Notably, the vesicles accumulate in embryos mutant for the MARVEL domain-containing protein Singles-bar (Sing), which is essential for myoblast fusion, suggesting that it may play a role in plaque formation (Estrada et al., 2007). The vesicles are thought to be recruited in response to activation of cell-surface receptors and to deliver fusion-associated molecules to points of cell contact, perhaps through targeted exocytosis mediated by the actin nucleation-promoting factor (NPF) Wiskott-Aldrich syndrome protein (WASp) (Kim et al., 2007). However, the exact role and composition of these vesicles remain unclear.
F-actin foci, F-actin nucleating complexes and fusion pores
A large body of work has established that actin polymerization plays a fundamental role in myoblast fusion in the fly embryo. Dynamic F-actin foci are formed and dissolve coincident with myoblast fusion in live embryos (Richardson et al., 2007), and actin foci are found at points of cell-cell contact in fixed tissue samples (Kesper et al., 2007; Richardson et al., 2007; Kim et al., 2007). Recent studies have shown that, in founder cells, F-actin forms a thin sheath underlying points of cell contact (Sens et al., 2010). By contrast, within the limits of confocal microscopy, the dense actin focus appears to reside exclusively in the FCMs (Sens et al., 2010; Haralalka et al., 2011), as depicted in Figs 4, 5 and 7. These F-actin foci are formed by the action of the NPFs Scar and WASp on the Arp2/3 complex as outlined below.
Fig. 7.
Protrusion of FCM in developing myotube. (A) A single confocal slice through the middle of a primary myoblast and its associated myofiber. Kirre protein (turquoise) and Sns protein (green) are highly enriched at the point of protrusion. Actin (red) is highly restricted to the fusion-competent myoblast, although faint actin fibers are visible in the myotube. A dotted line outlines the surface of the myotube and fusion-competent myoblast. Reproduced, with permission, from Haralalka et al. (Haralalka et al., 2011). (B) Schematic of the image shown in A.
The Scar and Kette subunits of the pentameric Scar complex (see Box 3) mediate actin polymerization via activation of Arp2/3 and are required for myoblast fusion (Schröter et al., 2004; Richardson et al., 2007; Berger et al., 2008; Gildor et al., 2009). Scar is required in both cell types, as expression in neither the founder cells nor the FCMs fully rescues the muscle defects of scar mutant embryos (Sens et al., 2010). The Kette subunit interacts genetically with the FCM-specific, PH-domain-containing Blown fuse (Blow) protein (Artero et al., 2001; Schröter et al., 2004). Interestingly, the actin foci are often larger in embryos mutant for scar or kette (Richardson et al., 2007), possibly owing to the number of FCMs that can simultaneously contact the founder cells in these mutants, or to less dense actin foci resembling those seen in the absence of Mbc, the Rac1 GEF (Haralalka et al., 2011). Thus, Kette may contribute in some way to organization of actin the foci and/or their breakdown prior to fusion. Interestingly, in this regard, mechanistic studies in vitro have shown that the actin cytoskeleton is an impediment to expansion of a fusion pore, consistent with this suggestion (Chen et al., 2008). Notably, the fusion plaques observed by transmission electron microscopy (TEM) (see below) are also larger in the absence of kette (Schröter et al., 2004).
The WASp NPF (Schäfer et al., 2007) and associated Verprolin1/Drosophila-WASp-interacting protein/Solitary (Vrp1/D-Wip/Sltr) protein (Paunola et al., 2002; Kim et al., 2007; Massarwa et al., 2007), also promote polymerization of F-actin at sites of cell-cell contact. In contrast to Scar, the Vrp1/WASp complex functions exclusively in the FCMs, owing to the FCM-specific expression of Vrp1 (Kim et al., 2007; Massarwa et al., 2007). Vrp1 can be recruited to Sns through interaction with the ubiquitously expressed SH2-SH3 adaptor protein Crk (Galletta et al., 1999) and, in turn, can recruit WASp to sites of myoblast contact coincident with Sns (Kim et al., 2007) (Fig. 5). Crk also binds to the FCM-specific Blow protein, and can recruit it to points of fusion (Jin et al., 2011). Blow functions in this location to modulate the stability of the Vrp1/WASp complex, by competing with WASp for Vrp1 binding (Jin et al., 2011). As in scar or kette mutants, embryos mutant for blow exhibit larger actin foci (Richardson et al., 2007; Jin et al., 2011). These large foci appear to reflect a slower exchange rate of G-actin, in addition to higher levels of Vrp1/WASp at points of cell-cell contact (Jin et al., 2011).
F-actin foci are nonetheless present in embryos mutant for WASp, vrp1, kette or scar (Kim et al., 2007; Richardson et al., 2007). Interestingly, embryos mutant for both vrp1 and scar exhibit more severe defects in myoblast fusion (Berger et al., 2008) and, with the strong sltr allele of vrp1 and absence of both maternal and zygotic scar, a significant reduction in the size of the actin foci (Sens et al., 2010). These data suggest functional redundancy between the NPFs. However, foci remain in embryos mutant for other combinations of NPF-associated proteins, including a strong allele of kette and WASp (Gildor et al., 2009). Thus, other actin polymerizing proteins may be present, or some of these mutant alleles require more detailed characterization. Despite the convergence of both WASp and Scar on Arp2/3, embryos zygotically null for Arp3 exhibit some fusion (Berger et al., 2008), possibly as a consequence of maternally provided protein that complicates this analysis.
Myoblast fusion is assumed to proceed from one or more small membrane pores that expand to engulf the fusing FCM. As assayed morphologically by conventional TEM or monitored by cytoplasmic transfer of GFP, myoblast membranes are intact and pores do not form in embryos lacking kette (Schröter et al., 2004; Gildor et al., 2009). Thus, the Scar pathway is essential for pore formation. By contrast, studies examining the role of Vrp1 and WASp in pore formation have resulted in somewhat disparate results. Using GFP to monitor the transfer of cytoplasm and TEM with conventional chemical fixation, studies have reported pore formation in vrp1 mutants and multiple gaps in the membrane of WASp mutant embryos (Massarwa et al., 2007; Gildor et al., 2009; Sens et al., 2010). Together, these data suggest that the Scar pathway mediates pore formation and that Vrp1/WASp functions subsequently in pore expansion (Massarwa et al., 2007; Gildor et al., 2009). By contrast, Kim and colleagues found no evidence of pores in vrp1 mutant embryos by cytoplasmic transfer or by TEM using high pressure freezing (Kim et al., 2007), thereby maintaining membrane integrity and avoiding membrane discontinuities associated with conventional fixation (Sens et al., 2010). These studies suggest that both NPFs are necessary for pore formation.
Molecular asymmetry and FCM-specific podosome-like membrane protrusions
The actin foci described above form in the center of a ring of Sns and Kirre in embryos and primary myoblasts (Kesper et al., 2007; Haralalka et al., 2011) (Figs 5, 7). Originally thought to be symmetric at points of cell contact (Haralalka and Abmayr, 2010; Rochlin et al., 2010), these dense actin foci or actin ‘plugs’ were suggested as the core of an adhesive, podosome-like structure termed the fusion restricted myogenic adhesive structure (FuRMAS) that colocalizes with the Rols and Blow proteins discussed earlier (Kesper et al., 2007; Onel, 2009; Onel et al., 2011). It is now clear that the foci are not symmetrically distributed between fusing partners but, instead, are very highly enriched in and possibly exclusive to the FCMs (Sens et al., 2010; Haralalka et al., 2011) (Figs 5, 7). These foci are part of a protrusive structure that pushes from the FCM into the developing founder cell or myotube, suggesting that fusion is a highly asymmetric event driven molecularly and morphologically in the FCMs (Sens et al., 2010; Chen, 2011; Haralalka et al., 2011; Sung and Weaver, 2011).
As evidenced by identification of the FCM-specific Vrp1/WASp complex (Kim et al., 2007; Massarwa et al., 2007) and by FCM-exclusive expression of Blow (Jin et al., 2011), studies have established the molecular asymmetry between the founder cells and FCMs. Recent studies have also shown that, despite ubiquitous expression (Erickson et al., 1997), the Rac1 GEF Mbc and active Rac1 are asymmetrically distributed in the FCMs at points of cell-cell contact (Haralalka et al., 2011). As mentioned earlier, Mbc is one subunit of a highly conserved bipartite guanine nucleotide exchange factor for the small GTPase Rac1 (Erickson et al., 1997; Balagopalan et al., 2006). Like its partner Cell death abnormality 12/Engulfment and cell motility protein (Ced12/Elmo) (Geisbrecht et al., 2008) and target Rac1 (Luo et al., 1994; Hakeda-Suzuki et al., 2002), Mbc is essential for myoblast fusion. It is required in the FCMs, but is not required in the founder cells for their fusion with these FCMs (Haralalka et al., 2011). Activated Rac1 is sufficient in the FCMs to rescue fusion in mbc mutant embryos (Haralalka et al., 2011), suggesting that the sole function of mbc is to activate Rac1 in these cells. As in other systems (Kobayashi et al., 1998; Lebensohn and Kirschner, 2009), the function of Rac1 may be to mediate the local activation of Scar prior to myoblast fusion (Fig. 5). Notably, in addition to its role in formation of the actin focus discussed earlier, Scar is also responsible for the thin sheath of actin present in the founder cells (Sens et al., 2010).
Given the high levels of Mbc and active Rac1 in the FCMs at sites of cell-cell contact, it remains to be determined how Scar is activated in the founder cells to generate this sheath (Sens et al., 2010), and whether undetectable levels of Rac1 are sufficient. Moreover, as Mbc is not required in the founder cells, a GEF other than Mbc must accomplish this goal (Haralalka et al., 2011). Interestingly, studies in mammalian cells have shown that Arf GTPases can, like Rac GTPases, also directly activate the Scar complex (Koronakis et al., 2011). Thus, perhaps the Arf GEF Schizo, described above, serves this purpose in the founder cells.
Analysis with high-pressure freezing TEM has revealed that sites of cell contact are also highly asymmetric morphologically, with finger-like projections from the FCM invading the syncytium (Sens et al., 2010). It seems likely that the dynamic F-actin foci seen in live embryos (Richardson et al., 2007; Sens et al., 2010; Haralalka et al., 2011) and in the actin core bounded by Sns and Kirre (Kesper et al., 2007; Sens et al., 2010; Haralalka et al., 2011) are associated with these invasive finger-like projections, which seem to be devoid of cytoplasmic content and filled with filaments (Sens et al., 2010). These protrusive structures precede the formation of fusion pores (Sens et al., 2010) and, like the WASp-dependent actin-rich podosomes defined in macrophages and osteoclasts (Murphy and Courtneidge, 2011), depend on the FCM-specific WASp-associated protein Vrp1 (Sens et al., 2010). Though membrane projections are present in embryos mutant for vrp1 or WASp (Sens et al., 2010), consistent with the presence of F-actin in these mutant embryos (Gildor et al., 2009), the finger-like projections appear to collapse, suggesting that WASp-dependent F-actin contributes to their rigidity but perhaps not their actual formation (Sens et al., 2010). Interestingly, studies in other systems have suggested that the protrusive activity associated with podosomes and invadopodia involves the coupling of actin polymerization with membrane deformation (Gimona et al., 2008; Murphy and Courtneidge, 2011). Moreover, WASp-dependent actin polymerization can be activated by proteins that cause membrane curvature, a feature commonly associated with destabilization of the membrane and membrane fusion (Albiges-Rizo et al., 2009). Thus, the mechanism responsible for initial formation of these invadopodia might involve unidentified membrane bending proteins at sites of fusion.
In contrast to the collapsed fingers in embryos mutant for the WASp complex noted above (Sens et al., 2010), the fingers are present and appear to be normal with regard to depth and rate of G-actin exchange in embryos lacking the pentameric Scar complex (Jin et al., 2011). These studies, in combination with additional studies described above that examine pore formation (Schröter et al., 2004; Kim et al., 2007; Gildor et al., 2009), suggest that Vrp1/WASp drives formation of the invasive fingers prior to pore formation, and that Vrp1/WASp and Kette/Scar function redundantly or sequentially in pore formation. Sens and colleagues (Sens et al., 2010) proposed that a single pore forms at the tip of one invasive projection and expands to engulf the FCM. Although these investigators have observed macro-pores ranging from 300 nm to 1.5 μm (Sens et al., 2010), smaller micropores have not been described. Thus, it remains to be determined whether fusion is accomplished by a single 25-50 μm micropore that expands too rapidly to be observed, or whether multiple micropores converge to form this large opening. Perhaps, as seen in mutations of other fusion systems (Gattegno et al., 2007), analysis of Drosophila mutants that block complete fusion will allow this issue to be addressed.
Relevant to this issue, TEM using conventional chemical fixation originally suggested the presence of multiple pores and extensive membrane vesiculation, and implicated several molecules in pore expansion (Doberstein et al., 1997; Massarwa et al., 2007; Berger et al., 2008). However, this membrane breakdown is not observed in samples processed by high-pressure freezing (Sens et al., 2010), leaving unresolved the issue of whether multiple pores can form and which genes mediate pore expansion.
Myoblast fusion in mammals
Much of the work in this area has been geared towards determining whether the specific molecules identified in flies that have a role in actin cytoskeletal rearrangements display conserved functions in mouse muscle cells in vitro and in vivo.
Actin cytoskeletal remodeling in mouse myoblasts
Extensive cytoskeletal reorganization occurs before and after fusion in cultured mouse myoblasts (Fulton et al., 1981). Visualization of the actin cytoskeleton revealed dynamic changes in fusing mouse myoblasts in vitro, as described for flies (Swailes et al., 2006; Duan and Gallagher, 2009; Nowak et al., 2009; Stadler et al., 2010). A dense actin wall forms in one cell, paralleling the long axis of aligned myoblasts (Duan and Gallagher, 2009), and is hypothesized to provide the membrane rigidity needed for cell fusion. As fusion proceeds, gaps appear in this actin wall at sites of vesicle accumulation, vesicles pair in both cells along the membrane, and fusion pores then form. Similar vesicles are also observed in regenerating muscles (Robertson et al., 1990). Non-muscle myosin 2A is associated with the actin wall and is required for its formation, as well as for the appearance of vesicles at the membrane and for myoblast fusion (Duan and Gallagher, 2009). In the absence of proper actin cytoskeletal remodeling, F-actin structures accumulate at the site of cell-cell contact and are correlated with a decrease in myoblast fusion (Nowak et al., 2009). Together, these studies demonstrate a functional role for the actin cytoskeleton in myoblast fusion in mice.
Signaling to the actin cytoskeleton
Similar to the situation in flies, actin regulatory molecules also play key roles in fusion of mouse myoblasts. Mutations in these proteins lead to defects in myoblast fusion in vitro and in vivo (Dalkilic et al., 2006; Pajcini et al., 2008; Vasyutina et al., 2009; O’Brien et al., 2000; Charrasse et al., 2007; Laurin et al., 2008). For example, the functions of several signaling molecules involved in actin dynamics in flies are evolutionarily conserved in mice. These include four GEFs: brefeldin A-resistant Arf GEF (Brag2; Iqsec1 – Mouse Genome Informatics); Dedicator of cytokinesis 1 and dedicator of cytokinesis 5 (Dock1 and Dock5); and Trio. Loss of either Brag2 (the mammalian ortholog of Schizo/Loner) or Dock1 (the mammalian ortholog of Mbc, also referred to as Dock180), results in myotubes with few nuclei in vitro (Pajcini et al., 2008). Furthermore, Dock1/Dock5 loss impairs primary myofiber formation during mouse embryonic development (Laurin et al., 2008). In addition, Trio is essential for secondary myofiber formation during development (O’Brien et al., 2000) as well as myoblast fusion in vitro (Dalkilic et al., 2006; Charrasse et al., 2007). Other signaling molecules with conserved functions in mouse myoblast fusion are the small GTPases Arf6 (Chen et al., 2003) and Rac1 (Charrasse et al., 2007; Vasyutina et al., 2009). Conserved roles also exist for actin regulatory molecules such as Nck-associated protein 1 (Nap1; Nckap1 – Mouse Genome Informatics) (the ortholog of Kette) (Nowak et al., 2009). Knockdown of Nap1 in cultured mouse myoblasts impairs cell fusion but not cell migration as observed in kette mutants in flies (Rochlin et al., 2010). Furthermore, defects in neural Wiskott-Aldrich syndrome protein [NWASP; Wasl – Mouse Genome Informatics (the ortholog of WASp) (Kim et al., 2007)] and Wip (Wipf – Mouse Genome Informatics; the ortholog of Vrp1/D-Wip/Sltr) (Kim et al., 2007) also lead to impaired cell fusion of cultured mouse myotubes.
Several molecules with a role in actin cytoskeletal changes and myoblast fusion have been studied only in mice. Among these are filamin C, a muscle-specific member of the filamin family, which is an actin crosslinking protein necessary for fusion of muscle cells with nascent myotubes in vitro as well as primary myofiber formation during development (Dalkilic et al., 2006). The small G-protein cell division cycle 42 homolog (Cdc42) also regulates myoblast fusion in vitro and in vivo, and is required to recruit F-actin and other molecules to cell-cell contact sites (Vasyutina et al., 2009). Recently, GTPase regulator associated with focal adhesion kinase (Graf1; Arhgap26 – Mouse Genome Informatics), a novel Rho-specific GTPase-activating protein that regulates actin remodeling, was demonstrated to be crucial for myoblast fusion (Doherty et al., 2011).
The best-described signaling pathway involved in actin dynamics in mice is dependent on M-cadherin. Engagement of M-cadherin mediates Rac1 activation via Trio (Charrasse et al., 2007). A multiprotein complex composed of M-cadherin, Rac1 and Trio exists during fusion along with Arf6 (Bach et al., 2010). Arf6 is responsible for formation of the M-cadherin/Rac1/Trio complex and regulates myoblast fusion through activation of phospholipase D and production of phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2] (Bach et al., 2010), a molecule that regulates actin cytoskeletal reorganization at the plasma membrane, vesicle trafficking and membrane curvature (Donaldson, 2008). Thus, although the cell-surface molecules involved in activation of fusion in mice and flies can differ, the downstream pathways engaged by these cell-surface receptors that impinge on the actin cytoskeleton are well conserved. Molecules with roles in signaling to and modification of the actin cytoskeleton are summarized in Fig. 6 and Tables 1 and 2.
Conclusions
One of the more unusual aspects of muscle differentiation, in contrast to other tissues, is the fusion of cells to form multinucleated muscle fibers. Research over the past two decades has revealed a striking convergence of genes and morphological events controlling this process in several model systems. Indeed, each of these systems brings with it significant strengths that have contributed to our progress. Drosophila embryos offer easier analysis of genetic mutants and genetic interactions. Morphological analysis of mutants in vivo using such methods as TEM and live imaging coupled with more universal biochemical methods have allowed us to examine mechanistic details. By contrast, mammalian tissue culture models allow us to examine and quantify the impact of genes affecting myoblast migration and adhesion, which can only be inferred in flies. Ultimately, as a fundamental goal of this research is to address defects in muscle development and muscle disease, the mouse provides a model system that better approximates humans. As such, mouse models more directly parallel myoblast fusion in humans, and its importance in regeneration in response to muscle aging and damage. In short, rapid advances in the last decade are, in part, the result of such a multipronged approach. Nevertheless, significant challenges remain for a full understanding of myoblast fusion. First and foremost is the existence and nature of the fusogen associated with formation of the fusion pore. Present models only infer the need to break down actin filaments that may be a barrier to integration of the myoblast into the myotube and/or accommodate recycling of membrane as fusion proceeds. The morphology and behavior of the cells themselves, and events at points of cell-cell contact, would benefit from advances in live-imaging on a rapid time scale with greater resolution. Finally, the mechanisms by which the process is regulated, preventing excess fusion, are poorly understood.
Additional questions remain unresolved from an evolutionary perspective. Among these is why mice, and probably humans, require so many additional molecules for processes such as migration and adhesion that precede the actual membrane fusion event. Certainly mammalian muscles are more complex (integrating both fast and slow fibers) and larger (requiring the fusion of many thousands of myoblasts in a single 30 cm myofiber). The myoblasts also migrate further distances than just one or two cell diameters. The scale of this process dwarfs that occurring in the fly, possibly accounting for additional complexity as well as redundancy. In this context, the increasing commonalities between flies and mice are surprising at the very least and, at best, an integrative approach using multiple model organisms will aid in identifying the fundamental basics of the process.
Acknowledgments
S.M.A. thanks Dr S. Haralalka for critical discussions and assistance in the writing this review.
Footnotes
Funding
S.M.A. is supported by the Stowers Institute for Medical Research and by the National Institutes of Heath. G.K.P. is supported by the National Institutes of Health and by the Muscular Dystrophy Association. Deposited in PMC for release after 12 months.
Competing interests statement
The authors declare no competing financial interests.
References
- Abramovici H., Gee S. H. (2007). Morphological changes and spatial regulation of diacylglycerol kinase-zeta, syntrophins, and Rac1 during myoblast fusion. Cell Motil. Cytoskel. 64, 549–567 [DOI] [PubMed] [Google Scholar]
- Albiges-Rizo C., Destaing O., Fourcade B., Planus E., Block M. R. (2009). Actin machinery and mechanosensitivity in invadopodia, podosomes and focal adhesions. J. Cell Sci. 122, 3037–3049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anton I. M., Jones G. E., Wandosell F., Geha R., Ramesh N. (2007). WASP-interacting protein (WIP): working in polymerisation and much more. Trends Cell Biol. 17, 555–562 [DOI] [PubMed] [Google Scholar]
- Arnold L., Henry A., Poron F., Baba-Amer Y., van Rooijen N., Plonquet A., Gherardi R. K., Chazaud B. (2007). Inflammatory monocytes recruited after skeletal muscle injury switch into antiinflammatory macrophages to support myogenesis. J. Exp. Med. 204, 1057–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artero R. D., Castanon I., Baylies M. K. (2001). The immunoglobulin-like protein Hibris functions as a dose-dependent regulator of myoblast fusion and is differentially controlled by Ras and Notch signaling. Development 128, 4251–4264 [DOI] [PubMed] [Google Scholar]
- Bach A. S., Enjalbert S., Comunale F., Bodin S., Vitale N., Charrasse S., Gauthier-Rouviere C. (2010). ADP-ribosylation factor 6 regulates mammalian myoblast fusion through phospholipase D1 and phosphatidylinositol 4,5-bisphosphate signaling pathways. Mol. Biol. Cell 21, 2412–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae G. U., Gaio U., Yang Y. J., Lee H. J., Kang J. S., Krauss R. S. (2008). Regulation of myoblast motility and fusion by the CXCR4-associated sialomucin, CD164. J. Biol. Chem. 283, 8301–8309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balagopalan L., Chen M., Geisbrecht E., Abmayr S. (2006). The CDM superfamily protein MBC directs myoblast fusion through a mechanism that requires phosphatidylinositol 3, 4, 5-triphosphate binding but is independent of direct interaction with DCrk. Mol. Cell. Biol. 26, 9442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandow K., Ohnishi T., Tamura M., Semba I., Daikuhara Y. (2004). Hepatocyte growth factor/scatter factor stimulates migration of muscle precursors in developing mouse tongue. J. Cell Physiol. 201, 236–243 [DOI] [PubMed] [Google Scholar]
- Bataillé L., Delon I., Da Ponte J. P., Brown N. H., Jagla K. (2010). Downstream of identity genes: muscle-type-specific regulation of the fusion process. Dev. Cell 19, 317–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bate M. (1990). The embryonic development of larval muscles in Drosophila. Development 110, 791–804 [DOI] [PubMed] [Google Scholar]
- Bate M. (1993). The mesoderm and its derivatives. In The Development of Drosophila melanogaster, Vol. II (ed. Bate M., Martinez-Arias A.), pp. 1013–1090 Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; [Google Scholar]
- Beckett K., Baylies M. K. (2007). 3D analysis of founder cell and fusion competent myoblast arrangements outlines a new model of myoblast fusion. Dev. Biol. 309, 113–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger S., Schafer G., Kesper D. A., Holz A., Eriksson T., Palmer R. H., Beck L., Klambt C., Renkawitz-Pohl R., Onel S. F. (2008). WASP and SCAR have distinct roles in activating the Arp2/3 complex during myoblast fusion. J. Cell Sci. 121, 1303–1313 [DOI] [PubMed] [Google Scholar]
- Birchmeier C., Brohmann H. (2000). Genes that control the development of migrating muscle precursor cells. Curr. Opin. Cell Biol. 12, 725–730 [DOI] [PubMed] [Google Scholar]
- Biressi S., Molinaro M., Cossu G. (2007a). Cellular heterogeneity during vertebrate skeletal muscle development. Dev. Biol. 308, 281–293 [DOI] [PubMed] [Google Scholar]
- Biressi S., Tagliafico E., Lamorte G., Monteverde S., Tenedini E., Roncaglia E., Ferrari S., Cusella-De Angelis M. G., Tajbakhsh S., Cossu G. (2007b). Intrinsic phenotypic diversity of embryonic and fetal myoblasts is revealed by genome-wide gene expression analysis on purified cells. Dev. Biol. 304, 633–651 [DOI] [PubMed] [Google Scholar]
- Bischoff R. (1997). Chemotaxis of skeletal muscle satellite cells. Dev. Dyn. 208, 505–515 [DOI] [PubMed] [Google Scholar]
- Bondesen B. A., Jones K. A., Glasgow W. C., Pavlath G. K. (2007). Inhibition of myoblast migration by prostacyclin is associated with enhanced cell fusion. FASEB J. 21, 3338–3345 [DOI] [PubMed] [Google Scholar]
- Bour B. A., O’Brien M. A., Lockwood W. L., Goldstein E. S., Bodmer R., Taghert P. H., Abmayr S. M., Nguyen H. T. (1995). Drosophila MEF2, a transcription factor that is essential for myogenesis. Genes Dev. 9, 730–741 [DOI] [PubMed] [Google Scholar]
- Bour B. A., Chakravarti M., West J. M., Abmayr S. M. (2000). Drosophila SNS, a member of the Immunoglobulin superfamily that is essential for myoblast fusion. Genes Dev. 14, 1498–1511 [PMC free article] [PubMed] [Google Scholar]
- Brand-Saberi B., Muller T. S., Wilting J., Christ B., Birchmeier C. (1996). Scatter factor/hepatocyte growth factor (SF/HGF) induces emigration of myogenic cells at interlimb level in vivo. Dev. Biol. 179, 303–308 [DOI] [PubMed] [Google Scholar]
- Bruusgaard J. C., Liestol K., Ekmark M., Kollstad K., Gundersen K. (2003). Number and spatial distribution of nuclei in the muscle fibres of normal mice studied in vivo. J. Physiol. 551, 467–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruusgaard J. C., Liestol K., Gundersen K. (2006). Distribution of myonuclei and microtubules in live muscle fibers of young, middle-aged, and old mice. J. Appl. Physiol. 100, 2024–2030 [DOI] [PubMed] [Google Scholar]
- Brzoska E., Bello V., Darribere T., Moraczewski J. (2006). Integrin alpha3 subunit participates in myoblast adhesion and fusion in vitro. Differentiation 74, 105–118 [DOI] [PubMed] [Google Scholar]
- Bulchand S., Menon S. D., George S. E., Chia W. (2010). The intracellular domain of Dumbfounded affects myoblast fusion efficiency and interacts with Rolling pebbles and Loner. PLoS ONE 5, e9374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter S., Karpati G. (1989). Segmental necrosis and its demarcation in experimental micropuncture injury of skeletal muscle fibers. J. Neuropathol. Exp. Neurol. 48, 154–170 [DOI] [PubMed] [Google Scholar]
- Charrasse S., Comunale F., Grumbach Y., Poulat F., Blangy A., Gauthier-Rouviere C. (2006). RhoA GTPase regulates M-cadherin activity and myoblast fusion. Mol. Biol. Cell 17, 749–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrasse S., Comunale F., Fortier M., Portales-Casamar E., Debant A., Gauthier-Rouviere C. (2007). M-cadherin activates Rac1 GTPase through the Rho-GEF trio during myoblast fusion. Mol. Biol. Cell 18, 1734–1743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chazaud B., Christov C., Gherardi R. K., Barlovatz-Meimon G. (1998). In vitro evaluation of human muscle satellite cell migration prior to fusion into myotubes. J. Muscle Res. Cell Motil. 19, 931–936 [DOI] [PubMed] [Google Scholar]
- Chen E. H. (2011). Invasive podosomes and myoblast fusion. In Current Topics in Membranes (ed. Chernomordik L. V., Kozlov M. M.), pp. 235–258 Amsterdam, The Netherlands: Elsevier; [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E. H., Olson E. N. (2001). Antisocial, an intracellular adaptor protein, is required for myoblast fusion in Drosophila. Dev. Cell 1, 705–715 [DOI] [PubMed] [Google Scholar]
- Chen E. H., Pryce B. A., Tzeng J. A., Gonzalez G. A., Olson E. N. (2003). Control of myoblast fusion by a guanine nucleotide exchange factor, loner, and its effector ARF6. Cell 114, 751–762 [DOI] [PubMed] [Google Scholar]
- Chen A., Leikina E., Melikov K., Podbilewicz B., Kozlov M. M., Chernomordik L. V. (2008). Fusion-pore expansion during syncytium formation is restricted by an actin network. J. Cell Sci. 121, 3619–3628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Borek D., Padrick S. B., Gomez T. S., Metlagel Z., Ismail A. M., Umetani J., Billadeau D. D., Otwinowski Z., Rosen M. K. (2010). Structure and control of the actin regulatory WAVE complex. Nature 468, 533–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou H.-C., Antón I. M., Holt M. R., Curcio C., Lanzardo S., Worth A., Burns S., Thrasher A. J., Jones G. E., Calle Y. (2006). WIP regulates the stability and localization of WASP to podosomes in migrating dendritic cells. Curr. Biol. 16, 2337–2344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christ B., Brand-Saberi B. (2002). Limb muscle development. Int. J. Dev. Biol. 46, 905–914 [PubMed] [Google Scholar]
- Cifuentes-Diaz C., Nicolet M., Alameddine H., Goudou D., Dehaupas M., Rieger F., Mege R. M. (1995). M-cadherin localization in developing adult and regenerating mouse skeletal muscle: possible involvement in secondary myogenesis. Mech. Dev. 50, 85–97 [DOI] [PubMed] [Google Scholar]
- Corti S., Salani S., Del Bo R., Sironi M., Strazzer S., D’Angelo M. G., Comi G. P., Bresolin N., Scarlato G. (2001). Chemotactic factors enhance myogenic cell migration across an endothelial monolayer. Exp. Cell Res. 268, 36–44 [DOI] [PubMed] [Google Scholar]
- D’Souza-Schorey C., Chavrier P. (2006). ARF proteins: roles in membrane traffic and beyond. Nat. Rev. Mol. Cell Biol. 7, 347–358 [DOI] [PubMed] [Google Scholar]
- Dalkilic I., Schienda J., Thompson T. G., Kunkel L. M. (2006). Loss of FilaminC (FLNc) results in severe defects in myogenesis and myotube structure. Mol. Cell. Biol. 26, 6522–6534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fuente M. A., Sasahara Y., Calamito M., Anton I. M., Elkhal A., Gallego M. D., Suresh K., Siminovitch K., Ochs H. D., Anderson K. C., et al. (2007). WIP is a chaperone for Wiskott-Aldrich syndrome protein (WASP). Proc. Natl. Acad. Sci. USA 104, 926–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derivery E., Gautreau A. (2010). Generation of branched actin networks: assembly and regulation of the N-WASP and WAVE molecular machines. BioEssays 32, 119–131 [DOI] [PubMed] [Google Scholar]
- Dietrich S. (1999). Regulation of hypaxial muscle development. Cell Tissue Res. 296, 175–182 [DOI] [PubMed] [Google Scholar]
- Doberstein S. K., Fetter R. D., Mehta A. Y., Goodman C. S. (1997). Genetic analysis of myoblast fusion: blown fuse is required for progression beyond the prefusion complex. J. Cell Biol. 136, 1249–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty J. T., Lenhart K. C., Cameron M. V., Mack C. P., Conlon F. L., Taylor J. M. (2011). Skeletal muscle differentiation and fusion are regulated by the BAR-containing Rho-GTPase-activating protein (Rho-GAP), GRAF1. J. Biol. Chem. 286, 25903–25921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson J. G. (2008). Arfs and membrane lipids: sensing, generating and responding to membrane curvature. Biochem. J. 414, e1–e2 [DOI] [PubMed] [Google Scholar]
- Dowling J. J., Vreede A. P., Kim S., Golden J., Feldman E. L. (2008). Kindlin-2 is required for myocyte elongation and is essential for myogenesis. BMC Cell Biol. 9, 36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan R., Gallagher P. J. (2009). Dependence of myoblast fusion on a cortical actin wall and nonmuscle myosin IIA. Dev. Biol. 325, 374–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan H., Skeath J. B., Nguyen H. T. (2001). Drosophila Lame duck, a novel member of the Gli superfamily, acts as a key regulator of myogenesis by controlling fusion-competent myoblast development. Development 128, 4489–4500 [DOI] [PubMed] [Google Scholar]
- Duchek P., Somogyi K., Jekely G., Beccari S., Rorth P. (2001). Guidance of cell migration by the Drosophila PDGF/VEGF receptor. Cell 107, 17–26 [DOI] [PubMed] [Google Scholar]
- Duxson M. J., Usson Y., Harris A. J. (1989). The origin of secondary myotubes in mammalian skeletal muscles: ultrastructural studies. Development 107, 743–750 [DOI] [PubMed] [Google Scholar]
- Dworak H. A., Charles M. A., Pellerano L. B., Sink H. (2001). Characterization of Drosophila hibris, a gene related to human nephrin. Development 128, 4265–4276 [DOI] [PubMed] [Google Scholar]
- Eden S., Rohatgi R., Podtelejnikov A. V., Mann M., Kirschner M. W. (2002). Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature 418, 790–793 [DOI] [PubMed] [Google Scholar]
- Engel L. C., Egar M. W., Przybylski R. J. (1986). Morphological characterization of actively fusing L6 myoblasts. Eur. J. Cell Biol. 39, 360–365 [PubMed] [Google Scholar]
- Enriquez J., Boukhatmi H., Dubois L., Philippakis A. A., Bulyk M. L., Michelson A. M., Crozatier M., Vincent A. (2010). Multi-step control of muscle diversity by Hox proteins in the Drosophila embryo. Development 137, 457–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson M. R. S., Galletta B. J., Abmayr S. M. (1997). Drosophila myoblast city encodes a conserved protein that is essential for myoblast fusion, dorsal closure and cytoskeletal organization. J. Cell Biol. 138, 589–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada B., Maeland A. D., Gisselbrecht S. S., Bloor J. W., Brown N. H., Michelson A. M. (2007). The MARVEL domain protein, Singles Bar, is required for progression past the pre-fusion complex stage of myoblast fusion. Dev. Biol. 307, 328–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasch M. (1999). Controls in patterning and diversification of somatic muscles during Drosophila embryogenesis. Curr. Opin. Genet. Dev. 9, 522–529 [DOI] [PubMed] [Google Scholar]
- Fulton A. B., Prives J., Farmer S. R., Penman S. (1981). Developmental reorganization of the skeletal framework and its surface lamina in fusing muscle cells. J. Cell Biol. 91, 103–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galletta B. J., Niu X. P., Erickson M. R., Abmayr S. M. (1999). Identification of a Drosophila homologue to vertebrate Crk by interaction with MBC. Gene 228, 243–252 [DOI] [PubMed] [Google Scholar]
- Galletta B. J., Chakravarti M., Banerjee R., Abmayr S. M. (2004). SNS: adhesive properties, localization requirements and ectodomain dependence in S2 cells and embryonic myoblasts. Mech. Dev. 121, 1455–1468 [DOI] [PubMed] [Google Scholar]
- Gattegno T., Mittal A., Valansi C., Nguyen K. C. Q., Hall D. H., Chernomordik L. V., Podbilewicz B. (2007). Genetic control of fusion pore expansion in the epidermis of Caenorhabditis elegans. Mol. Biol. Cell 18, 1153–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisbrecht E. R., Haralalka S., Swanson S. K., Florens L., Washburn M. P., Abmayr S. M. (2008). Drosophila ELMO/CED-12 interacts with Myoblast city to direct myoblast fusion and ommatidial organization. Dev. Biol. 314, 137–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gildor B., Massarwa R., Shilo B. Z., Schejter E. D. (2009). The SCAR and WASp nucleation-promoting factors act sequentially to mediate Drosophila myoblast fusion. EMBO Rep. 10, 1043–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimona M., Buccione R., Courtneidge S. A., Linder S. (2008). Assembly and biological role of podosomes and invadopodia. Curr. Opin. Cell Biol. 20, 235–241 [DOI] [PubMed] [Google Scholar]
- Griffin C. A., Kafadar K. A., Pavlath G. K. (2009). MOR23 promotes muscle regeneration and regulates cell adhesion and migration. Dev. Cell 17, 649–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin C. A., Apponi L. H., Long K. K., Pavlath G. K. (2010). Chemokine expression and control of muscle cell migration during myogenesis. J. Cell Sci. 123, 3052–3060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grounds M. D. (1987). Phagocytosis of necrotic muscle in muscle isografts is influenced by the strain, age, and sex of host mice. J. Pathol. 153, 71–82 [DOI] [PubMed] [Google Scholar]
- Hakeda-Suzuki S., Ng J., Tzu J., Dietzl G., Sun Y., Harms M., Nardine T., Luo L., Dickson B. J. (2002). Rac function and regulation during Drosophila development. Nature 416, 438–442 [DOI] [PubMed] [Google Scholar]
- Hall J. K., Banks G. B., Chamberlain J. S., Olwin B. B. (2010). Prevention of muscle aging by myofiber-associated satellite cell transplantation. Sci. Transl. Med. 2, 57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haralalka S., Abmayr S. M. (2010). Myoblast fusion in Drosophila. Exp. Cell Res. 316, 3007–3013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haralalka S., Shelton C., Cartwright H. N., Katzfey E., Janzen E., Abmayr S. M. (2011). Asymmetric Mbc, active Rac1 and F-actin foci in the fusion-competent myoblasts during myoblast fusion in Drosophila. Development 138, 1551–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollnagel A., Grund C., Franke W. W., Arnold H. H. (2002). The cell adhesion molecule M-cadherin is not essential for muscle development and regeneration. Mol. Cell. Biol. 22, 4760–4770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsley V., Jansen K. M., Mills S. T., Pavlath G. K. (2003). IL-4 acts as a myoblast recruitment factor during mammalian muscle growth. Cell 113, 483–494 [DOI] [PubMed] [Google Scholar]
- Hutcheson D. A., Zhao J., Merrell A., Haldar M., Kardon G. (2009). Embryonic and fetal limb myogenic cells are derived from developmentally distinct progenitors and have different requirements for beta-catenin. Genes Dev. 23, 997–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishido M., Kasuga N. (2011). In situ real-time imaging of the satellite cells in rat intact and injured soleus muscles using quantum dots. Histochem. Cell Biol. 135, 21–26 [DOI] [PubMed] [Google Scholar]
- Ismail A. M., Padrick S. B., Chen B., Umetani J., Rosen M. K. (2009). The WAVE regulatory complex is inhibited. Nat. Struct. Mol. Biol. 16, 561–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai Y., Usui T., Hirano S., Steward R., Takeichi M., Uemura T. (1997). Axon patterning requires DN-cadherin, a novel neuronal adhesion receptor, in the Drosophila embryonic CNS. Neuron 19, 77–89 [DOI] [PubMed] [Google Scholar]
- Jansen K. M., Pavlath G. K. (2006). Mannose receptor regulates myoblast motility and muscle growth. J. Cell Biol. 174, 403–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin P., Duan R., Luo F., Zhang G., Hong S. N., Chen E. H. (2011). Competition between Blown Fuse and WASP for WIP binding regulates the dynamics of WASP-dependent actin polymerization in vivo. Dev. Cell 20, 623–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jockusch H., Voigt S. (2003). Migration of adult myogenic precursor cells as revealed by GFP/nLacZ labelling of mouse transplantation chimeras. J. Cell Sci. 116, 1611–1616 [DOI] [PubMed] [Google Scholar]
- Kelly A. M., Zacks S. I. (1969). The histogenesis of rat intercostal muscle. J. Cell Biol. 42, 135–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesper D. A., Stute C., Buttgereit D., Kreiskother N., Vishnu S., Fischbach K. F., Renkawitz-Pohl R. (2007). Myoblast fusion in Drosophila melanogaster is mediated through a fusion-restricted myogenic-adhesive structure (FuRMAS). Dev. Dyn. 236, 404–415 [DOI] [PubMed] [Google Scholar]
- Kim A. S., Kakalis L. T., Abdul-Manan N., Liu G. A., Rosen M. K. (2000). Autoinhibition and activation mechanisms of the Wiskott-Aldrich syndrome protein. Nature 404, 151–158 [DOI] [PubMed] [Google Scholar]
- Kim S., Shilagardi K., Zhang S., Hong S. N., Sens K. L., Bo J., Gonzalez G. A., Chen E. H. (2007). A critical function for the actin cytoskeleton in targeted exocytosis of prefusion vesicles during myoblast fusion. Dev. Cell 12, 571–586 [DOI] [PubMed] [Google Scholar]
- Kitityakara A., Angevine D. M. (1963). A study of the pattern of postembryonic growth of M. gracilis in mice. Dev. Biol. 8, 322–340 [DOI] [PubMed] [Google Scholar]
- Kobayashi K., Kuroda S., Fukata M., Nakamura T., Nagase T., Nomura N., Matsuura Y., Yoshida-Kubomura N., Iwamatsu A., Kaibuchi K. (1998). p140Sra-1 (specifically Rac1-associated protein) is a novel specific target for Rac1 small GTPase. J. Biol. Chem. 273, 291–295 [DOI] [PubMed] [Google Scholar]
- Kocherlakota K. S., Wu J. M., McDermott J., Abmayr S. M. (2008). Analysis of the cell adhesion molecule Sticks-and-Stones reveals multiple redundant functional domains, protein-interaction motifs and phosphorylated tyrosines that direct myoblast fusion in Drosophila melanogaster. Genetics 178, 1371–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koronakis V., Hume P. J., Humphreys D., Liu T., Hørning O., Jensen O. N., McGhie E. J. (2011). WAVE regulatory complex activation by cooperating GTPases Arf and Rac1. Proc. Natl. Acad. Sci. USA 108, 14449–14454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss R. S., Cole F., Gaio U., Takaesu G., Zhang W., Kang J. S. (2005). Close encounters: regulation of vertebrate skeletal myogenesis by cell-cell contact. J. Cell Sci. 118, 2355–2362 [DOI] [PubMed] [Google Scholar]
- Kurisu S., Takenawa T. (2010). WASP and WAVE family proteins: friends or foes in cancer invasion? Cancer Sci. 101, 2093–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafreniere J. F., Mills P., Bouchentouf M., Tremblay J. P. (2006). Interleukin-4 improves the migration of human myogenic precursor cells in vitro and in vivo. Exp. Cell Res. 312, 1127–1141 [DOI] [PubMed] [Google Scholar]
- Lafuste P., Sonnet C., Chazaud B., Dreyfus P. A., Gherardi R. K., Wewer U. M., Authier F. J. (2005). ADAM12 and alpha9beta1 integrin are instrumental in human myogenic cell differentiation. Mol. Biol. Cell 16, 861–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurin M., Fradet N., Blangy A., Hall A., Vuori K., Cote J. F. (2008). The atypical Rac activator Dock180 (Dock1) regulates myoblast fusion in vivo. Proc. Natl. Acad. Sci. USA 105, 15446–15451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebensohn A. M., Kirschner M. W. (2009). Activation of the WAVE complex by coincident signals controls actin assembly. Mol. Cell 36, 512–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. K., Wong C. C., Webb S. E., Tang M. K., Leung A. K., Kwok P. F., Cai D. Q., Chan K. M. (1999). Hepatocyte growth factor stimulates chemotactic response in mouse embryonic limb myogenic cells in vitro. J. Exp. Zool. 283, 170–180 [DOI] [PubMed] [Google Scholar]
- Lepper C., Fan C. M. (2010). Inducible lineage tracing of Pax7-descendant cells reveals embryonic origin of adult satellite cells. Genesis 48, 424–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepper C., Partridge T. A., Fan C. M. (2011). An absolute requirement for Pax7-positive satellite cells in acute injury-induced skeletal muscle regeneration. Development 138, 3639–3646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Lemay S., Aoudjit L., Kawachi H., Takano T. (2004). SRC-family kinase Fyn phosphorylates the cytoplasmic domain of nephrin and modulates its interaction with podocin. J. Am. Soc. Nephrol. 15, 3006–3015 [DOI] [PubMed] [Google Scholar]
- Luo L., Liao Y. J., Jan L. Y., Jan Y. N. (1994). Distinct morphogenetic functions of similar small GTPases: Drosophila Drac1 is involved in axonal outgrowth and myoblast fusion. Genes Dev. 8, 1787–1802 [DOI] [PubMed] [Google Scholar]
- Lyons G. E., Ontell M., Cox R., Sassoon D., Buckingham M. (1990). The expression of myosin genes in developing skeletal muscle in the mouse embryo. J. Cell Biol. 111, 1465–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky L. M., Insall R. H. (1998). Scar1 and the related Wiskott-Aldrich syndrome protein, WASP, regulate the actin cytoskeleton through the Arp2/3 complex. Curr. Biol. 8, 1347–1356 [DOI] [PubMed] [Google Scholar]
- Martinez-Quiles N., Rohatgi R., Antón I. M., Medina M., Saville S. P., Miki H., Yamaguchi H., Takenawa T., Hartwig J. H., Geha R. S., et al. (2001). WIP regulates N-WASP-mediated actin polymerization and filopodium formation. Nat. Cell Biol. 3, 484–491 [DOI] [PubMed] [Google Scholar]
- Massarwa R., Carmon S., Shilo B. Z., Schejter E. D. (2007). WIP/WASp-based actin-polymerization machinery is essential for myoblast fusion in Drosophila. Dev. Cell 12, 557–569 [DOI] [PubMed] [Google Scholar]
- McLennan I. S. (1996). Degenerating and regenerating skeletal muscles contain several subpopulations of macrophages with distinct spatial and temporal distributions. J. Anat. 188, 17–28 [PMC free article] [PubMed] [Google Scholar]
- Menon S. D., Chia W. (2001). Drosophila rolling pebbles: a multidomain protein required for myoblast fusion that recruits D-Titin in response to the myoblast attractant Dumbfounded. Dev. Cell 1, 691–703 [DOI] [PubMed] [Google Scholar]
- Menon S. D., Osman Z., Chenchill K., Chia W. (2005). A positive feedback loop between Dumbfounded and Rolling pebbles leads to myotube enlargement in Drosophila. J. Cell Biol. 169, 909–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai A., Hashimoto N. (2008). Localized cyclic AMP-dependent protein kinase activity is required for myogenic cell fusion. Exp. Cell Res. 314, 387–397 [DOI] [PubMed] [Google Scholar]
- Mukai A., Kurisaki T., Sato S. B., Kobayashi T., Kondoh G., Hashimoto N. (2009). Dynamic clustering and dispersion of lipid rafts contribute to fusion competence of myogenic cells. Exp. Cell Res. 315, 3052–3063 [DOI] [PubMed] [Google Scholar]
- Murphy D. A., Courtneidge S. A. (2011). The ins and outs of podosomes and invadopodia: characteristics, formation and function. Nat. Rev. Mol. Cell Biol. 12, 413–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy M. M., Lawson J. A., Mathew S. J., Hutcheson D. A., Kardon G. (2011). Satellite cells, connective tissue fibroblasts and their interactions are crucial for muscle regeneration. Development 138, 3625–3637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedachi T., Hatakeyama H., Kono T., Sato M., Kanzaki M. (2009). Characterization of contraction-inducible CXC chemokines and their roles in C2C12 myocytes. Am. J. Physiol. Endocrinol. Metab. 297, E866–E878 [DOI] [PubMed] [Google Scholar]
- Nowak S. J., Nahirney P. C., Hadjantonakis A. K., Baylies M. K. (2009). Nap1-mediated actin remodeling is essential for mammalian myoblast fusion. J. Cell Sci. 122, 3282–3293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien S. P., Seipel K., Medley Q. G., Bronson R., Segal R., Streuli M. (2000). Skeletal muscle deformity and neuronal disorder in Trio exchange factor-deficient mouse embryos. Proc. Natl. Acad. Sci. USA 97, 12074–12078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odemis V., Boosmann K., Dieterlen M. T., Engele J. (2007). The chemokine SDF1 controls multiple steps of myogenesis through atypical PKCzeta. J. Cell Sci. 120, 4050–4059 [DOI] [PubMed] [Google Scholar]
- Onel S. F. (2009). Actin regulators take the reins in Drosophila myoblast fusion. Cent. Eur. J. Biol. 4, 11–18 [Google Scholar]
- Onel S., Bolke L., Klambt C. (2004). The Drosophila ARF6-GEF Schizo controls commissure formation by regulating Slit. Development 131, 2587–2594 [DOI] [PubMed] [Google Scholar]
- Onel S., Dottermusch C., Sickmann A., Buttgereit D., Renkawitz-Pohl R. (2011). Role of the actin cytoskeleton within FuRMAS during Drosophila myoblast fusion and first functionally conserved factors in vertebrates. In Cell Fusions (ed. Larsson L. I.), pp. 139–170 Berlin, Germany: Springer; [Google Scholar]
- Ontell M., Kozeka K. (1984). Organogenesis of the mouse extensor digitorum logus muscle: a quantitative study. Am. J. Anat. 171, 149–161 [DOI] [PubMed] [Google Scholar]
- Padrick S. B., Doolittle L. K., Brautigam C. A., King D. S., Rosen M. K. (2011). Arp2/3 complex is bound and activated by two WASP proteins. Proc. Natl. Acad. Sci. USA 108, E472–E479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajcini K. V., Pomerantz J. H., Alkan O., Doyonnas R., Blau H. M. (2008). Myoblasts and macrophages share molecular components that contribute to cell-cell fusion. J. Cell Biol. 180, 1005–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadimitriou J. M., Robertson T. A., Mitchell C. A., Grounds M. D. (1990). The process of new plasmalemma formation in focally injured skeletal muscle fibers. J. Struct. Biol. 103, 124–134 [DOI] [PubMed] [Google Scholar]
- Paunola E., Mattila P. K., Lappalainen P. (2002). WH2 domain: a small, versatile adapter for actin monomers. FEBS Lett. 513, 92–97 [DOI] [PubMed] [Google Scholar]
- Pavlath G. K. (2010). Spatial and functional restriction of regulatory molecules during mammalian myoblast fusion. Exp. Cell Res. 316, 3067–3072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips G. D., Hoffman J. R., Knighton D. R. (1990). Migration of myogenic cells in the rat extensor digitorum longus muscle studied with a split autograft model. Cell Tissue Res. 262, 81–88 [DOI] [PubMed] [Google Scholar]
- Pimorady-Esfahani A., Grounds M. D., McMenamin P. G. (1997). Macrophages and dendritic cells in normal and regenerating murine skeletal muscle. Muscle Nerve 20, 158–166 [DOI] [PubMed] [Google Scholar]
- Powell J. A. (1973). Development of normal and genetically dystrophic mouse muscle in tissue culture. I. Prefusion and fusion activities of muscle cells: phase contrast and time lapse study. Exp. Cell Res. 80, 251–264 [DOI] [PubMed] [Google Scholar]
- Prakash S., Caldwell J. C., Eberl D. F., Clandinin T. R. (2005). Drosophila N-cadherin mediates an attractive interaction between photoreceptor axons and their targets. Nat. Neurosci. 8, 443–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokop A., Martín-Bermudo M. D., Bate M., Brown N. H. (1998). Absence of PS integrins or laminin A affects extracellular adhesion, but not intracellular assembly, of hemiadherens and neuromuscular junctions in Drosophila embryos. Dev. Biol. 196, 58–76 [DOI] [PubMed] [Google Scholar]
- Quach N. L., Biressi S., Reichardt L. F., Keller C., Rando T. A. (2009). Focal adhesion kinase signaling regulates the expression of caveolin 3 and beta1 integrin, genes essential for normal myoblast fusion. Mol. Biol. Cell 20, 3422–3435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rash J. E., Fambrough D. (1973). Ultrastructural and electrophysiological correlates of cell coupling and cytoplasmic fusion during myogenesis in vitro. Dev. Biol. 30, 166–186 [DOI] [PubMed] [Google Scholar]
- Rau A., Buttgereit D., Holz A., Fetter R., Doberstein S. K., Paululat A., Staudt N., Skeath J., Michelson A. M., Renkawitz-Pohl R. (2001). rolling pebbles (rols) is required in Drosophila muscle precursors for recruitment of myoblasts for fusion. Development 128, 5061–5073 [DOI] [PubMed] [Google Scholar]
- Richardson B. E., Beckett K., Nowak S. J., Baylies M. K. (2007). SCAR/WAVE and Arp2/3 are crucial for cytoskeletal remodeling at the site of myoblast fusion. Development 134, 4357–4367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson T. A., Grounds M. D., Mitchell C. A., Papadimitriou J. M. (1990). Fusion between myogenic cells in vivo: an ultrastructural study in regenerating murine skeletal muscle. J. Struct. Biol. 105, 170–182 [DOI] [PubMed] [Google Scholar]
- Robertson T. A., Maley M. A., Grounds M. D., Papadimitriou J. M. (1993a). The role of macrophages in skeletal muscle regeneration with particular reference to chemotaxis. Exp. Cell Res. 207, 321–331 [DOI] [PubMed] [Google Scholar]
- Robertson T. A., Papadimitriou J. M., Grounds M. D. (1993b). Fusion of myogenic cells to the newly sealed region of damaged myofibres in skeletal muscle regeneration. Neuropathol. Appl. Neurobiol. 19, 350–358 [DOI] [PubMed] [Google Scholar]
- Rochlin K. M., Yu S., Roy S., Baylies M. K. (2010). Myoblast fusion: when it takes more to make one. Dev. Biol. 341, 66–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roote C. E., Zusman S. (1995). Functions for PS integrins in tissue adhesion, migration, and shape changes during early embryonic development in Drosophila. Dev. Biol. 169, 322–336 [DOI] [PubMed] [Google Scholar]
- Ross J. J., Duxson M. J., Harris A. J. (1987). Formation of primary and secondary myotubes in rat lumbrical muscles. Development 100, 383–394 [DOI] [PubMed] [Google Scholar]
- Ruiz-Gomez M., Coutts N., Price A., Taylor M. V., Bate M. (2000). Drosophila Dumbfounded: a myoblast attractant essential for fusion. Cell 102, 189–198 [DOI] [PubMed] [Google Scholar]
- Ruiz-Gomez M., Coutts N., Suster M. L., Landgraf M., Bate M. (2002). myoblasts incompetent encodes a zinc finger transcription factor required to specify fusion-competent myoblasts in Drosophila. Development 129, 133–141 [DOI] [PubMed] [Google Scholar]
- Sambasivan R., Yao R., Kissenpfennig A., Van Wittenberghe L., Paldi A., Gayraud-Morel B., Guenou H., Malissen B., Tajbakhsh S., Galy A. (2011). Pax7-expressing satellite cells are indispensable for adult skeletal muscle regeneration. Development 138, 3647–3656 [DOI] [PubMed] [Google Scholar]
- Schäfer G., Weber S., Holz A., Bogdan S., Schumacher S., Müller A., Renkawitz-Pohl R., Onel S.-F. (2007). The Wiskott-Aldrich syndrome protein (WASP) is essential for myoblast fusion in Drosophila. Dev. Biol. 304, 664–674 [DOI] [PubMed] [Google Scholar]
- Schmalbruch H. (1976). The morphology of regeneration of skeletal muscles in the rat. Tissue Cell 8, 673–692 [DOI] [PubMed] [Google Scholar]
- Schröter R. H., Lier S., Holz A., Bogdan S., Klämbt C., Beck L., Renkawitz-Pohl R. (2004). kette and blown fuse interact genetically during the second fusion step of myogenesis in Drosophila. Development 131, 4501–4509 [DOI] [PubMed] [Google Scholar]
- Schwander M., Leu M., Stumm M., Dorchies O. M., Ruegg U. T., Schittny J., Muller U. (2003). Beta1 integrins regulate myoblast fusion and sarcomere assembly. Dev. Cell 4, 673–685 [DOI] [PubMed] [Google Scholar]
- Sens K. L., Zhang S., Jin P., Duan R., Zhang G., Luo F., Parachini L., Chen E. H. (2010). An invasive podosome-like structure promotes fusion pore formation during myoblast fusion. J. Cell Biol. 191, 1013–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelton C., Kocherlakota K. S., Zhuang S., Abmayr S. M. (2009). The immunoglobulin superfamily member Hbs functions redundantly with Sns in interactions between founder and fusion-competent myoblasts. Development 136, 1159–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada Y. (1971). Electron microscope observations on the fusion of chick myoblasts in vitro. J. Cell Biol. 48, 128–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel A. L., Atchison K., Fisher K. E., Davis G. E., Cornelison D. D. (2009). 3D timelapse analysis of muscle satellite cell motility. Stem Cells 27, 2527–2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simionescu A., Pavlath G. K. (2011). Molecular mechanisms of myoblast fusion across species. Adv. Exp. Med. Biol. 713, 113–135 [DOI] [PubMed] [Google Scholar]
- Snow M. H. (1977). Myogenic cell formation in regenerating rat skeletal muscle injured by mincing. I. A fine structural study. Anat. Rec. 188, 181–199 [DOI] [PubMed] [Google Scholar]
- Sohn R. L., Huang P., Kawahara G., Mitchell M., Guyon J., Kalluri R., Kunkel L. M., Gussoni E. (2009). A role for nephrin, a renal protein, in vertebrate skeletal muscle cell fusion. Proc. Natl. Acad. Sci. USA 106, 9274–9279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler B., Blattler T. M., Franco-Obregon A. (2010). Time-lapse imaging of in vitro myogenesis using atomic force microscopy. J. Microsc. 237, 63–69 [DOI] [PubMed] [Google Scholar]
- Strunkelnberg M., Bonengel B., Moda L. M., Hertenstein A., de Couet H. G., Ramos R. G., Fischbach K. F. (2001). rst and its paralogue kirre act redundantly during embryonic muscle development in Drosophila. Development 128, 4229–4239 [DOI] [PubMed] [Google Scholar]
- Sung B. H., Weaver A. (2011). Cell-cell fusion: a new function for invadosomes. Curr. Biol. 21, R121–R123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swailes N. T., Colegrave M., Knight P. J., Peckham M. (2006). Non-muscle myosins 2A and 2B drive changes in cell morphology that occur as myoblasts align and fuse. J. Cell Sci. 119, 3561–3570 [DOI] [PubMed] [Google Scholar]
- Tixier V., Bataillé L., Jagla K. (2010). Diversification of muscle types: recent insights from Drosophila. Exp. Cell Res. 316, 3019–3027 [DOI] [PubMed] [Google Scholar]
- Tokura Y., Nakayama Y., Fukada S., Nara N., Yamamoto H., Matsuda R., Hara T. (2011). Muscle injury-induced thymosin beta4 acts as a chemoattractant for myoblasts. J. Biochem. 149, 43–48 [DOI] [PubMed] [Google Scholar]
- van den Eijnde S. M., van den Hoff M. J., Reutelingsperger C. P., van Heerde W. L., Henfling M. E., Vermeij-Keers C., Schutte B., Borgers M., Ramaekers F. C. (2001). Transient expression of phosphatidylserine at cell-cell contact areas is required for myotube formation. J. Cell Sci. 114, 3631–3642 [DOI] [PubMed] [Google Scholar]
- Vasyutina E., Stebler J., Brand-Saberi B., Schulz S., Raz E., Birchmeier C. (2005). CXCR4 and Gab1 cooperate to control the development of migrating muscle progenitor cells. Genes Dev. 19, 2187–2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasyutina E., Martarelli B., Brakebusch C., Wende H., Birchmeier C. (2009). The small G-proteins Rac1 and Cdc42 are essential for myoblast fusion in the mouse. Proc. Natl. Acad. Sci. USA 106, 8935–8940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villena J., Brandan E. (2004). Dermatan sulfate exerts an enhanced growth factor response on skeletal muscle satellite cell proliferation and migration. J. Cell Physiol. 198, 169–178 [DOI] [PubMed] [Google Scholar]
- Watt D. J., Morgan J. E., Clifford M. A., Partridge T. A. (1987). The movement of muscle precursor cells between adjacent regenerating muscles in the mouse. Anat. Embryol. 175, 527–536 [DOI] [PubMed] [Google Scholar]
- Watt D. J., Karasinski J., England M. A. (1993). Migration of lacZ positive cells from the tibialis anterior to the extensor digitorum longus muscle of the X-linked muscular dystrophic (mdx) mouse. J. Muscle Res. Cell Motil. 14, 121–132 [DOI] [PubMed] [Google Scholar]
- Williams P. E., Goldspink G. (1971). Longitudinal growth of striated muscle fibres. J. Cell Sci. 9, 751–767 [DOI] [PubMed] [Google Scholar]
- Yamazaki D., Oikawa T., Takenawa T. Y. (2007). Rac-WAVE-mediated actin reorganization is required for organization and maintenance of cell-cell adhesion. J. Cell Sci. 120, 86–100 [DOI] [PubMed] [Google Scholar]
- Yoon S., Molloy M. J., Wu M. P., Cowan D. B., Gussoni E. (2007). C6ORF32 is upregulated during muscle cell differentiation and induces the formation of cellular filopodia. Dev. Biol. 301, 70–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeschnigk M., Kozian D., Kuch C., Schmoll M., Starzinski-Powitz A. (1995). Involvement of M-cadherin in terminal differentiation of skeletal muscle cells. J. Cell Sci. 108, 2973–2981 [DOI] [PubMed] [Google Scholar]
- Zhang M., McLennan I. S. (1995). During secondary myotube formation, primary myotubes preferentially absorb new nuclei at their ends. Dev. Dyn. 204, 168–177 [DOI] [PubMed] [Google Scholar]
- Zusman S., Grinblat Y., Yee G., Kafatos F. C., Hynes R. O. (1993). Analyses of PS integrin functions during Drosophila development. Development 118, 737–750 [DOI] [PubMed] [Google Scholar]