Abstract
Gene duplication has been proposed to drive the evolution of novel morphologies. After gene duplication, it is unclear whether changes in the resulting paralogs’ coding-regions, or in their cis-regulatory elements, contribute most significantly to the assembly of novel gene regulatory networks. The Transcription Factor Activator Protein 2 (Tfap2) was duplicated in the chordate lineage and is essential for development of the neural crest, a tissue that emerged with vertebrates. Using a tfap2-depleted zebrafish background, we test the ability of available gnathostome, agnathan, cephalochordate and insect tfap2 paralogs to drive neural crest development. With the exception of tfap2d (lamprey and zebrafish), all are able to do so. Together with expression analyses, these results indicate that sub-functionalization has occurred among Tfap2 paralogs, but that neo-functionalization of the Tfap2 protein did not drive the emergence of the neural crest. We investigate whether acquisition of novel target genes for Tfap2 might have done so. We show that in neural crest cells Tfap2 directly activates expression of sox10, which encodes a transcription factor essential for neural crest development. The appearance of this regulatory interaction is likely to have coincided with that of the neural crest, because AP2 and SoxE are not co-expressed in amphioxus, and because neural crest enhancers are not detected proximal to amphioxus soxE. We find that sox10 has limited ability to restore the neural crest in Tfap2-deficient embryos. Together, these results show that mutations resulting in novel Tfap2-mediated regulation of sox10 and other targets contributed to the evolution of the neural crest.
Keywords: Neural crest, Evolution, Tfap2, Transcription factor
INTRODUCTION
The neural crest (NC) and cranial placodes are present in all vertebrates (including the basal lamprey and hagfish) but are absent from the basal chordate amphioxus, suggesting that these structures were not present in the first chordates. Although recent work suggests that evolutionary precursors of these tissues were probably present in the proximate vertebrate ancestor, bona fide NC and a full complement of cranial placodes are nevertheless considered to be vertebrate synapomorphies (Canestro et al., 2003; Mazet and Shimeld, 2005). Derivatives of the NC and cranial placodes, including jaws and paired sensory structures, contribute to the predatory behavior of vertebrates, which is not exhibited by protochordates (Gans and Northcutt, 1983; Northcutt and Gans, 1983). These NC-derived features were key to the success of vertebrates, which adds interest to identifying the genomic changes that resulted in the emergence of this novel cell type.
The development of the NC is controlled by a gene regulatory network (GRN) composed of transcription factors and other regulatory molecules (Sauka-Spengler and Bronner-Fraser, 2008). Importantly, at least some regulatory interactions within the NC GRN must be evolutionarily novel because many of the transcription factors contributing to the NC GRN were recruited to the neural-plate border from other tissues (Meulemans and Bronner-Fraser, 2005). The evolution of novel regulatory interactions can result from mutations causing ‘protein neo-functionalization’, or those causing ‘regulatory neo-functionalization’. In the first scenario, mutations in protein-coding sequence would imbue an existing transcription factor with new functionality (e.g. new DNA-binding specificity or affinity for new co-factors), permitting it to make new regulatory connections. A long-standing hypothesis is that protein neo-functionalization is facilitated by gene duplication events, which would relieve constraints on transcription factors imposed by pleiotropy. Indeed, gene duplication followed by protein neo-functionalization has been proposed as a driving force in vertebrate evolution (Ohno, 1970). According to this model, duplicated proteins with initially identical biochemical functions diverge as one copy performs the ancestral functions and the other gains the ability to carry out additional functions (Wagner, 1998). There is support for this model because many examples of paralogous transcription factors with distinct functions have been documented (reviewed by Wagner and Lynch, 2008). However, it remains to be tested whether gene duplication followed by protein neo-functionalization contributed to the emergence of the NC GRN, or to any other vertebrate novelty.
In the alternative scenario, regulatory neo-functionalization, mutations lead to the appearance of a transcription factor’s cognate DNA-binding site in cis-regulatory sequences of a gene not previously regulated by that transcription factor. In many cases, a transcription factor from one species can replace the function of its ortholog in another species. This has often been shown in the context of evolutionarily conserved GRNs. For example, Pax6 function is conserved in the photoreceptor GRN across all phyla (Onuma et al., 2002). However, evidence for regulatory neo-functionalization requires a transcription factor from one species to replace functionally its ortholog in another species in an evolutionarily novel GRN. The best example to date is that Drosophila melanogaster SoxE (Sox100B) can partially replace vertebrate Sox10 in the NC GRN (Cossais et al., 2010); however, whether this example is typical or exceptional remains unclear.
The transcription factor Activator Protein 2 (Tfap2) family, members of which promote growth or differentiation in a variety of cell types (Eckert et al., 2005), presents an excellent opportunity to test whether the formation of one node in the NC GRN required protein neo-functionalization. This family is optimal for such a test because Tfap2 proteins have been duplicated in the chordate lineage, and because they are essential for the early steps of NC development (Hoffman et al., 2007; Li and Cornell, 2007; Luo et al., 2003). Our results support the model that sub-functionalization has contributed to retention of the Tfap2 paralogs in evolution. However, they do not support a role for protein neo-functionalization of Tfap2 in the assembly of the NC GRN. Instead of gaining novel protein function, Tfap2 acquired novel targets, including sox10 and other genes, and this contributed to the evolution of the NC.
MATERIALS AND METHODS
Fish maintenance
Zebrafish embryos and adults were reared as described previously (Westerfield, 1993). Embryos were staged by hours or days post-fertilization (hpf or dpf, respectively) at 28.5°C (Kimmel et al., 1995). Because of the poor effectiveness of tfap2a morpholinos (MOs) beyond 48 hpf (our unpublished observations), we used homozygous tfap2a mutants, generated from heterozygous adults harboring a presumed null allele of tfap2a (lockjaw, tfap2ats213) (Knight et al., 2003), in analyses of melanophores.
Generation of cDNAs for in situ hybridization and mis-expression experiments
First-strand cDNA was generated from total RNA extracted from zebrafish embryos at 24 hpf as described (O’Brien et al., 2004) or lamprey and amphioxus embryos/adults as described (Meulemans and Bronner-Fraser, 2002). Partial or full length cDNA clones were amplified (supplementary material Table S1). Digoxigenin (DIG)-labeled probes (Roche Diagnostics, Mannheim, Germany) were synthesized, and whole-mount in situ hybridization was performed following standard methods (Thisse and Thisse, 2008).
For mis-expression experiments, full-length coding regions of each zebrafish gene studied were amplified from a pool of 24 hpf first-strand cDNA. PCR products were cloned into pSCA (Stratagene/Agilent, Santa Clara, CA, USA), pCR4-TOPO (Invitrogen, Carlsbad, CA, USA) or pENTR/D-TOPO (Invitrogen) shuttling vectors, as indicated (supplementary material Table S1). Coding regions were then shuttled into pCS2+ either using standard cloning methods (pSCA and pCR4-TOPO) (Sambrook et al., 1989) or Gateway cloning using LR Clonase II (pENTR-D/TOPO, Invitrogen). All final plasmids were confirmed by sequencing. Plasmids were linearized and capped mRNA synthesized in vitro using the SP6 mMessage mMachine kit (Ambion, Austin, TX, USA) and concentrated using Microcon Spin Columns (Millipore, Billerica, MA, USA).
For forced expression of sox10, embryos were injected with hsp70:sox10 (Elworthy et al., 2003). At 80% epiboly, embryos were transferred to 37°C water and incubated at this temperature for 60 minutes then allowed to develop at room temperature.
Amphioxus soxE enhancer analysis
Regions of amphioxus genomic DNA proximal to soxE (see supplementary material Table S2 for genomic coordinates) were amplified and, with the exception of soxE promoter region 2, cloned into a tol-2 based GFP reporter vector described previously (Fisher et al., 2006). Several F0 embryos injected with construct containing the soxE region 4 were raised, and two independent stable transgenic strains were isolated that had identical GFP expression to transient transgenic embryos. The amphioxus soxE promoter region 2 was amplified and ligated into the sox10:eGFP plasmid (Wada et al., 2005) in place of the zebrafish sox10 element using standard cloning methods (Sambrook et al., 1989).
For analysis of conserved transcription factor binding sites the online tool ConSite (Sandelin et al., 2004) was used. Search parameters included a conservation cut-off of 38%, a window size of 50 and a TF score threshold of 80%.
Morpholinos and microinjection
Plasmids used for in vitro mRNA synthesis are described above. Plasmids and MOs were injected at the one-cell stage and mRNA at the two- to four-cell stage. Approximately 125 pg of tfap2 mRNAs, 125 pg of all plasmids and 5 ng of MOs were injected. MOs targeting tfap2a (tfap2a e2i2) and tfap2c (tfap2c e3i3) used here have been described (Li and Cornell, 2007). Supplementary material Table S1 contains sequences.
Chromatin immunoprecipitation (ChIP)
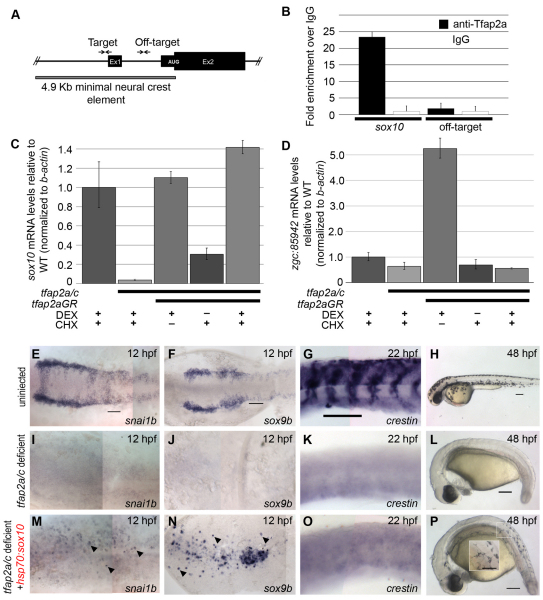
A modified ChIP protocol for fish embryos was used (Lindeman et al., 2009). Chromatin was sheared on ice using a probe-tip sonicator (VirTis Virsonic 600) with the following settings: power setting, 5; pulse time, 20 seconds; number of pulses, ten; break between pulses, 2 minutes. Anti-Tfap2a(CT) (Anaspec, Fremont, CA, USA) or Rabbit-IgG (Millipore) were used for immunoprecipitation. ChIP experiments were performed in triplicate, on newly isolated embryos. PCR reactions (10 μl) following ChIP were prepared with immunoprecipitated DNA, using the SYBR Green kit (Applied Biosystems, Foster City, CA, USA) following the manufacturer’s instructions. Primers were used at a final concentration of 200 nM in separate PCR reactions. Quantitative real-time PCR in Low 96-well plates (Bio-Rad, Hercules, CA, USA) was conducted using a Bio-Rad thermal cycler (CFX96 Real-Time PCR Detection System) following the default protocol; triplicates of each reaction were carried out simultaneously and mean and standard error were calculated. Melt-curve analysis was also performed to confirm specificity of primers. Figure 5B shows a representative run from one of the three repeats.
Fig. 5.
sox10 is directly regulated by Tfap2a in zebrafish embryos, and forced expression of sox10 can restore melanophores in tfap2a/c-deficient embryos. (A) Schematic of sox10 upstream element sufficient to drive NC-specific expression (Wada et al., 2005). Ex1, exon 1; Ex2, exon 2. Arrows indicate primers used to quantify results of ChIP experiments. (B) Representative quantitative real-time PCR (qPCR) results from anti-Tfap2a antibody ChIP conducted on lysates of 12 hpf zebrafish embryos (mean ± s.e.m. in technical replicates from a single representative experiment). In two additional biological replicates, qPCR revealed ∼12-fold enrichment at the target site with anti-Tfap2a relative to IgG, and minimal enrichment at the off-target site (data not shown). (C,D) Representative results of qRT-PCR analysis of sox10 (C) or the ednra homolog zgc:85942 (D) mRNA expression in lysates of embryos that were uninjected or injected with MOs targeting tfap2a and tfap2c and with tfap2a-GR mRNA, as indicated, and treated with cycloheximide (CHX) to inhibit protein translation, and with dexamethasone (DEX) to trigger nuclear localization of Tfap2a-GR, as indicated. The results are shown as fold-enrichment relative to levels in uninjected embryos treated with CHX/DEX, after normalization to b-actin. These experiments were performed three times with similar results observed (graphs represent mean ± s.e.m. of one repeat). (E-P) Dorsal (E,F,I,J,M,N) or lateral (G,H,K,L,O,P) views of embryos at the times indicated stage processed with the indicated marker. Embryos are either uninjected or injected with tfap2a and tfap2c MOs (or tfap2c MO injected into tfap2a mutant; K,L,O,P) and hsp70:sox10 plasmid, as indicated. All embryos exposed to heat-shock for 60 minutes at 90% epiboly. Rescue numbers: snai1b, uninjected, 20/23; tfap2a/c-deficient, 0/10; tfap2a/c-deficient plus hsp70:sox10, 19/27; sox9b, uninjected, 24/25; tfap2a/c-deficient, 0/10; tfap2a/c-deficient plus hsp70:sox10, 26/27; crestin, uninjected, 20/20; tfap2a/c-deficient, 0/5; tfap2a/c-deficient plus hsp70:sox10, 0/17; melanophores, uninjected, 59/59; tfap2a/c-deficient, 0/21; tfap2a/c-deficient plus hsp70:sox10,22/30. Embryos shown with anterior to the left, unless otherwise indicated. Scale bars: 100 μm.
Cycloheximide experiments
Embryos at 90% epiboly were incubated for ∼2 hours in 100 μg/ml cycloheximide (diluted in fish water from a 40 mg/ml stock dissolved in DMSO) (Sigma-Aldrich), a dose several times the level previously shown to strongly reduce protein translation in 1.5 hpf zebrafish embryos (Leung et al., 2003), or in diluted DMSO as a negative control. Subsequently, embryos were incubated for an additional 2.5 hours in dexamethasone (100 μM in fish water from a 40 mM stock dissolved in ethanol) (Sigma-Aldrich), or in diluted ethanol as a negative control. Next, RNA was isolated (Trizol, Invitrogen) following the manufacturer’s instructions and treated with DNaseI to remove genomic DNA. Quantitative reverse-transcriptase PCR (qRT-PCR) analysis of mRNA levels was conducted, starting with 1 μg of total RNA, as previously described (Van Otterloo et al., 2010). Primer efficiencies were confirmed using a tenfold series dilution of cDNA and generating a standard curve. All primers were designed to span a large intron, preventing amplification of genomic DNA. The 2ΔΔCt method was used to calculate fold-enrichment over wild-type samples and all groups were normalized to β-actin (Dussault and Pouliot, 2006). Each experiment was repeated in triplicate, starting from injections. Results are shown from a representative experiment.
Western blotting
Western blotting of zebrafish embryos was carried out essentially as previously described (Link et al., 2006). Antibodies 9E 10 (anti-myc, 1:100) and AA4.3 (alpha-tubulin 1:100) were developed by J. Michael Bishop and Charles Walsh, respectively, and were obtained from the Developmental Studies Hybridoma Bank (Iowa, USA).
RESULTS
Sequential sub-functionalization of tfap2 duplicates in the vertebrate lineage
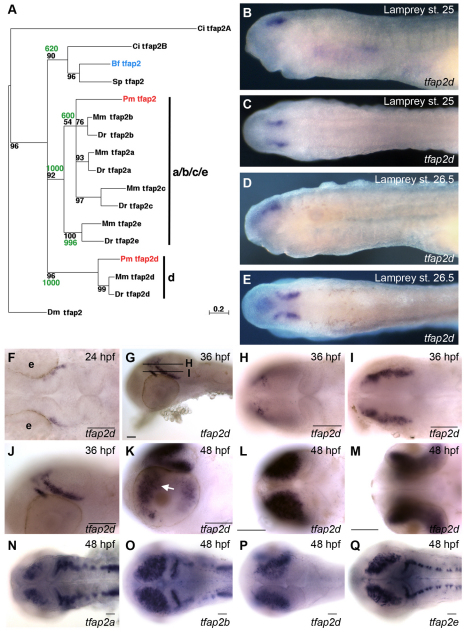
We first sought to establish the relationships among Tfap2 paralogs present in amphioxus, lamprey, mammals and zebrafish. Previously, we had identified five tfap2 homologs in zebrafish (Danio rerio, tfap2a-e), one in lamprey (Petromyzon marinus, Pm-tfap2) and one in amphioxus (Branchiostoma floridae, Bf-tfap2) (Li and Cornell, 2007; Meulemans and Bronner-Fraser, 2002). However, after the pre-assembly lamprey genome became available, we found a second lamprey tfap2 gene. Phylogenetic analysis of the Drosophila (D. melanogaster) and deuterostome Tfap2 proteins grouped this new lamprey Tfap2 with gnathostome Tfap2d genes. The previously described lamprey Tfap2 fell within a clade that includes the genes encoding gnathostome Tfap2a, Tfap2b, Tfap2c and Tfap2e (Fig. 1A). Given the evidence for two genome duplication events in the vertebrate lineage (Holland et al., 1994), these results support a scenario in which whole-genome duplication in the common ancestor of lamprey and gnathostomes generated two paralogs, Tfap2d and Tfap2a/b/c/e, from an ancestral Tfap2. Subsequent duplication events, including a second round of whole-genome duplication, generated all of the gnathostome Tfap2 paralogs (except Tfap2d) from the Tfap2a/b/c/e ancestor. By contrast, any duplicates of Tfap2d were lost. Although there is evidence for a genome duplication event within the teleost lineage (Postlethwait et al., 2004), given the one-to-one correspondence of tetrapod and zebrafish Tfap2 homologs, it appears that Tfap2 duplicates generated in this genome duplication event were lost. In summary, these findings provide an evolutionary scenario for the origin of Tfap2 paralogs present in vertebrates.
Fig. 1.
Characterization of embryonic tfap2d expression in zebrafish and lamprey. (A) Phylogenetic tree of deuterostome and Drosophila melanogaster tfap2 genes, constructed using the Maximum Likelihood method (Schmidt et al., 2002). A similar tree topology was obtained using the Neighbor-Joining method (not shown) (Saitou and Nei, 1987). Confidence values for both methods are shown at branch points with quartet-puzzling reliability scores in black and Neighbor-Joining bootstrap values in green. Each gene name includes a prefix with the initials of the corresponding species name. Ci, Ciona intestinalis; Pm, Petromyzon marinus; Mm, Mus musculus; Dr, Danio rerio; Bf, Branchiostoma floridae; Sp, Strongylocentrotus purpuratus; Dm, Drosophila melanogaster. (B-E) Lamprey embryos, fixed at the stage indicated and processed to reveal tfap2d expression. (B,C) Expression of tfap2d is first observed in bilateral spots in the presumed forebrain after neurulation (Tahara st. 25) in lateral (B) and dorsal (C) views. (D,E) Expression in the brain at late larval stage (st. 26.5) in (D) lateral and (E) dorsal views. (F-Q) Wild-type zebrafish embryos, fixed at the stages indicated and processed to reveal expression of tfap2d (F-M) or the indicated tfap2 family member (N-Q) by RNA in situ hybridization. (F) Dorsal view of the head showing tfap2d expression in a distinct structure within the presumptive mesencephalon. (G) Lateral view showing tfap2d expression within two distinct domains of the tegmentum; lines represent field of view for optical sections shown in H and I. (H,I) Dorsal views of the head, expression is in presumed tegmentum. (J) Higher magnification of embryo shown in G. (K) Lateral view of the head of a 48 hpf embryo, showing expression in tegmentum and optic tectum and retina (arrow). (L,M) Dorsal (L) and ventral (M) views of embryo shown in K, focusing on tfap2d expression within the optic tectum and retina, respectively. (N-Q) Embryos at 48 hpf, revealing shared and distinct expression domains of tfap2 family members in the brain. Embryos shown with anterior to the left, unless otherwise indicated. e, eye. Scale bars: 50 μm.
We asked whether the duplication-degeneration-complementation model explains the retention of vertebrate tfap2d paralogs (Force et al., 1999). Under this model, sub-functionalization refers to the partitioning of an ancestral gene’s expression pattern, or its functional domains, between its duplicates, leading to selective pressure to retain both duplicates (Force et al., 1999). For this analysis, we consider amphioxus to represent the vertebrate ancestor. Amphioxus tfap2 is expressed in the anterior neural tube and in the non-neural ectoderm (Meulemans and Bronner-Fraser, 2002). Similarly, lamprey tfap2 is expressed in the brain and the non-neural ectoderm, including the neural plate border/NC (Meulemans and Bronner-Fraser, 2002). However, lamprey tfap2d is expressed only in the brain (Fig. 1B-E) [tfap2d expression persists from larval stages into adulthood as observed by RT-PCR (data not shown)]. Similarly, mouse Tfap2d (Zhao et al., 2003) and zebrafish tfap2d are expressed solely within the brain (Fig. 1F-M,P). Together, these data are consistent with sub-functionalization of an ancestral tfap2 expression domain between two duplicates (tfap2 and tfap2d) pre-dating the split of agnathans and gnathostomes.
We next investigated whether sub-functionalization might also explain the retention of tfap2a, tfap2b, tfap2c and tfap2e in gnathostomes. In this analysis, we considered lamprey tfap2 expression to represent the ancestral condition. Zebrafish tfap2a, tfap2b and tfap2c are also expressed in this tissue but with distinct timing or spatial restrictions (supplementary material Fig. S1). Thus, during the gastrula stage zebrafish tfap2a and tfap2c are strongly expressed in the ventral ectoderm (presumptive skin) but by 24 hpf tfap2c expression remains high in embryonic skin, whereas tfap2a expression is relatively low, particularly in the periderm (Hoffman et al., 2007; Li and Cornell, 2007). Expression of tfap2b is absent from ventral ectoderm during gastrulation but is present in discrete regions of anterior skin at 24 hpf (Knight et al., 2005). In addition, lamprey tfap2 is expressed in premigratory and migratory NC (Meulemans and Bronner-Fraser, 2002). In zebrafish embryos at the gastrula stage, tfap2a and tfap2c are expressed in lateral neural plate cells, which later give rise to the NC (Li and Cornell, 2007). By the neurula stage and somitogenesis stages, however, only tfap2a is expressed at high levels in the premigratory NC. In NC derivatives, tfap2e is expressed in melanoblasts, whereas tfap2b is expressed in spinal sensory neurons, some of which are derived from the NC (Knight et al., 2005; Knight et al., 2003; O’Brien et al., 2004; Van Otterloo et al., 2010) (supplementary material Fig. S1 and Table S3). Finally, lamprey tfap2 is expressed in the brain, and zebrafish tfap2a, tfap2b, tfap2c and tfap2e are also all expressed in the brain, but in imperfectly overlapping patterns (Fig. 1N-Q) [tfap2c is expressed in the brain at 24 hpf (Li and Cornell, 2007)]. In summary, expression of modern tfap2a, tfap2b, tfap2c and tfap2e can be viewed as a partitioning of the ancestral pattern. These results are consistent with a model in which multiple rounds of duplication and sub-functionalization have resulted in retention of these four tfap2 duplicates.
Tfap2d is functionally distinct from other Tfap2 family members
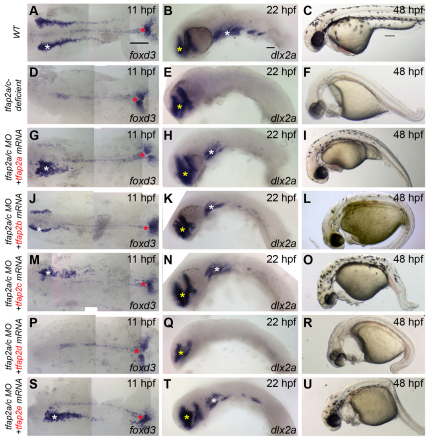
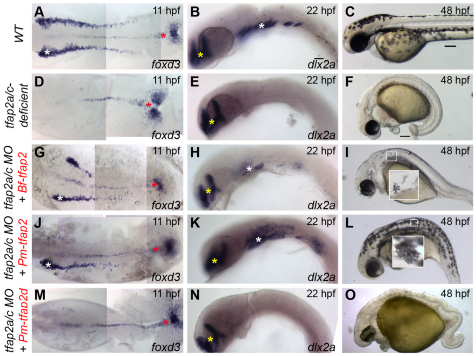
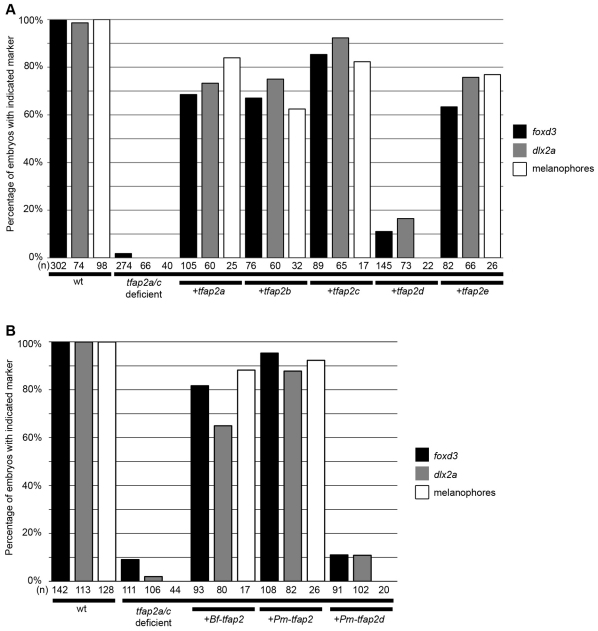
The proteins encoded by paralogs that have undergone sub-functionalization are under relaxed selection. Specifically, they are freed to lose functions appropriate for domains in which they are no longer expressed and to become functionally optimized for expression domains they retain (Conant and Wagner, 2003; Lynch and Wagner, 2008). We tested whether paralogs Tfap2b, Tfap2d and Tfap2e, which have all lost ancestral expression in the lateral neural border, have also lost a function appropriate for this domain, i.e. the ability to generate the NC. Starting with tfap2a/c-deficient embryos (which are embryos depleted of both tfap2a and tfap2c by mutation or by morpholino), we injected mRNAs encoding the various Tfap2 paralogs. We raised such embryos to appropriate stages then fixed and processed them to reveal a marker of premigratory NC, foxd3 (Kelsh et al., 2000; Odenthal and Nusslein-Volhard, 1998), or migratory NC, dlx2a (Akimenko et al., 1994). As expected, mRNAs encoding Tfap2a or Tfap2c efficiently rescued foxd3 expression (Fig. 2G,M), dlx2a expression (Fig. 2H,N) and also melanophores (Fig. 2I,O). Interestingly, injection of the tfap2b and tfap2e mRNAs also rescued foxd3 expression (Fig. 2J,S), dlx2a expression (Fig. 2K,T) and melanophores (Fig. 2L,U) in tfap2a/c-deficient embryos. Further tests revealed that overexpression of each of these Tfap2 paralogs similarly rescued in tfap2a/c-deficient embryos the NC derivatives peripheral glia, marked by foxd3 at 28 hpf (Kelsh et al., 2000), and sensory neuron precursors, marked by expression of tlxa at 22 hpf (Andermann and Weinberg, 2001) (supplementary material Fig. S2). By contrast, injection of a similar dose of the tfap2d mRNA into tfap2a/c-deficient embryos failed to rescue expression of foxd3, dlx2a or melanophores (Fig. 2P-R). Western blot analysis of epitope-tagged variants of Tfap2d and Tfap2a revealed similar stability in zebrafish embryos (supplementary material Fig. S3). Finally, we found that injection of lamprey tfap2 (56% and 55% protein similarity to Tfap2a and Tfap2c, respectively) also efficiently rescued foxd3 expression, dlx2a expression and melanophores in tfap2a/c-deficient embryos (Fig. 3J-L), whereas injection of the lamprey tfap2d (45% and 39% protein similarity to Tfap2a and Tfap2c, respectively) at a similar dose failed to do so (Fig. 3M-O) (all rescue experiments are summarized in Fig. 4). In summary, Tfap2b and Tfap2e, retain the ability to promote NC development, while Tfap2d virtually lacks this ability. As discussed above, it appears that tfap2d lost enhancers possessed by the tfap2 ancestor that drive expression in the non-neural ectoderm, and did so prior to the split of agnathans and gnathostomes. However, the functional experiments just described do not resolve whether Tfap2d subsequently lost the ability to induce NC, i.e. because of the absence of selective pressure to retain this function, or alternatively, whether the other Tfap2 paralog, which retained expression in non-neural ectoderm, gained this ability, i.e. via protein neo-functionalization.
Fig. 2.
Assessment of the ability of gnathostome Tfap2 paralogs to restore neural crest in tfap2a/c-deficient zebrafish embryos. (A,D,G,J,M,P,S) Dorsal views of flat-mounted wild-type zebrafish embryos (A) or embryos injected with the indicated mRNA and/or MO (D,G,J,M,P,S), fixed at 11 hpf and processed to reveal foxd3 expression. Restored foxd3 expression was found on left, right, or both sides as a result of mosaic injection of mRNA. White asterisks indicate premigratory neural crest. Red asterisks indicate non-neural crest-derived tailbud. (B,E,H,K,N,Q,T) Lateral views of wild-type zebrafish embryos (B) or embryos injected with the indicated mRNA and/or MO (E,H,K,N,Q,T), fixed at 22 hpf and processed to reveal dlx2a expression. With the exception of tfap2d, no trend was seen in the spatial extent of dlx2a expression and the tfap2 paralog used. White asterisks indicate migratory neural crest. Yellow asterisks indicate brain. (C,F,I,L,O,R,U) Lateral views of live embryos at 48 hpf that are wild type (C) or tfap2a mutants injected with tfap2c MO (F,I,L,O,R,U), and injected with the indicated mRNA. Embryos shown with anterior to the left, unless otherwise indicated. Scale bars: in A, 100 μm for A,D,G,J,M,P,S; in B, 50 μm for B,E,H,K,N,Q,T; in C, 100 μm for C,F,I,L,O,R,U.
Fig. 3.
Assessment of the ability of lamprey and amphioxus Tfap2 paralogs to restore neural crest in tfap2a/c-deficient zebrafish embryos. (A,D,G,J,M) Dorsal views of flat-mounted wild-type zebrafish embryos (A) or embryos injected with the indicated mRNA and/or MO (D,G,J,M), fixed at 11 hpf and processed to reveal foxd3 expression. White asterisks indicate premigratory neural crest. Red asterisks indicate non-neural crest-derived tailbud. (B,E,H,K,N) Lateral views of wild-type zebrafish embryos (B) or embryos injected with the indicated mRNA and/or MO (E,H,K,N), fixed at 22 hpf and processed to reveal dlx2a expression. White asterisks indicate migratory neural crest. Yellow asterisks indicate brain. (C,F,I,L,O) Lateral views of live embryos at 48 hpf that are wild type (C) or tfap2a mutants injected with tfap2c MO (F,I,L,O), and injected with the indicated mRNA. No trend was seen in the ability of Bf-tfap2 versus Pm-tfap2 to rescue more melanophores per embryos. Insets in figure I and L are higher magnification images of boxed melanophores. Embryos shown with anterior to the left, unless otherwise indicated. Scale bars: in A, 100 μm for A,D,G,J,M; in B, 50 μm for B,E,H,K,N; in C, 100 μm for C,F,I,L,O.
Fig. 4.
Summary of Tfap2 rescue results. (A,B) Histograms summarizing neural crest rescue results in tfap2a/c-deficient embryos injected with zebrafish tfap2 paralogs (A), or amphioxus (Bf) or lamprey (Pm) tfap2 homologs (B), as indicated. Number of embryos (n) per group is displayed below bars.
Tfap2 from amphioxus can induce neural crest
To determine whether the ability of Tfap2 to induce NC resulted from protein neo-functionalization, we generated a full-length amphioxus tfap2 cDNA and generated mRNA from it in vitro. Interestingly, injection of amphioxus tfap2 mRNA (48% and 46% protein similarity to Tfap2a and Tfap2c, respectively) into tfap2a/c-deficient embryos efficiently rescued expression of foxd3 (in NC and in peripheral glia), expression of dlx2a, expression of tlxa and melanophores (Fig. 3G-I; supplementary material Fig. S2). Testing an even more ancient AP2 homolog, we injected Drosophila tfap2 mRNA (40% and 39% protein similarity to Tfap2a and Tfap2c, respectively) into tfap2a/c-deficient zebrafish embryos, and observed robust foxd3 expression in the lateral neural plate at 11 hpf (supplementary material Fig. S4G); however, in such embryos, expression of dlx2a at 22 hpf and melanophores at 48 hpf were rarely present (supplementary material Fig. S4H,I, legend). The less robust rescue of NC by Drosophila tfap2 might reflect instability of Drosophila Tfap2 relative to other Tfap2 proteins tested, or possibly the absence in Drosophila Tfap2 of a function essential for NC maintenance or other later events in NC development. Nonetheless, because amphioxus efficiently rescued NC in tfap2a/c-deficient embryos, we conclude that recruitment of Tfap2 into a GRN that controls induction of NC at the neural plate border did not require Tfap2 to gain new protein function.
sox10 is targeted directly by Tfap2a and forced expression of sox10 partially restores trunk neural crest in tfap2a/c-deficient embryos
We sought to identify the novel regulatory connections gained by Tfap2 in the context of the NC GRN. In amphioxus, the orthologs of multiple NC regulatory proteins (e.g. SoxE, FoxD, Twist, ID) are expressed in the mesoderm but not in the neural plate border (Sauka-Spengler et al., 2007). Mutations in the regulatory regions of the genes encoding such regulatory proteins might have introduced Tfap2 binding sites, thereby recruiting their expression to the neural plate border. We investigated this possibility for sox10, a gene necessary for early steps of NC induction (Dutton et al., 2001; Honore et al., 2003) and expression of which in NC is lost in tfap2a/c-deficient embryos (Hoffman et al., 2007; Li and Cornell, 2007). We conducted anti-Tfap2a chromatin immunoprecipitation (ChIP) on 4.9 kb upstream of the start codon of sox10, a region previously shown to drive reporter expression in the NC (Wada et al., 2005). We started with lysates of zebrafish embryos at 12 hpf, a stage at which both tfap2a and sox10 are expressed at high levels in the NC, using an antibody generated against zebrafish Tfap2a, specificity of which was confirmed in tfap2a mutants (supplementary material Fig. S5). A pair of predicted Tfap2 binding sites are present proximal to the transcription start site (Fig. 5A). On the precipitated chromatin we performed quantitative PCR (qPCR) with primer pairs positioned near these predicted Tfap2a binding sites, or with primer pairs positioned 1.2 kb downstream, as a negative control (off-target site) (Fig. 5A). We observed 12- to 22-fold enrichment at the target site in anti-Tfap2a-precipitated chromatin versus IgG-precipitated chromatin, and minimal enrichment at the off-target site (Fig. 5B). These findings indicate that Tfap2a binds the zebrafish sox10 promoter at a time when sox10 is expressed in the NC.
Next, we investigated whether Tfap2a overexpression will elevate sox10 expression when protein translation is inhibited. We co-injected embryos with tfap2a MO, tfap2c MO, and mRNA encoding a dexamethasone-inducible variant of Tfap2a (Tfap2a fused to glucocorticoid-receptor, tfap2a-GR) (Luo et al., 2002). At 10 hpf, we treated the embryos with cycloheximide to block translation (Leung et al., 2003). Two hours later, we added dexamethasone to induce nuclear transport of Tfap2a-GR. Finally, 2.5 hours later (∼14 hpf), we harvested RNA from these embryos, generated first-strand cDNA and used qPCR to measure the levels of sox10 and zgc:85942 (ednrab – Zebrafish Information Network), an ednra homolog which, like sox10, is expressed at high levels in the NC at 13 hpf (Thisse and Thisse, 2004). As expected, in tfap2a/c-deficient embryos the levels of both sox10 and zgc:85942 mRNA were reduced relative to those in control embryos (Fig. 5C,D). In tfap2a/c-deficient embryos injected with tfap2a-GR and not treated with dexamethasone, sox10 and zgc:85942 expression were barely elevated above the levels detected in tfap2a/c-deficient embryos (Fig. 5C,D), confirming that Tfap2a-GR is only minimally active in the absence of dexamethasone. By contrast, in embryos treated with dexamethasone but not cycloheximide, the expression of both NC markers was significantly elevated (Fig. 5C,D). Interestingly, in embryos treated with dexamethasone in the presence of cycloheximide, the expression of sox10, but not that of zgc:85942, was raised to levels significantly higher than those in tfap2a/c-deficient embryos (Fig. 5C,D). Together, these findings strongly suggest that Tfap2 activates sox10 expression directly.
To assess whether activation of sox10 is the sole requirement of Tfap2a in NC development, we re-introduced sox10 into tfap2a/c-deficient embryos. To do so, we used the hsp70:sox10 plasmid, which contains a heat-shock-inducible promoter upstream of the sox10 coding region (Elworthy et al., 2003). In hsp70:sox10-injected, tfap2a/c-deficient embryos exposed to heat-shock at 9 hpf and fixed at 12 hpf, snai1b and sox9b were detected in a scattered pattern around the embryo (mosaic expression is typical of transient transgenesis) (Thisse et al., 1995; Yan et al., 2005) (Fig. 5M,N). However, foxd3 expression was absent in these embryos, and crestin expression was absent from similarly treated embryos fixed at 22 hpf (Rubinstein et al., 2000) (Fig. 5O; data not shown). Interestingly, in similarly treated embryos melanophores were present at 48 hpf, but only in the trunk, not in the head (Fig. 5P). Control experiments revealed the requirement for both the hsp70:sox10 plasmid and the heat-shock for the results described in this section. Because forced expression of sox10 was insufficient to restore a full NC program in tfap2a/c-deficient embryos, it is very likely that there are Tfap2 regulatory targets other than sox10 that are essential in the NC GRN.
Genomic elements proximal to amphioxus SoxE lack neural crest enhancer activity
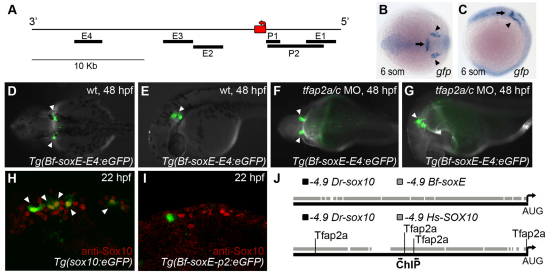
If regulation of the soxE gene by Tfap2 emerged in the context of the novel NC GRN then amphioxus soxE should lack regulatory elements sufficient to drive expression in the NC. To test this notion, we amplified 4.9 kb upstream of the amphioxus SoxE coding region and engineered it into the same GFP reporter vector used to show that the analogous 4.9 kb element upstream of zebrafish sox10 drives NC-specific expression (Wada et al., 2005). In addition, because canonical Wnt signaling promotes NC development in zebrafish (Lewis et al., 2004), we amplified several regions proximal to amphioxus SoxE that contain TCF/LEF sites, and cloned them into a reporter vector with a minimal promoter (Fisher et al., 2006) (supplementary material Table S2). We injected each of these constructs, or, as a positive control, a reporter construct containing 4.9 kb upstream of zebrafish sox10, into zebrafish embryos (Wada et al., 2005). However, only in embryos injected with the zebrafish construct did we detect GFP expression concentrated in the NC, as recognized by anti-Sox10 immunoreactivity (Fig. 6H,I). Interestingly, although amphioxus embryos are believed to lack ears, zebrafish embryos injected with a construct containing an element downstream of amphioxus soxE (Fig. 6A, soxE region 4) exhibited GFP expression in the ear and presumed midbrain-hindbrain boundary; embryos from two independent stable transgenic lines made from this reporter revealed the same expression (Fig. 6B-G). Similar to endogenous sox10 expression in the ear, expression of GFP in these lines was insensitive to knockdown of Tfap2a and Tfap2c (Fig. 6F,G).
Fig. 6.
Amphioxus soxE elements fail to drive neural crest specific expression in zebrafish embryos. (A) Schematic of amphioxus soxE locus showing location of DNA elements tested in zebrafish reporter analysis. Red box, coding region of soxE; red arrow, initial AUG. Genomic coordinates are shown in supplementary material Table S2. (B,C) Anterior dorsal (B) and lateral (C) views of a Tg(Bf-soxE-E4:eGFP) zebrafish embryo fixed at 12 hpf and processed for gfp expression. gfp expression in midbrain-hindbrain (arrows) and otic placodes (arrowheads) is apparent. (D-G) Dorsal (D,F) and lateral (E,G) views of live, 48 hpf Tg(Bf-soxE-E4:eGFP) embryos. Arrowheads in D and E indicate GFP expression in otic vesicle (observed in five of 20 injected embryos). (F,G) A Tg(Bf-soxE-E4:eGFP) embryo injected with MOs targeting tfap2a and tfap2c, revealing eGFP expression in otic vesicle (arrowheads) despite an absence of melanophores (otic vesicle expression seen in three of 14 similarly injected transgenic embryos). (H,I) Lateral views of 22 hpf fixed embryos injected with (H) sox10:egfp plasmid (Wada et al., 2005), showing many GFP-positive, anti-Sox10-positive NC cells (arrowheads) in the dorsum (18/22 injected embryos showed dorsal GFP-positive cells), or (I) an analogous plasmid harboring 4.9 kb upstream of amphioxus soxE (Bf-soxE-p2:eGFP), with very few such cells (4/22 embryos showed few dorsal GFP-positive cells all of which were negative for anti-Sox10 immunoreactivity). (J) Modified output of ConSite software showing conservation of sequence and Tfap2 transcription factor binding sites in 4.9 kb of genomic DNA upstream of the start of the initiator AUG in human SOX10 (–4.9 Hs-SOX10) and zebrafish sox10 (–4.9 Dr-sox10). Arrows, region tested in ChIP analysis. An identical comparison of amphioxus soxE (–4.9 Bf-soxE) and zebrafish sox10 using identical parameters fails to detect conserved Tfap2 binding sites. Embryos shown with anterior to the left, unless otherwise indicated.
Finally, we examined sequences upstream of the Sox10 or SoxE start codon in human, fish and amphioxus for conserved Tfap2 binding sites using the online analysis software ConSite (Sandelin et al., 2004). This analysis revealed multiple conserved predicted Tfap2 binding sites between human and zebrafish sequences (Fig. 6J). Importantly, two of these conserved binding sites were located precisely within the region we showed by ChIP analysis to be enriched with Tfap2a. Using identical parameters, no conserved Tfap2 binding sites were detected when comparing human with amphioxus sequences or zebrafish with amphioxus sequences (Fig. 6J; data not shown). In combination with the enhancer analysis of soxE discussed above, and the absence of co-expression of soxE and tfap2 in amphioxus (and Drosophila), these findings support the hypothesis that sox10 became a regulatory target of Tfap2 after the split of the cephalochordates from the lineage leading to vertebrates.
DISCUSSION
Selective pressure to retain Tfap2 paralogs resulted from sub-functionalization
Here, we began by showing that fish and mammalian Tfap2d paralogs resulted from an ancient duplication of an ancestral Tfap2 prior to the split of agnathans and gnathostomes, potentially during a genome-duplication event. Interestingly, both Maximum Likelihood and Neighbor Joining (not shown) analyses grouped amphioxus Tfap2, sea urchin Tfap2 and ascidian Tfap2B together in one clade, and placed ascidian Tfap2A as an outgroup to all other chordate Tfap2s with moderate to high support. This grouping contradicts recent phylogenomic analyses showing that urochordates are the vertebrate sister group (Delsuc et al., 2006). These results probably reflect the rapid evolution of ascidian tfap2 genes, as has been seen with many other urochordate developmental regulators (Holland and Gibson-Brown, 2003). However, regardless of the phylogenetic position of the ascidian genes, a comparison of the expression pattern of the single tfap2 homolog in amphioxus (a basal chordate) and the two homologs in lamprey (a basal vertebrate) supports the duplication-degeneration-complementation model (Force et al., 1999). This model posits that selective pressure to retain duplicated, otherwise redundant, genes emerges through a complementary loss of gene-regulatory elements inherited from the ancestral gene (a process termed sub-functionalization) (Force et al., 1999). Sub-functionalization also appears to have occurred among the tfap2a/b/c/e genes in gnathostomes. Like lamprey tfap2, the collective expression pattern of zebrafish tfap2a/b/c/e paralogs includes the presumptive skin, the neural plate border, NC and NC derivatives. However, as described here and elsewhere (Hoffman et al., 2007; Knight et al., 2005; Knight et al., 2003; Li and Cornell, 2007; Meulemans and Bronner-Fraser, 2002; O’Brien et al., 2004; Van Otterloo et al., 2010), the individual patterns of these paralogs are not identical. Thus, it appears that through the loss of tissue-specific enhancers associated with an ancestral tfap2a/b/c/e, initially redundant tfap2 duplicates have acquired distinct expression territories, resulting in selective pressure to retain all four paralogs.
Relaxed selective pressure on Tfap2d permitted the loss of its neural crest-promoting function
We have also tested the notion that expression-domain partitioning relaxes selective pressure on duplicated proteins. We showed that Tfap2d has lost, to a great degree, the ability to generate NC. Whether Tfap2d has also become optimized for its brain function, relative to the ancestral Tfap2, will require further tests. However, the DNA-binding preference of TFAP2D is distinct from that of the other homologs, supporting this possibility (Zhao et al., 2001). Of note, sub-functionalization usually refers to complementary loss of regulatory elements possessed by an ancestral gene, but it can also apply to complementary loss of subfunctions possessed by the ancestral protein (Force et al., 1999). Indeed, the loss of NC-inducing function by Tfap2d might have preceded the loss of its expression in the non-neural ectoderm and neural border. This loss of functionality would have released selective pressure to retain expression of tfap2d in these tissues. Although not expressed broadly in surface ectoderm, Tfap2b and Tfap2e retain NC-inducing ability, in contrast to Tfap2d. These proteins have a more recent origin than Tfap2d and might have not had time to lose this ability; alternatively, the functions for which they have been selected in their current expression domains are not easily separated from the surface ectoderm function [for a general discussion, see Force et al. (Force et al., 1999)].
Changes in the cis-regulatory elements of Tfap2 targets contributed to the origins of the neural crest
Our results favor the importance of regulatory versus protein neo-functionalization in the insertion of Tfap2 into the evolving NC GRN. Protein neo-functionalization, facilitated by gene duplication, has been proposed to drive new morphologies over the course of evolution (Ohno, 1970). However, although the Tfap2 family has been duplicated and is required for NC development, we found no evidence that protein neo-functionalization conferred a novel ‘NC-inducing’ function to a Tfap2 paralog. Specifically, we found that Tfap2 from basal animals that lack NC nonetheless harbored NC-promoting function. By contrast, we did find evidence for regulatory neo-functionalization of the sox10 gene. Thus, here we show that Tfap2a directly activates sox10 in zebrafish, a vertebrate. The evidence that this regulatory interaction emerged after the split of the lineage leading to vertebrates from cephalochordates is that soxE and tfap2 are not co-expressed in amphioxus nor is AP2 co-expressed with sox100B in flies (Hui Yong Loh and Russell, 2000; Meulemans and Bronner-Fraser, 2002; Meulemans and Bronner-Fraser, 2007; Monge and Mitchell, 1998; Yu et al., 2008). Moreover, multiple DNA elements proximal to amphioxus soxE, including the 4.9 kb upstream, lack NC enhancer function in zebrafish. The discovery of NC-like features in tunicates is consistent with the possibility that the novel regulation of sox10 by Tfap2 appeared prior to the split of urochordates and vertebrates (Jeffery et al., 2004).
In the evolution of the NC GRN, Tfap2 probably acquired targets in addition to sox10, as judged by the failure of forced expression of sox10 in tfap2a/c deficient embryos to fully restore the gene expression profile of premigratory NC. The absence of foxd3 expression in this paradigm is consistent with earlier findings that Sox10 is downstream of Foxd3 in the NC GRN (Arduini et al., 2009). The presence of sox9b and snai1b expression in this paradigm is noteworthy because although sox10, sox9b and snai1b are all expressed in the NC, they are also co-expressed in mesoderm (Chimal-Monroy et al., 2003; Thisse et al., 1995; Yan et al., 2005). NC shares qualities with the mesoderm, such as migratory properties and the ability to make bone. The novel activation of sox10 by Tfap2 might have served to recruit part of the mesoderm GRN to the NC, conferring mesoderm-like qualities to the NC. Interestingly, forcing sox10 expression in tfap2a/c-deficient embryos restored trunk melanophores. It has recently been shown that dynamic regulation of Sox10 is essential for differentiation of melanophores, and it remains to be seen whether trunk melanophores rescued by sox10 in this study are fully normal (Greenhill et al., 2011). Clearly, there are many details of the NC GRN and the melanophore GRN that remain to be discovered. However, the failure of sox10 to fully restore NC in this paradigm strongly suggests that other Tfap2 targets, whether novel or ancient, contribute to the NC GRN.
Finally, the presence of an apparent otic placode enhancer adjacent to amphioxus soxE serves to demonstrate that regulatory neo-functionalization is not the only mechanism by which novel GRNs are assembled. Amphioxus lack cranial placodes, indicating that at least some regulatory connections within the otic placode GRN are novel. Our findings reveal that cis-regulatory information sufficient to drive soxE expression in the ear was present prior to the split of cephalochordates and the lineage leading to vertebrates. Therefore regulatory neo-functionalization did not necessarily play a role in recruitment of SoxE into the otic placode GRN. Instead, soxE might have been recruited to the otic placode GRN via protein neo-functionalization, or as part of a conserved sub-network co-opted from another GRN. This unexpected finding supports other evidence that both protein neo-functionalization and regulatory neo-functionalization contribute to assembly of novel GRNs (Wagner and Lynch, 2008).
Supplementary Material
Acknowledgments
We are grateful to Greg Bonde for technical assistance with many aspects of this work and to Chris Blaumueller for editing the manuscript. We thank Dr Jean-Pierre Saint-Jeannet and Dr Paul Henion for plasmids.
Footnotes
Funding
This work was supported by a grant from the National Institute of General Medical Sciences (NIGMS) [R01GM067841]. Deposited in PMC for release after 12 months.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.071308/-/DC1
References
- Akimenko M. A., Ekker M., Wegner J., Lin W., Westerfield M. (1994). Combinatorial expression of three zebrafish genes related to distal-less: part of a homeobox gene code for the head. J. Neurosci. 14, 3475–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andermann P., Weinberg E. S. (2001). Expression of zTlxA, a Hox11-like gene, in early differentiating embryonic neurons and cranial sensory ganglia of the zebrafish embryo. Dev. Dyn. 222, 595–610 [DOI] [PubMed] [Google Scholar]
- Arduini B. L., Bosse K. M., Henion P. D. (2009). Genetic ablation of neural crest cell diversification. Development 136, 1987–1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canestro C., Bassham S., Postlethwait J. H. (2003). Seeing chordate evolution through the Ciona genome sequence. Genome Biol. 4, 208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimal-Monroy J., Rodriguez-Leon J., Montero J. A., Ganan Y., Macias D., Merino R., Hurle J. M. (2003). Analysis of the molecular cascade responsible for mesodermal limb chondrogenesis: Sox genes and BMP signaling. Dev. Biol. 257, 292–301 [DOI] [PubMed] [Google Scholar]
- Conant G. C., Wagner A. (2003). Asymmetric sequence divergence of duplicate genes. Genome Res. 13, 2052–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossais F., Sock E., Hornig J., Schreiner S., Kellerer S., Bosl M. R., Russell S., Wegner M. (2010). Replacement of mouse Sox10 by the Drosophila ortholog Sox100B provides evidence for co-option of SoxE proteins into vertebrate-specific gene-regulatory networks through altered expression. Dev. Biol. 341, 267–281 [DOI] [PubMed] [Google Scholar]
- Delsuc F., Brinkmann H., Chourrout D., Philippe H. (2006). Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature 439, 965–968 [DOI] [PubMed] [Google Scholar]
- Dussault A. A., Pouliot M. (2006). Rapid and simple comparison of messenger RNA levels using real-time PCR. Biol. Proced. Online 8, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutton K. A., Pauliny A., Lopes S. S., Elworthy S., Carney T. J., Rauch J., Geisler R., Haffter P., Kelsh R. N. (2001). Zebrafish colourless encodes sox10 and specifies non-ectomesenchymal neural crest fates. Development 128, 4113–4125 [DOI] [PubMed] [Google Scholar]
- Eckert D., Buhl S., Weber S., Jager R., Schorle H. (2005). The AP-2 family of transcription factors. Genome Biol. 6, 246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elworthy S., Lister J. A., Carney T. J., Raible D. W., Kelsh R. N. (2003). Transcriptional regulation of mitfa accounts for the sox10 requirement in zebrafish melanophore development. Development 130, 2809–2818 [DOI] [PubMed] [Google Scholar]
- Fisher S., Grice E. A., Vinton R. M., Bessling S. L., Urasaki A., Kawakami K., McCallion A. S. (2006). Evaluating the biological relevance of putative enhancers using Tol2 transposon-mediated transgenesis in zebrafish. Nat. Protoc. 1, 1297–1305 [DOI] [PubMed] [Google Scholar]
- Force A., Lynch M., Pickett F. B., Amores A., Yan Y. L., Postlethwait J. (1999). Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151, 1531–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gans C., Northcutt R. G. (1983). Neural crest and the origin of vertebrates: a new head. Science 220, 268–273 [DOI] [PubMed] [Google Scholar]
- Greenhill E. R., Rocco A., Vibert L., Nikaido M., Kelsh R. N. (2011). An iterative genetic and dynamical modelling approach identifies novel features of the gene regulatory network underlying melanocyte development. PLoS Genet. 7, e1002265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman T. L., Javier A. L., Campeau S. A., Knight R. D., Schilling T. F. (2007). Tfap2 transcription factors in zebrafish neural crest development and ectodermal evolution. J. Exp. Zool. B Mol. Dev. Evol. 308, 679–691 [DOI] [PubMed] [Google Scholar]
- Holland L. Z., Gibson-Brown J. J. (2003). The Ciona intestinalis genome: when the constraints are off. BioEssays 25, 529–532 [DOI] [PubMed] [Google Scholar]
- Holland P. W., Garcia-Fernandez J., Williams N. A., Sidow A. (1994). Gene duplications and the origins of vertebrate development. Development Suppl. 125–133 [PubMed] [Google Scholar]
- Honore S. M., Aybar M. J., Mayor R. (2003). Sox10 is required for the early development of the prospective neural crest in Xenopus embryos. Dev. Biol. 260, 79–96 [DOI] [PubMed] [Google Scholar]
- Hui Yong Loh S., Russell S. (2000). A Drosophila group E Sox gene is dynamically expressed in the embryonic alimentary canal. Mech. Dev. 93, 185–188 [DOI] [PubMed] [Google Scholar]
- Jeffery W. R., Strickler A. G., Yamamoto Y. (2004). Migratory neural crest-like cells form body pigmentation in a urochordate embryo. Nature 431, 696–699 [DOI] [PubMed] [Google Scholar]
- Kelsh R. N., Dutton K., Medlin J., Eisen J. S. (2000). Expression of zebrafish fkd6 in neural crest-derived glia. Mech. Dev. 93, 161–164 [DOI] [PubMed] [Google Scholar]
- Kimmel C. B., Ballard W. W., Kimmel S. R., Ullmann B., Schilling T. F. (1995). Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310 [DOI] [PubMed] [Google Scholar]
- Knight R. D., Nair S., Nelson S. S., Afshar A., Javidan Y., Geisler R., Rauch G. J., Schilling T. F. (2003). lockjaw encodes a zebrafish tfap2a required for early neural crest development. Development 130, 5755–5768 [DOI] [PubMed] [Google Scholar]
- Knight R. D., Javidan Y., Zhang T., Nelson S., Schilling T. F. (2005). AP2-dependent signals from the ectoderm regulate craniofacial development in the zebrafish embryo. Development 132, 3127–3138 [DOI] [PubMed] [Google Scholar]
- Leung T., Bischof J., Soll I., Niessing D., Zhang D., Ma J., Jackle H., Driever W. (2003). bozozok directly represses bmp2b transcription and mediates the earliest dorsoventral asymmetry of bmp2b expression in zebrafish. Development 130, 3639–3649 [DOI] [PubMed] [Google Scholar]
- Lewis J. L., Bonner J., Modrell M., Ragland J. W., Moon R. T., Dorsky R. I., Raible D. W. (2004). Reiterated Wnt signaling during zebrafish neural crest development. Development 131, 1299–1308 [DOI] [PubMed] [Google Scholar]
- Li W., Cornell R. A. (2007). Redundant activities of Tfap2a and Tfap2c are required for neural crest induction and development of other non-neural ectoderm derivatives in zebrafish embryos. Dev. Biol. 304, 338–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeman L. C., Vogt-Kielland L. T., Alestrom P., Collas P. (2009). Fish’n ChIPs: chromatin immunoprecipitation in the zebrafish embryo. Methods Mol. Biol. 567, 75–86 [DOI] [PubMed] [Google Scholar]
- Link V., Shevchenko A., Heisenberg C. P. (2006). Proteomics of early zebrafish embryos. BMC Dev. Biol. 6, 1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo T., Matsuo-Takasaki M., Thomas M. L., Weeks D. L., Sargent T. D. (2002). Transcription factor AP-2 is an essential and direct regulator of epidermal development in Xenopus. Dev. Biol. 245, 136–144 [DOI] [PubMed] [Google Scholar]
- Luo T., Lee Y. H., Saint-Jeannet J. P., Sargent T. D. (2003). Induction of neural crest in Xenopus by transcription factor AP2alpha. Proc. Natl. Acad. Sci. USA 100, 532–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch V. J., Wagner G. P. (2008). Resurrecting the role of transcription factor change in developmental evolution. Evolution 62, 2131–2154 [DOI] [PubMed] [Google Scholar]
- Mazet F., Shimeld S. M. (2005). Molecular evidence from ascidians for the evolutionary origin of vertebrate cranial sensory placodes. J. Exp. Zool. B Mol. Dev. Evol. 304, 340–346 [DOI] [PubMed] [Google Scholar]
- Meulemans D., Bronner-Fraser M. (2002). Amphioxus and lamprey AP-2 genes: implications for neural crest evolution and migration patterns. Development 129, 4953–4962 [DOI] [PubMed] [Google Scholar]
- Meulemans D., Bronner-Fraser M. (2005). Central role of gene cooption in neural crest evolution. J. Exp. Zool. B Mol. Dev. Evol. 304, 298–303 [DOI] [PubMed] [Google Scholar]
- Meulemans D., Bronner-Fraser M. (2007). The amphioxus SoxB family: implications for the evolution of vertebrate placodes. Int. J. Biol. Sci. 3, 356–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monge I., Mitchell P. J. (1998). DAP-2, the Drosophila homolog of transcription factor AP-2. Mech. Dev. 76, 191–195 [DOI] [PubMed] [Google Scholar]
- Northcutt R. G., Gans C. (1983). The genesis of neural crest and epidermal placodes: a reinterpretation of vertebrate origins. Q. Rev. Biol. 58, 1–28 [DOI] [PubMed] [Google Scholar]
- O’Brien E. K., d’Alencon C., Bonde G., Li W., Schoenebeck J., Allende M. L., Gelb B. D., Yelon D., Eisen J. S., Cornell R. A. (2004). Transcription factor Ap-2alpha is necessary for development of embryonic melanophores, autonomic neurons and pharyngeal skeleton in zebrafish. Dev. Biol. 265, 246–261 [DOI] [PubMed] [Google Scholar]
- Odenthal J., Nusslein-Volhard C. (1998). fork head domain genes in zebrafish. Dev. Genes Evol. 208, 245–258 [DOI] [PubMed] [Google Scholar]
- Ohno S. (1970). Evolution by Gene Duplication. Berlin, New York: Springer-Verlag; [Google Scholar]
- Onuma Y., Takahashi S., Asashima M., Kurata S., Gehring W. J. (2002). Conservation of Pax 6 function and upstream activation by Notch signaling in eye development of frogs and flies. Proc. Natl. Acad. Sci. USA 99, 2020–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postlethwait J., Amores A., Cresko W., Singer A., Yan Y. L. (2004). Subfunction partitioning, the teleost radiation and the annotation of the human genome. Trends Genet. 20, 481–490 [DOI] [PubMed] [Google Scholar]
- Rubinstein A. L., Lee D., Luo R., Henion P. D., Halpern M. E. (2000). Genes dependent on zebrafish cyclops function identified by AFLP differential gene expression screen. Genesis 26, 86–97 [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 [DOI] [PubMed] [Google Scholar]
- Sambrook J., Maniatis T., Fritsch E. F. (1989). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; [Google Scholar]
- Sandelin A., Wasserman W. W., Lenhard B. (2004). ConSite: web-based prediction of regulatory elements using cross-species comparison. Nucleic Acids Res. 32, W249–W252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauka-Spengler T., Bronner-Fraser M. (2008). A gene regulatory network orchestrates neural crest formation. Nat. Rev. Mol. Cell. Biol. 9, 557–568 [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T., Meulemans D., Jones M., Bronner-Fraser M. (2007). Ancient evolutionary origin of the neural crest gene regulatory network. Dev. Cell 13, 405–420 [DOI] [PubMed] [Google Scholar]
- Schmidt H. A., Strimmer K., Vingron M., von Haeseler A. (2002). TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18, 502–504 [DOI] [PubMed] [Google Scholar]
- Thisse B., Thisse C. (2004). Fast Release Clones: A High Throughput Expression Analysis. ZFIN Direct Data Submission (http://www.zfin.org).
- Thisse C., Thisse B. (2008). High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3, 59–69 [DOI] [PubMed] [Google Scholar]
- Thisse C., Thisse B., Postlethwait J. H. (1995). Expression of snail2, a second member of the zebrafish snail family, in cephalic mesendoderm and presumptive neural crest of wild-type and spadetail mutant embryos. Dev. Biol. 172, 86–99 [DOI] [PubMed] [Google Scholar]
- Van Otterloo E., Li W., Bonde G., Day K. M., Hsu M. Y., Cornell R. A. (2010). Differentiation of zebrafish melanophores depends on transcription factors AP2 alpha and AP2 epsilon. PLoS Genet. 6, e1001122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada N., Javidan Y., Nelson S., Carney T. J., Kelsh R. N., Schilling T. F. (2005). Hedgehog signaling is required for cranial neural crest morphogenesis and chondrogenesis at the midline in the zebrafish skull. Development 132, 3977–3988 [DOI] [PubMed] [Google Scholar]
- Wagner A. (1998). The fate of duplicated genes: loss or new function? BioEssays 20, 785–788 [DOI] [PubMed] [Google Scholar]
- Wagner G. P., Lynch V. J. (2008). The gene regulatory logic of transcription factor evolution. Trends Ecol. Evol. 23, 377–385 [DOI] [PubMed] [Google Scholar]
- Westerfield M. (1993). The Zebrafish Book. Eugene, OR: University of Oregon Press; [Google Scholar]
- Yan Y. L., Willoughby J., Liu D., Crump J. G., Wilson C., Miller C. T., Singer A., Kimmel C., Westerfield M., Postlethwait J. H. (2005). A pair of Sox: distinct and overlapping functions of zebrafish sox9 co-orthologs in craniofacial and pectoral fin development. Development 132, 1069–1083 [DOI] [PubMed] [Google Scholar]
- Yu J. K., Meulemans D., McKeown S. J., Bronner-Fraser M. (2008). Insights from the amphioxus genome on the origin of vertebrate neural crest. Genome Res. 18, 1127–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F., Satoda M., Licht J. D., Hayashizaki Y., Gelb B. D. (2001). Cloning and characterization of a novel mouse AP-2 transcription factor, AP-2delta, with unique DNA binding and transactivation properties. J. Biol. Chem. 276, 40755–40760 [DOI] [PubMed] [Google Scholar]
- Zhao F., Lufkin T., Gelb B. D. (2003). Expression of Tfap2d, the gene encoding the transcription factor Ap-2 delta, during mouse embryogenesis. Gene Expr. Patterns 3, 213–217 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.