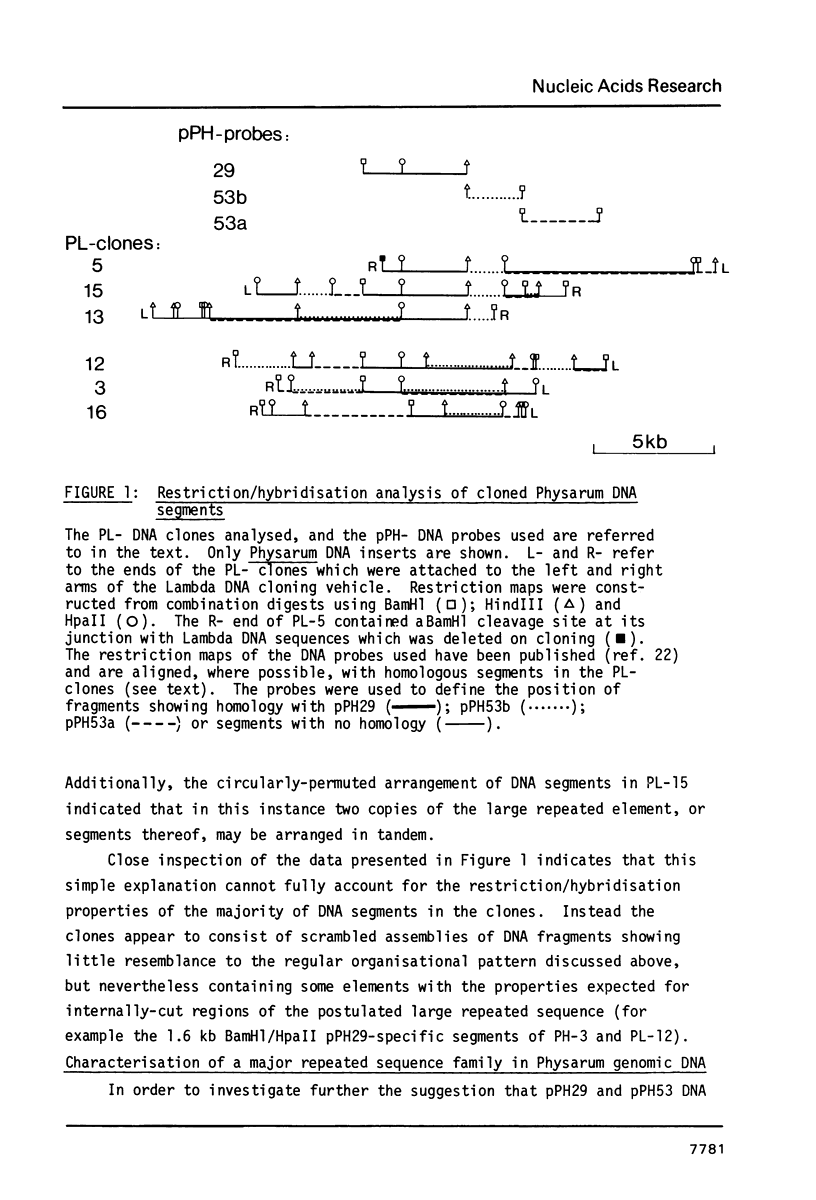
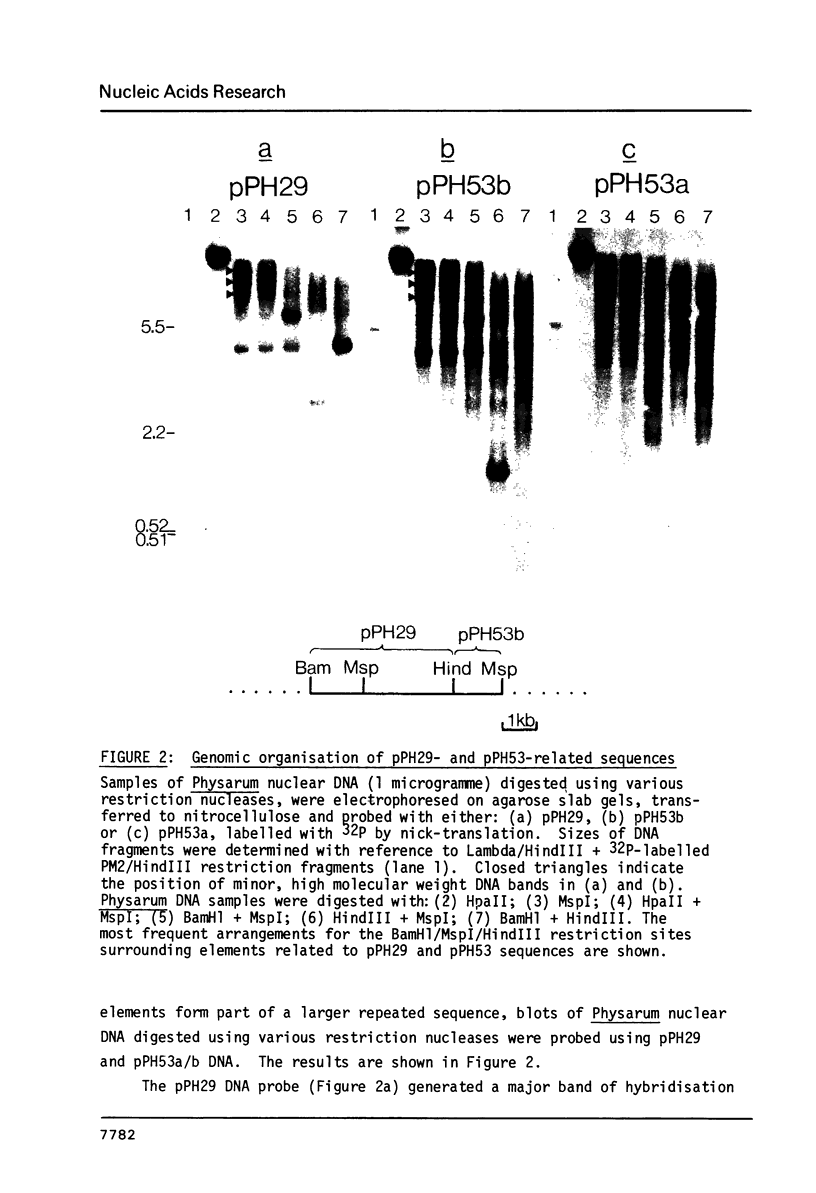
Abstract
A significant portion (20%) of the Physarum genome can be isolated as a HpaII-resistant, methylated fraction. Cloned DNA probes containing highly-repeated sequences derived from this fraction were used to define the pattern of structural organisation of homologous repeats in Physarum genomic DNA. It is shown that the probes detect an abundant, methylated family of sequences with an estimated genomic repetition frequency greater than 2100, derived from a large repeated element whose length exceeds 5.8kb. Sequences comprising the long repetitive element dominate the HpaII-resistant compartment and account for between 4-20% of the Physarum genome. Detailed restriction/hybridisation analysis of cloned DNA segments derived from this compartment shows that HpaII/MspI restriction sites within some copies of the long repeated sequence are probably deleted by mutation. Additionally, segments of the repeat are often found in different organisational patterns that represent scrambled versions of its basic structure, and which are presumed to have arisen as a result of recombinational rearrangement in situ in the Physarum genome. Preliminary experiments indicate that the sequences are transcribed and that the structural properties of the repeat bear some resemblance to those of transposable genetic elements defined in other eukaryotic species.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Hardman N., Jack P. L., Brown A. J., McLachlan A. Distribution of inverted repeat sequences in nuclear DNA from Physarum polycephalum. Eur J Biochem. 1979 Feb 15;94(1):179–187. doi: 10.1111/j.1432-1033.1979.tb12884.x. [DOI] [PubMed] [Google Scholar]
- Houck C. M., Rinehart F. P., Schmid C. W. A ubiquitous family of repeated DNA sequences in the human genome. J Mol Biol. 1979 Aug 15;132(3):289–306. doi: 10.1016/0022-2836(79)90261-4. [DOI] [PubMed] [Google Scholar]
- Kole L. B., Haynes S. R., Jelinek W. R. Discrete and heterogeneous high molecular weight RNAs complementary to a long dispersed repeat family (a possible transposon) of human DNA. J Mol Biol. 1983 Apr 5;165(2):257–286. doi: 10.1016/s0022-2836(83)80257-5. [DOI] [PubMed] [Google Scholar]
- Levis R., Dunsmuir P., Rubin G. M. Terminal repeats of the Drosophila transposable element copia: nucleotide sequence and genomic organization. Cell. 1980 Sep;21(2):581–588. doi: 10.1016/0092-8674(80)90496-1. [DOI] [PubMed] [Google Scholar]
- Manuelidis L., Biro P. A. Genomic representation of the Hind II 1.9 kb repeated DNA. Nucleic Acids Res. 1982 May 25;10(10):3221–3239. doi: 10.1093/nar/10.10.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLachlan A., Hardman N. Analysis of foldback sequences and repetitive sequences in cloned segments of nuclear DNA from Physarum polycephalum. Biochim Biophys Acta. 1982 Apr 26;697(1):89–100. doi: 10.1016/0167-4781(82)90049-5. [DOI] [PubMed] [Google Scholar]
- Musti A. M., Sobieski D. A., Chen B. B., Eden F. C. Repeated deoxyribonucleic acid clusters in the chicken genome contain homologous sequence elements in scrambled order. Biochemistry. 1981 May 26;20(11):2889–2899. doi: 10.1021/bi00514a001. [DOI] [PubMed] [Google Scholar]
- Potter S. S., Brorein W. J., Jr, Dunsmuir P., Rubin G. M. Transposition of elements of the 412, copia and 297 dispersed repeated gene families in Drosophila. Cell. 1979 Jun;17(2):415–427. doi: 10.1016/0092-8674(79)90168-5. [DOI] [PubMed] [Google Scholar]
- Potter S. S. DNA sequence of a foldback transposable element in Drosophila. Nature. 1982 May 20;297(5863):201–204. doi: 10.1038/297201a0. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Singer M. F. SINEs and LINEs: highly repeated short and long interspersed sequences in mammalian genomes. Cell. 1982 Mar;28(3):433–434. doi: 10.1016/0092-8674(82)90194-5. [DOI] [PubMed] [Google Scholar]
- Smith G. E., Summers M. D. The bidirectional transfer of DNA and RNA to nitrocellulose or diazobenzyloxymethyl-paper. Anal Biochem. 1980 Nov 15;109(1):123–129. doi: 10.1016/0003-2697(80)90019-6. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Long range periodicities in mouse satellite DNA. J Mol Biol. 1975 May 5;94(1):51–69. doi: 10.1016/0022-2836(75)90404-0. [DOI] [PubMed] [Google Scholar]
- Wensink P. C., Tabata S., Pachl C. The clustered and scrambled arrangement of moderately repetitive elements in Drosophila DNA. Cell. 1979 Dec;18(4):1231–1246. doi: 10.1016/0092-8674(79)90235-6. [DOI] [PubMed] [Google Scholar]
- Whittaker P. A., McLachlan A., Hardman N. Sequence organisation in nuclear DNA from Physarum polycephalum: methylation of repetitive sequences. Nucleic Acids Res. 1981 Feb 25;9(4):801–814. doi: 10.1093/nar/9.4.801. [DOI] [PMC free article] [PubMed] [Google Scholar]