Abstract
BDNF is a well-characterized neurotrophin that mediates a wide variety of activities in the central nervous system (CNS), including neuronal differentiation, neuroprotection and synaptic plasticity. The canonical Wnt signaling pathway is a critical regulator of embryonic development and homeostasis in adult tissues. Our group and others recently demonstrated that Wnt signaling induces BDNF expression in neurons and glia. However, the precise relationship between BDNF and Wnt signaling pathways is not understood. Here, we investigated Wnt signaling regulation of BDNF at the transcriptional level by using a combination of bioinformatics and molecular analyses. Analysis of the BDNF gene promoter identified seven binding motifs for Wnt-dependent TCF/LEF transcription factors. Furthermore, specific BDNF promoters were induced by the Wnt3a ligand using chloramphenicol acetyl transferase (CAT) reporter assays, and a dominant-negative TCF4 gene reduced Wnt3a-mediated induction. Finally, Wnt3a induced expression of BDNF and other members of the BDNF signaling pathway in glia cells. Therefore, these data indicate that BDNF is a direct target of Wnt signaling, which provides new insight into the interaction between two essential signaling pathways.
Keywords: Wnt signaling, TCF/LEF, glia, retina, BDNF
Introduction
BDNF is a neurotrophic factor that plays important roles in neuronal survival, neurogenesis, differentiation and neurite growth throughout the central nervous system (CNS) [1]. Multiple types of stimuli are reported to increase BDNF transcript levels, including neuronal activity and tissue injury [1]. During CNS injury, upregulation of BDNF and other pro-survival factors is an intrinsic tissue response that is proposed to protect remaining neurons from further damage [2,3]. Glia cells are an important source of BDNF and targeted over-expression of BDNF in glia protects adjacent neurons from degeneration [4,5]. Because of the significant role of BDNF in neuronal survival, characterizing signaling pathways that regulate BDNF expression in glia will have important implications for understanding neuronal-glia interactions. Additionally, identifying cross-talk between BDNF and other pro-survival pathways will lead to new insights into potential mechanisms of neuroprotection.
The canonical Wnt signaling pathway regulates a wide range of essential processes in embryonic and adult tissues, including neuroprotection, neuronal differentiation and synapse formation. Similar to BDNF, Wnt signaling increases in glia during injury [6], suggesting coordinated regulation. Indeed, the non-typical Wnt activator Norrin induced BDNF in retina glia during NMDA-induced damage, and the Wnt3a ligand induced BDNF in a retina ganglion cell line [7,8]. However, the precise relationship between BDNF and Wnt signaling has not been examined.
The biological effects of Wnt signaling are mediated by TCF/LEF transcriptional activators that bind to specific elements in target genes. The N-terminal domain of Wnt-regulated TCF/LEF proteins associates with β-catenin, which allows interaction with specific DNA sequence motifs. There are four TCF genes in vertebrates, TCF1 (also known as TCF7), TCF3 (TCF7L1), TCF4 (TCF7L2) and LEF1, and several isoforms of each protein are created by using alternative translation start sites and differential splicing. The minimal consensus DNA sequence for binding of all TCF/LEF complexes is A/T-A/T-C-A-A-A/T-G [9,10]. TCF/LEF binding sites have been identified proximal to and large distances away from the transcription start site of target genes and multiple binding sites are often found clustered together within a regulatory element of a gene [11]. There is a significant correlation between TCF binding to a target gene and upregulation of the corresponding transcripts [11], indicating that identification of TCF/LEF binding motifs within a gene is a good predictor of regulation by the canonical Wnt pathway.
To investigate the relationship between Wnt signaling and BDNF expression, we focused on the regulation of BDNF by Wnt signaling at the transcriptional level. We used a combination of in silico genomic analyses and experimental confirmations. Our findings indicate that BDNF is a direct target of the canonical Wnt pathway in glia. Therefore, these results characterize the interaction between two important neuroprotective pathways and increase our understanding of the regulation of the BDNF in the CNS.
Methods
Reagents
The MIO-M1 Muller glia cell line [12], a retina-specific glial type, was cultured inDMEM growth medium supplemented with 10% fetal bovine serum, 100 units/ml of penicillin and 100 μg/ml of streptomycin at 37°C in 5 % CO2. Purified recombinant Wnt3a was obtained from R&D (Manassas, VA) and lithium chloride (LiCl) was from Sigma (St. Louis, MO). Wnt3a conditioned media was from mouse L-cells stably expressing Wnt3a (ATCC, Manassas, VA) and control conditioned media lacking Wnt3a was obtained from parental L-cells (ATCC, Manassas, VA). Both conditioned media were filtered and mixed in a 1:1 ratio with normal media prior to use [7]. All experiments used conditioned media from the same preparation batch to minimize variability.
Promoter analyses
The rat BDNF gene (NM_012513) contains promoters within several of its 5’ exons. Therefore, the entire 3.9 kb gene and the 1kb sequences upstream of the transcription start site (TSS) were downloaded from the UCSC database and both positive and negative strands were considered. The analysis was implemented by a Perl program, as described previously [13]. Poisson distribution was used to calculate the probability that a motif occurs in a particular sequence [13].
Quantitative PCR
Canonical Wnt signaling was induced in MIO-M1 cells with recombinant Wnt3a (100 ng/ml) for 24 hrs. The cells were harvested and total RNA was isolated using Trizol phenol-based extraction (Invitrogen), cDNA was synthesized using Thermoscript (Invitrogen) and QPCR was performedusing the iCycler thermocycler (BioRad). To analyze expression of the BDNF pathway and receptor genes we used the RT2 Profiler PCR Array containing Human Neurotrophin and Receptors (SA Biosciences, Frederick, MD), according to the manufacturer’s directions. The primers amplified the following regions: BDNF: nucleotide position 3538-3729, TrkB: position 2062-2198, Bcl2: position 956-1039.
Chloramphenicol acetyl transferase (CAT) assay
The glia MIO-M1 cell line was transfected with the rat BDNF promoter constructs described in [14]. Pilot assays showed the greatest induction for constructs 2 (rBDNF II 3.7 CAT, using the annotation in [14]), 4 (rBDNF IV 4.4 CAT), 5 (rBDNF IV 0.4 CAT) and 7 (rBDNF I-III minigene), and these were chosen for further analysis. Twenty-four hours after transfection, the cells were treated with Wnt3a-containing conditioned media, control conditioned media or 40 mM LiCl for 24 hrs. For the TCF4 over-expression experiments, the cells were transfected with a TCF4 or dominant-negative TCF4 expression plasmid [15] using lipofectamine, and Wnt signaling was induced by Wnt3a or LiCl for 24 hr. The CAT assays were performed using the FAST CAT green (deoxy) Chloramphenicol Acetyltransferase Assay kit (Invitrogen), according to the manufacturer’s directions. Briefly, the cells were washed with PBS and scraped into lysis solution (40 mM Tris-HCl pH 7.4, 1 mM EDTA, 180 mM NaCl), centrifuged and resuspended in 0.25 M Tris-HCl pH 7.4. The cells were lysed by multiple rounds of freeze-thaws, centrifuged, and the supernatant was collected. FAST CAT substrate was added followed by addition of freshly prepared acetyl CoA and the reactions were incubated for 5 hrs at 37 °C. After the reactions were terminated using ice-cold ethyl acetate, the solvent was evaporated and the residues were dissolved in a small volume of ethyl acetate. Chromatography was performed using a silica gel TLC plate in a chromatography chamber filled with chloroform:methanol solution (85:15 v/v). After the solvent ascended, the plate was air dried and scanned with a fluorescence scanner (Typhoon, Amersham Bioscience). Spot intensity was measured and calculated using NIH ImageJ software and the amount of product was calculated, and compared with a promoter-less CAT plasmid.
Statistical Analysis
Values are reported as mean plus standard deviation. For the CAT assays, unpaired t-test or one-way analysis of variance and Tukey post-test were used for statistical analyses. The differences were considered statistically significant when p<0.05.
Results
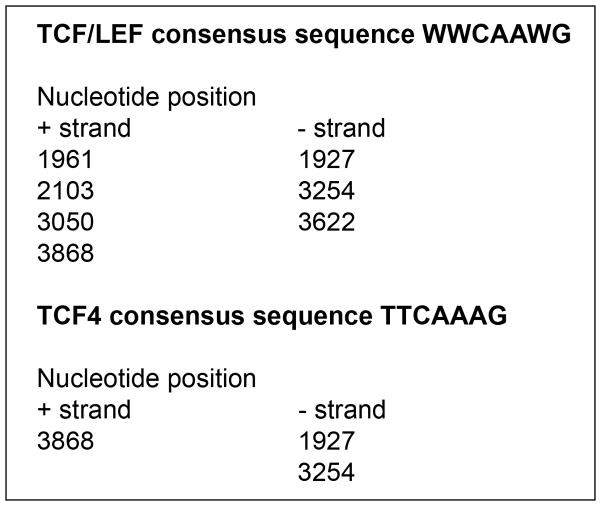
The focus of this study is characterizing BDNF regulation at the transcriptional level. To determine whether BDNF is a direct target Wnt signaling, our approach was to first examine whether TCF/LEF elements are present within the BDNF gene, and then to confirm Wnt-dependent regulation by expression analyses and promoter assays. The rat BDNF transcript contains eight 5’ untranslated exons and one protein-coding 3’ exon; four of the untranslated exons contain promoters that control gene expression [14,16]. TCF/LEF binding motifs were searched for in the entire 3.9 kb coding region of the rat BDNF gene as well as the the 1kb sequences upstream of the transcription start site. The 7-base consensus motif for TCF/LEF (WWCAAWG) was analyzed first [9,10]. As shown in Fig. 1, there are seven copies of the TCF/LEF motif in the positive and negative strands and this number was higher than predicted due to chance (significance level p=0.042). We also analyzed the BDNF gene for the presence of the 7-base consensus motif for TCF4 (TTCAAAG) [9–11] and identified three predicted binding sites in BDNF (significance level p=0.00696). Therefore, the identification of TCF/LEF sites within the BDNF gene provide the first line of evidence that BDNF is a potential direct target of Wnt signaling, with the possibility that even more TCF/LEF sites may lie outside the analyzed region.
Fig. 1.
Tcf binding site prediction in the rat BDNF gene. The positions of the TCF/LEF general consensus and the specific TCF4 consensus motifs are shown in the plus and minus strands.
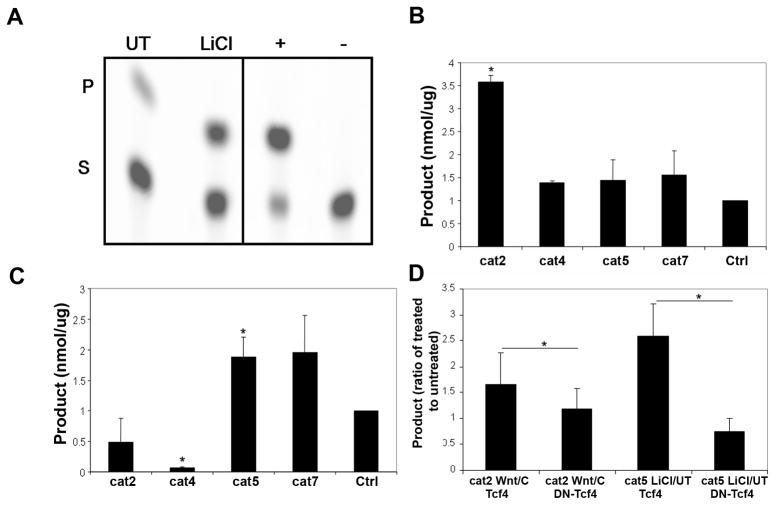
Muller glia cells are a retina-specific glial type that is essential to tissue differentiation, neuronal survival and homeostasis. During injury to the retina, Muller glia secrete BDNF and other growth factors that protect neurons from further damage. We previously demonstrated robust Wnt3a-mediated Wnt signaling in the Muller glia cell line MIO-M1 [6], indicating that they are an appropriate in vitro model to study the relationship between Wnt and BDNF pathways. We used CAT reporter promoter assays to quantitate induction of the BDNF promoters. The CAT constructs contain specific BDNF promoter regions, as described above and in [14]. Each CAT construct was transfected into the glia cell line and the cells were treated with Wnt3a, 40 mM LiCl or control. As shown in Fig. 2, construct 2, which contains a 3.7 kb fragment of exon II and its 5’ flanking region (promoter II), had the highest induction with Wnt3a (ratio of Wnt3a/control was 3.6, n=4, p=0.023). In contrast, LiCl, another activator of the Wnt pathway, showed the highest induction of construct 5, which contains promoter III (LiCl/control ratio was 1.9, n=4, p=0.044). Interestingly, there was a large repression of construct 4, which contains a 5.5 kb fragment of exon IV (LiCl/control 0.07, p=0.005). The difference between the response to Wnt3a and LiCl, especially the lack of activation of construct 2 by LiCl, is unknown but we speculate that it may involve their different mechanisms of action: Wnt3a is an upstream ligand and is specific to the Wnt pathway, whereas LiCl induces Wnt signaling by inhibiting GSK3β and can stimulate other signaling pathways, potentially including pathways that could represses construct 2.
Fig. 2.
The Wnt pathway activators Wnt3a ligand and LiCl induce the BDNF promoter in the glia MIO-M1 cell line. (A) Representative CAT assay of the BDNF promoter construct 7. The upper spot is the product (P), the lower spot is the substrate (S). CAT construct 7 was transfected into glia cells and the cells were untreated or treated with LiCl. The positive control (+) is purified CAT enzyme, the negative control (−) is the CAT vector without an adjacent promoter sequence. (B) Quantification of CAT assays in Wnt3a-treated cells demonstrates that Wnt3a has the strongest effect on construct 2, which contains the exon II promoter and its 5’ flanking sequence. (C) CAT assays on LiCl-treated cells shows the strongest induction on constructs 5, which contains the exon III promoter, and construct 7, which contains the exon I and II promoters and flanking genes. There is also significant repression of construct 4, which contains the exon III promoter and flanking sequences. (D) The dominant-negative TCF4 (DN-TCF4) decreased the Wnt3a and LiCl-induced regulation of constructs 2 and 5, respectively. The assays were normalized to a promoter-less plasmid that contained the CAT enzyme alone (Ctrl), and are expressed as a ratio of Wnt3a or LiCl to their respective control treatments (“C” for Wnt3a, “UT” for LiCl). Mean + SEM. *p<0.05, comparison of DN-TCF4 with TCF4.
To test the specificity of Wnt-dependent induction of BDNF, we transfected the glia cells with dominant-negative TCF4 (DN-TCF4) [15] followed by treatment with Wnt3a and LiCl. The control transfection was wild-type TCF4 [15]. DN-TCF4 reduced Wnt3a-dependent induction of BDNF construct 2 (p<0.01) and also reduced LiCl-dependent induction (p<0.01) of BDNF construct 5.
Finally, to quantify the effect of Wnt signaling on BDNF transcript levels, the glia cells were treated with the Wnt ligand Wnt3a to induce Wnt signaling and gene expression levels were compared between Wnt3a-treated and untreated cells. The expression of BDNF, its receptor TrkB and the downstream BDNF-regulated pro-survival gene Bcl-2 were examined by quantitative PCR. The BDNF primers amplified the 3’ untranslated region, which is common to all splice variants [16] and will detect all BDNF transcripts. As shown in Fig. 3, expression of BDNF and several other genes in the BDNF signaling pathway were induced by Wnt3a. Therefore, the expression analysis, together with the TCF/LEF motif identification and inhibition by DN-TCF4, indicate that BDNF is a direct target of the Wnt pathway.
Fig. 3.
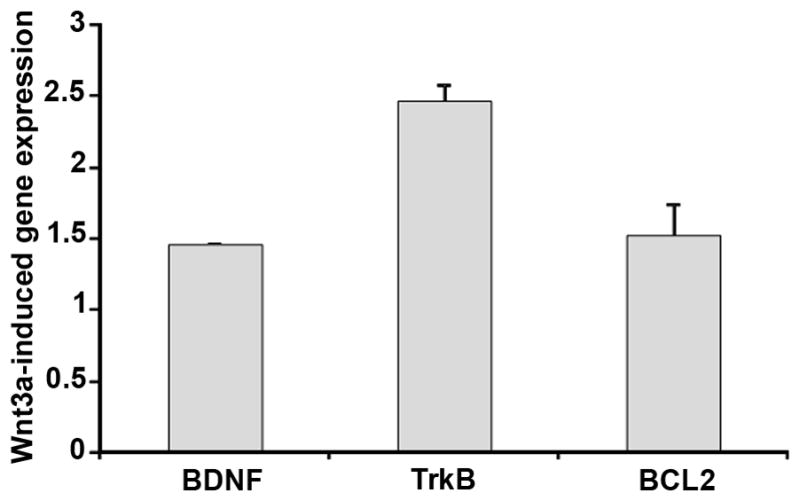
Wnt3a induces BDNF, TrkB and the downstream pro-survival gene Bcl2 in a glia cell line. Quantitative PCR analysis of gene expression in glia treated for 24 hr with recombinant Wnt3a. PCR measurements of each gene were normalized to the housekeeping genes GAPDH and β-actin. The gene expression is shown as a ratio to cells not treated with Wnt3a.
Discussion
BDNF enhances survival of neurons exposed to multiple injuries, including oxidative stress, axotomy and hypoxic ischemic injury [1,2]. However, prolonged exposure to BDNF can directly induce death of neurons and non-neuronal cells [17,18] and increases vulnerability to oxygen/glucose deprivation [19]. Therefore, precise control of BDNF levels and activity is critical. Although much effort has focused on the downstream signaling cascades induced by BDNF and other growth factors, a key question is the identity of the upstream signaling pathways that regulate growth factor induction. In this study, we demonstrated that the Wnt signaling pathway directly induces BDNF expression in Muller glia, which are the principle supportive glia in the retina that are the central mediators of growth factor-mediated protection in the retina [3]. Therefore, our data suggest that Wnt signaling contributes to, or even amplifies, the intrinsic neurotrophic protective tissue response during injury. Our results add to the understanding of how BDNF is regulated in glia and highlight cross-talk between pro-survival pathways.
Despite the number of TCF/LEF binding sites identified in BDNF, the overall expression increase measured by QPCR was moderate. This finding may indicate that cellular injury is required for a higher induction level, or it could be a consequence of the in vitro assay system. Although we found that BDNF was induced by Wnt signaling without the requirement for injury in cell culture, induction of BDNF by the non-typical Wnt activator Norrin in rat retinal glia was only observed when Norrin was combined with NMDA injury, although this was also at moderate levels (approximately 3-fold) [8].
Other factors are also known to regulate BDNF, including upstream stimulatory factors (USF) 1 and 2 bind to Ca2+-responsive E-box elements in the BDNF promoter in response to calcium signaling [20]. Our data do not exclude the possibility that indirect regulation of BDNF expression by Wnt signaling may occur, via Wnt-dependent induction of other signaling pathways. The finding that USF1 is a direct target of TCF4 [11] suggests that Wnt may also control BDNF expression indirectly, as a secondary level of control. Similarly, dopamine-induced transcription of BDNF involves cAMP/CREB binding to the BDNF promoter III [14], and we identified TCF/LEF motifs in CREB proteins (Yi and Hackam, unpublished observations).
Combining expression analyses with identification of TCF binding sites within the promoters of candidate genes gives an indication of potential direct targets of Wnt signaling. TCF/LEF proteins are central to the regulation of gene expression by canonical Wnt signaling, although Wnt-independent activity of TCF/LEF proteins has been reported [21]. For example, recent evidence suggests that TCF proteins act independently of Wnt/β-catenin in the central retina during embryonic development in the mouse [22]. TCF/LEF may also control chromatin structure through their binding motifs [11]. Therefore, the possibility exists that BDNF could also be regulated by TCF4 in the absence of Wnt pathway activation.
A study by Hatzis et al demonstrated significant correlation between TCF4 binding to a target gene and upregulation of the corresponding transcripts [11]. Therefore, identifying TCF/LEF binding sites within a promoter region predicts direct regulation by Wnt signaling. Identification of the specific binding elements that are used will require extensive chromatin immunoprecipitiation and mutagenesis studies. The number of putative TCF/LEF sites within BDNF may have been underestimated in our analysis because many transcription factors bind to regions other than the proximal promoter [11]. Additional sites outside the 1 kb region may be used and may more precisely regulate BDNF expression. Extending our search to cover more than the proximal region would likely identify more sites, but would not alter our overall conclusions.
The canonical Wnt pathway regulates neuronal viability and differentiation, stem cell proliferation and regeneration in the retina and elsewhere in the CNS. Identifying genes regulated by Wnt/TCF/LEF, such as BDNF, provides important clues into how the Wnt pathway exerts its phenotypic effects. Recent reports have demonstrated that Wnt signaling regulates expression of other growth factors, including NT-3 in the limb bud [23], GDNF during kidney development and TGFβ during chondrogenesis [24]. Wnt3a also upregulated NT3, NGF and BDNF in a rat retinal ganglion cell line [25]. Wnt signaling pathway proteins and BDNF are both present in Muller glia, implying that BDNF can be considered a potential target of Wnt ligands in vivo. Confirming that BDNF is a factor in Wnt-dependent activities will require analysis in more complex systems, including animal models, to assess the regulation of BDNF at the protein and activity level, and to determine the importance of Wnt-induced BDNF in specific situations such as differentiation, injury or regeneration.
Acknowledgments
This work was supported by a Research for the Prevention of Blindness Career Development Award, the Karl Kirchgessner Foundation, and the National Institutes of Health (EY017837 to A.S.H., 5R01EY017589 to J.Q.). Institutional support was provided by an unrestricted grant to Bascom Palmer Eye Institute from the Research to Prevent Blindness and National Eye Institute Core Grant P30 EY014801.
The authors wish to thank Robin Smith and Xueping Yu for their assistance. Thanks also to Drs. Tönis Timmusk, Hans Clevers and Astrid Limb for generously providing reagents for use in this study.
Footnotes
Conflicts of interest: none declared
Author contributions
HY performed the molecular experimental analyses, JH and JQ performed the bioinformatics analyses, ASH conceived the study and wrote the manuscript.
References
- 1.Binder DK, Scharfman HE. Brain-derived neurotrophic factor. Growth factors (Chur, Switzerland) 2004;22:123–131. doi: 10.1080/08977190410001723308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hughes PE, Alexi T, Walton M, Williams CE, Dragunow M, Clark RG, et al. Activity and injury-dependent expression of inducible transcription factors, growth factors and apoptosis-related genes within the central nervous system. Progress in neurobiology. 1999;57:421–450. doi: 10.1016/s0301-0082(98)00057-4. [DOI] [PubMed] [Google Scholar]
- 3.Harada T, Harada C, Nakayama N, Okuyama S, Yoshida K, Kohsaka S, et al. Modification of glial-neuronal cell interactions prevents photoreceptor apoptosis during light-induced retinal degeneration. Neuron. 2000;26:533–541. doi: 10.1016/s0896-6273(00)81185-x. [DOI] [PubMed] [Google Scholar]
- 4.Gauthier R, Joly S, Pernet V, Lachapelle P, Di Polo A. Brain-derived neurotrophic factor gene delivery to muller glia preserves structure and function of light-damaged photoreceptors. Invest Ophthalmol Vis Sci. 2005;46:3383–3392. doi: 10.1167/iovs.05-0362. [DOI] [PubMed] [Google Scholar]
- 5.Okoye G, Zimmer J, Sung J, Gehlbach P, Deering T, Nambu H, et al. Increased expression of brain-derived neurotrophic factor preserves retinal function and slows cell death from rhodopsin mutation or oxidative damage. J Neurosci. 2003;23:4164–4172. doi: 10.1523/JNEUROSCI.23-10-04164.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yi H, Nakamura RE, Mohamed O, Dufort D, Hackam AS. Characterization of Wnt signaling during photoreceptor degeneration. Investigative ophthalmology & visual science. 2007;48:5733–5741. doi: 10.1167/iovs.07-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fragoso MA, Yi H, Nakamura RE, Hackam AS. The Wnt signaling pathway protects retinal ganglion cell 5 (RGC-5) cells from elevated pressure. Cellular and molecular neurobiology. 2011;31:163–173. doi: 10.1007/s10571-010-9603-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seitz R, Hackl S, Seibuchner T, Tamm ER, Ohlmann A. Norrin mediates neuroprotective effects on retinal ganglion cells via activation of the Wnt/beta-catenin signaling pathway and the induction of neuroprotective growth factors in Muller cells. J Neurosci. 2010;30:5998–6010. doi: 10.1523/JNEUROSCI.0730-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van de Wetering M, Oosterwegel M, Dooijes D, Clevers H. Identification and cloning of TCF-1, a T lymphocyte-specific transcription factor containing a sequence-specific HMG box. Embo J. 1991;10:123–132. doi: 10.1002/j.1460-2075.1991.tb07928.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, et al. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC−/− colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 11.Hatzis P, van der Flier LG, van Driel MA, Guryev V, Nielsen F, Denissov S, et al. Genome-wide pattern of TCF7L2/TCF4 chromatin occupancy in colorectal cancer cells. Mol Cell Biol. 2008;28:2732–2744. doi: 10.1128/MCB.02175-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Limb GA, Salt TE, Munro PM, Moss SE, Khaw PT. In vitro characterization of a spontaneously immortalized human Muller cell line (MIO-M1) Invest Ophthalmol Vis Sci. 2002;43:864–869. [PubMed] [Google Scholar]
- 13.Hu J, Hu H, Li X. MOPAT: a graph-based method to predict recurrent cis-regulatory modules from known motifs. Nucleic Acids Res. 2008;36:4488–4497. doi: 10.1093/nar/gkn407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Timmusk T, Palm K, Metsis M, Reintam T, Paalme V, Saarma M, et al. Multiple promoters direct tissue-specific expression of the rat BDNF gene. Neuron. 1993;10:475–489. doi: 10.1016/0896-6273(93)90335-o. [DOI] [PubMed] [Google Scholar]
- 15.van de Wetering M, Sancho E, Verweij C, de Lau W, Oving I, Hurlstone A, et al. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111:241–250. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- 16.Aid T, Kazantseva A, Piirsoo M, Palm K, Timmusk T. Mouse and rat BDNF gene structure and expression revisited. J Neurosci Res. 2007;85:525–535. doi: 10.1002/jnr.21139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giehl KM, Rohrig S, Bonatz H, Gutjahr M, Leiner B, Bartke I, et al. Endogenous brain-derived neurotrophic factor and neurotrophin-3 antagonistically regulate survival of axotomized corticospinal neurons in vivo. J Neurosci. 2001;21:3492–3502. doi: 10.1523/JNEUROSCI.21-10-03492.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim SH, Won SJ, Sohn S, Kwon HJ, Lee JY, Park JH, et al. Brain-derived neurotrophic factor can act as a pronecrotic factor through transcriptional and translational activation of NADPH oxidase. J Cell Biol. 2002;159:821–831. doi: 10.1083/jcb.200112131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koh JY, Gwag BJ, Lobner D, Choi DW. Potentiated necrosis of cultured cortical neurons by neurotrophins. Science. 1995;268:573–575. doi: 10.1126/science.7725105. [DOI] [PubMed] [Google Scholar]
- 20.Chen WG, West AE, Tao X, Corfas G, Szentirmay MN, Sawadogo M, et al. Upstream stimulatory factors are mediators of Ca2+-responsive transcription in neurons. J Neurosci. 2003;23:2572–2581. doi: 10.1523/JNEUROSCI.23-07-02572.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoppler S, Kavanagh CL. Wnt signalling: variety at the core. J Cell Sci. 2007;120:385–393. doi: 10.1242/jcs.03363. [DOI] [PubMed] [Google Scholar]
- 22.Fuhrmann S, Riesenberg AN, Mathiesen AM, Brown EC, Vetter ML, Brown NL. Characterization of a transient TCF/LEF-responsive progenitor population in the embryonic mouse retina. Invest Ophthalmol Vis Sci. 2009;50:432–440. doi: 10.1167/iovs.08-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patapoutian A, Backus C, Kispert A, Reichardt LF. Regulation of neurotrophin-3 expression by epithelial-mesenchymal interactions: the role of Wnt factors. Science. 1999;283:1180–1183. doi: 10.1126/science.283.5405.1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tuli R, Tuli S, Nandi S, Huang X, Manner PA, Hozack WJ, et al. Transforming growth factor-beta-mediated chondrogenesis of human mesenchymal progenitor cells involves N-cadherin and mitogen-activated protein kinase and Wnt signaling cross-talk. J Biol Chem. 2003;278:41227–41236. doi: 10.1074/jbc.M305312200. [DOI] [PubMed] [Google Scholar]
- 25.Fragoso M, Yi H, Nakamura R, Hackam A. The Wnt Signaling Pathway Protects Retinal Ganglion Cell 5 (RGC-5) Cells from Elevated Pressure. Cellular and Molecular Neurobiology. 2010 doi: 10.1007/s10571-010-9603-z. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]