Abstract
Melatonin exerts its biological effects through at least two transmembrane G-protein-coupled receptors, MT1 and MT2, and a lower-affinity cytosolic binding site, designated MT3. MT3 has recently been identified as QR2 (quinone reductase 2) (EC 1.10.99.2) which is of significance since it links the antioxidant effects of melatonin to a mechanism of action. Initially, QR2 was believed to function analogously to QR1 in protecting cells from highly reactive quinones. However, recent studies indicate that QR2 may actually transform certain quinone substrates into more highly reactive compounds capable of causing cellular damage. Therefore it is hypothesized that inhibition of QR2 in certain cases may lead to protection of cells against these highly reactive species. Since melatonin is known to inhibit QR2 activity, but its binding site and mode of inhibition are not known, we determined the mechanism of inhibition of QR2 by melatonin and a series of melatonin and 5-hydroxytryptamine (serotonin) analogues, and we determined the X-ray structures of melatonin and 2-iodomelatonin in complex with QR2 to between 1.5 and 1.8 Å (1 Å =0.1 nm) resolution. Finally, the thermodynamic binding constants for melatonin and 2-iodomelatonin were determined by ITC (isothermal titration calorimetry). The kinetic results indicate that melatonin is a competitive inhibitor against N-methyldihydronicotinamide (Ki = 7.2 μM) and uncompetitive against menadione (Ki = 92 μM), and the X-ray structures shows that melatonin binds in multiple orientations within the active sites of the QR2 dimer as opposed to an allosteric site. These results provide new insights into the binding mechanisms of melatonin and analogues to QR2.
Keywords: 2-iodomelatonin, melatonin, melatonin-binding site 3 (MT3), quinone reductase 2 (QR2)
INTRODUCTION
Melatonin is a neurohormone circulating in the blood of mammals at low levels during the day, and at higher concentrations during the night [1]. Melatonin is synthesized mainly in the pineal gland from a precursor, N-acetyltryptamine, that melatonin shares with 5-hydroxytryptamine (serotonin). Melatonin relays circadian information to peripheral tissues by at least two high-affinity binding sites, MT1 and MT2, which are both G-protein-coupled receptors, and by one lower-affinity soluble binding site designated MT3 [2]. MT3 was first described by Dubocovich [3] and Molinari et al. [4] by using the synthetic ligands 2-iodo-melatonin (2-125I-melatonin) and 2-iodo-MCA-NAT (2-125I-5-methoxycarbonylamino-N-acetyltryptamine). Although both ligands bind to MT3 with nanomolar affinity, 2-iodo-MCA-NAT was also found to be very selective towards the newly identified MT3 compared with MT1 and MT2.
Recently, MT3 was purified to homogeneity and identified as QR2 (quinone reductase 2) {also known as NQO2 [NRH (N-ribosyldihydronicotinamide):quinone oxidoreductase 2)]} (EC 1.10.99.2) [5]. QR2 was initially described as a flavoprotein with unexpected co-substrate specificity. Indeed, this enzyme, apparently an analogue of the well-known NAD(P)H-dependent quinone reductase 1 (QR1, NQO1 or DT-diaphorase (EC 1.6.5.2), does not recognize NADH or NAD(P)H as co-substrates. Instead, QR2 recognizes substrates such as reduced N-ribosylnicotin-amide or N-methylnicotinamide which are the breakdown products of these cofactors [6–9]. The activity of QR2 is typically measured with synthetic substrates such as the vitamin K3 derivative menadione, or cytotoxic compounds such as CB1954 or mytomycin C. Because of its close similarity to QR1, it was believed that QR2 might also serve as a detoxification enzyme that can produce hydroquinones which are less toxic to cells compared with the parent quinones [10].
QR2 is a cytosolic enzyme with many enigmatic features [11]. It is one of the most potently inhibited (nanomolar) molecular targets of resveratrol, a natural polyphenol found in wine and peanuts, and the X-ray structure of QR2 in complex with resveratrol has been determined [12]. Mice that have been fed resveratrol have been shown to be less sensitive to menadione-induced toxicity compared with control mice, and this observation parallels the observation that QR2−/− mice are also less sensitive to menadione-induced toxicity compared with their wild-type littermates. The toxicity of menadione in the wild-type animals has been shown to increase upon co-administration of a QR2 co-substrate [13], and cells derived from QR2−/− animals or QR2-RNAi (RNA interference) knockdown cells have been shown to be less sensitive to menadione toxicity [12,14].
QR2 also functions as an atypical melatonin-binding site, known as MT3, which was initially identified and subsequently characterized in membrane homogenates of animal tissues via the synthetic ligands 2-iodomelatonin and 2-iodo-MCA-NAT. The higher specific activity of radio-iodinated ligands, compared with that of [3H]melatonin, allowed the identification and characterization of this low-affinity melatonin-binding site. Evidence that QR2 may correspond to MT3 came from a study in which organs from mice that were deprived of the NQO2 gene (QR2−/− mice), were shown to lack MT3 [15]. The possibility that QR2 is also MT3 would help to shed new light on the antioxidant properties of melatonin at high concentrations, i.e. above 1 μM (see [16] for a review). For instance, in a study whereby a large number of structurally unrelated quinones were tested as inhibitors or substrates of QR2, coenzyme Q0, a recognized antioxidant compound [17,18], was found to be a potent substrate [2]. This observation suggests a link between cellular antioxidant status and QR2.
In the present paper, we report a series of studies aimed at probing the structural, kinetic and thermodynamic properties of melatonin and melatonin analogues binding to human QR2 in order to ascertain the mechanism of inhibition of QR2 and the precise location of the melatonin-binding site. Together, the data lend support to the hypothesis that QR2 and MT3 are the same protein, and that melatonin and its analogues bind to and inhibit QR2 within the active site and not at an allosteric site.
EXPERIMENTAL
Reagents
All chemicals were purchased from Sigma, unless stated otherwise. Melatonin and IPTG (isopropyl β-D-thiogalactoside) were purchased from Acros Organics. 2-Iodomelatonin was obtained from Tocris.
Cloning of the human NQO2 gene
Human NQO2 cDNA (MGC-12729) was purchased from the A.T.C.C. (Manassas, VA, U.S.A.). The cloning procedure reported by Wang et al. [19] was followed. The primers used in cloning were: 5′-AACCATGGCAGGTAAGAAAGTACTC-3′ (added NcoI site is underlined) and 5′-AACTCGAGTTATTGCC-CGAAGTGCCAGTG-3′ (added XhoI site is underlined). The PCR product was digested with NcoI and XhoI restriction enzymes, purified and then ligated into a pET-23d expression vector (Novagen).
Expression and purification of human QR2
Human QR2 was purified from 3 litres of Escherichia coli BL21(DE3) cells grown in Luria–Bertani medium supplemented with 100 μg/ml ampicillin (Fisher Scientific). Cells were grown at 37°C until a D600 of 0.7–0.8 was achieved. At this point, expression of QR2 was induced by addition of IPTG to a final concentration of 1 mM, and cultures were transferred to 16°C for 12–16 h. The cells were collected by centrifugation at 4300 rev./min for 15 min in a Sorvall Super-T21 centrifuge (ST-H750 rotor) and stored at −80°C. The frozen cell pellet was thawed on ice and then resuspended in ice-cold buffer A [25 mM Tris/HCl (pH 8.0), 10 mM NaCl and 1 mM DTT (dithiothreitol)] containing two tablets of EDTA-free Complete™ mini-protease inhibitor cocktail tablets (Roche Diagnostics) and 0.5 mM PMSF. The cell suspension was sonicated at 600 W for a total of 6 min using a high-intensity ultrasonic processor (Sonic processor VCX600), with a pulse of 6.6 s on and 9.9 s off. The lysed cells were pelleted by centrifugation at 18 000 rev./min for 45 min at 4°C in a Sorvall Super T21 centrifuge (SL-50T rotor). The supernatant was applied to a 30 ml XK26 DEAE-Sepharose column (GE Healthcare) equilibrated with buffer A. The enzyme was obtained by washing with 5%buffer B (25 mM Tris/HCl, pH 8.0, 1 M NaCl and 1 mM DTT) in 8 ml fractions. Fractions containing the enzyme (detected by SDS/PAGE) were pooled and concentrated to greater than 10 mg/ml using an 80 ml Centricon Plus-20 concentration device from Millipore (molecular mass cut-off of 10 000 Da) to a final volume of 3–5 ml.
The enzyme (3–5 ml) was then applied to a Hiload 26/60 Superdex 75 (GE Healthcare) column (300 ml) equilibrated with buffer A. The enzyme was eluted with buffer A, and the fractions containing pure enzyme were pooled and finally applied to a 10/100 GL MonoQ (GE Healthcare) strong anion-exchange column (8 ml) equilibrated with buffer A. The protein was eluted using a gradient ranging from 5 to 50% buffer B. The purest protein fractions were pooled, buffer-exchanged with 50 mM Tris/HCl (pH 8.0), 150 mM NaCl, and 10% glycerol, and concentrated using the Centricon Plus-20 device to a final concentration of ~7–30 mg/ml. Freshly prepared, i.e. non-frozen, QR2 was used to set up crystallization trays. For kinetic and other biochemical assays, QR2 that was stored at −80°C after buffer-exchange and concentration was used. During the purification, the protein concentration was measured using the Bradford assay using BSA as a standard.
Co-crystallization of QR2 with melatonin and 2-iodomelatonin
Melatonin and 2-iodomelatonin were dissolved to concentrations of 100 mM in ethanol and DMSO respectively. Aliquots of the protein were mixed separately with the inhibitors and then incubated for 15 min on ice. The final protein concentration was 16 mg/ml, and the final concentration of the inhibitors was 1 mM. Crystallization trays were prepared the same day using the hanging-drop vapour-diffusion method, by mixing 2 μl of protein with 2 μl of reservoir solution on a siliconized coverslip. The reservoir solution contained 0.1 M Hepes (pH 6.0–7.0), 0.1 M NaCl, 12 μM FAD, 5 mM DTT and 1.3–1.7 M ammonium sulfate. Crystals suitable for X-ray data collection grew to dimensions of approx. 0.2–0.5 mm after 7–14 days.
X-ray data collection and structure refinement
All diffraction data were collected at 100 K on an ADSC Quantum-315 detector at Bio-CARS beamline 14 BM-C at the Advanced Photon Source, Argonne National Laboratory, Argonne, IL, U.S.A. Complete datasets on three separate crystals were collected for QR2 in complex with melatonin. The crystals diffracted between 1.5 and 1.8 Å (1 Å =0.1 nm) resolution. Complete datasets on two separate crystals were collected for the QR2–2-iodomelatonin complex. These crystals diffracted to 1.6 and 2.3 Å. X-ray data were integrated and scaled using either XDS [20] or the HKL2000 [21] suite of programs. All crystals belonged to space group P212121 with a dimer in the asymmetric unit. The initial phases for QR2 in complex with the inhibitors were determined by the method of molecular replacement using PHASER in the CCP4 suite (Collaborative Computational Project) [22]. The search model used for molecular replacement was the structure of human QR2 in complex with resveratrol (PDB entry 1SG0 [12]).
Alternate cycles of model building using O [23] and refinement using CNS (Crystallography and NMR System) [24] were performed until the parameter shifts converged and until no large peaks in the σA-weighted Fo − Fc electron-density maps were observed. Co-ordinates and molecular topology and parameter files for FAD were obtained from the HIC-Up website [25]. Structures for melatonin and 2-iodomelatonin were built using Sybyl7.0 (Tripos). Dihedral angles and torsion bonds of the ligands were determined, and the topology and parameter files were generated using the program Xplor2D [26–28]. These ligands were manually adjusted to fit the electron-density map using O, and their positions were refined using CNS. Water molecules were added to the structural model followed by further positional and individual B-factor refinement. The final model was validated using Procheck [29] and MolProbity [30]. X-ray data processing and refinement statistics are summarized in Supplementary Table S1 (http://www.BiochemJ.org/bj/413/bj4130081add.htm). Figures were produced using the program PyMOL (http://pymol.sourceforge.net). The atomic co-ordinates and structure factors have been deposited in the Protein Data Bank [31] under codes 2QWX, 2QX4 and 2QX6 for the QR2–melatonin complex, and 2QX8 and 2QX9 for the QR2–2-iodomelatonin complex.
ITC (isothermal titration calorimetry) experiments
Calorimetric titrations of QR2 with melatonin, 2-iodomelatonin and resveratrol were performed using a VP-ITC microcalorimeter (MicroCal). Samples of QR2 were extensively dialysed against ITC buffer consisting of 50 mM Hepes (pH 7.5), 100 mM NaCl and 1 mM TCEP [tris-(2-carboxyethyl)phosphine]. All samples were filtered through a 0.22 μm sterile filter membrane (Millipore), and thoroughly degassed by gentle stirring under vacuum. The 1.35 ml sample cell was filled with a 20 μM solution of protein and the 250 μl syringe was filled with either 500 μM melatonin or 400 μM 2-iodomelatonin. For the titration with resveratrol, 10 μM solution of the protein and 250 μM ligand were used. Each titration was performed by performing an initial injection with 1 μl of titrant followed by 60 subsequent 2 μl injections, which were all spaced by 5 min intervals. All ITC titration data were fitted to the single-site binding model using the program Origin 7.0 (MicroCal).
Steady-state kinetics and inhibitor studies
The activity of QR2 under steady-state conditions was evaluated on a Molecular Devices SpectraMax Plus 384 UV–visible spectrophotometer by monitoring the decrease in absorbance of the enzyme co-substrate NMeH (N-methyldihydronicotinamide) at 360 nm at 25°C. Reactions were carried out in a 96-well plate and were initiated by the addition of QR2 to the assay buffer (50 mM Tris/HCl, pH 8.0, 100 mM NaCl and 0.1% Triton X-100) containing various concentrations of menadione (5–75 μM), and various concentrations of NMeH (10–140 μM). Melatonin concentrations in the assay ranged from 50 to 500 μM. Stock QR2 enzyme concentrations were determined using the Bio-Rad protein assay. The final enzyme concentration was 5 nM in a reaction volume of 200 μl. The plate was shaken vigorously for 5 s to mix reagents, and the loss in absorbance upon oxidation of NMeH was monitored until the reaction reached completion. Reaction rates were converted into specific activity using ε360 = 7060 M−1 · cm−1 for NMeH [32], with a well pathlength of 0.445 cm. The specific activity of QR2 is expressed in μmol of NMeH oxidized per ml per min per mg of QR2 added (units/mg). One unit of activity is defined as 1 μmol of NMeH oxidized per min.
The inhibition of QR2 activity by resveratrol, melatonin and melatonin analogues was determined using the same assay conditions described above, except for the following differences. The reactions were initiated by adding the enzyme to a final concentration of 5 nM and to a reaction mixture containing 100 μM NMeH, 30 μM menadione and different concentrations of inhibitor (resveratrol, melatonin and analogues). The inhibitory concentration that produces 50%inhibition (IC50 value) for each inhibitor was calculated by fitting the kinetic data to the following equation:
Kinetic data analysis
All initial velocity data were measured in triplicate, and the mean ± S.D. rate was plotted as a function of substrate or inhibitor concentration. Kinetic data were fitted to either the Michaelis–Menten equation or to standard equations describing competitive, uncompetitive and non-competitive inhibition models using the Enzyme Kinetics Module of the SigmaPlot (version 9.0) software package. Kinetic parameters Km, Ki and Vmax were derived from the best-fit curves obtained by non-linear regression. The reaction mechanisms were initially determined qualitatively by inspection of the double-reciprocal plots. Inhibition models were excluded if the errors in the kinetic parameters were large or the fitting algorithm failed to converge. The model that fitted the kinetic data best was selected using the goodness-of-fit criterion AIC (Akaike Information Criterion) and the coefficient of determination (r2) derived from SigmaPlot 9.0 (Supplementary Table S2 at http://www.BiochemJ.org/bj/413/bj4130081add.htm). The best-fitting models have the lowest AIC values and r2 values close to 1.
MCA-NAT docking
MCA-NAT docking was performed using the crystal structure of QR2 in complex with resveratrol (Protein Data Bank code 1SG0). Before docking, water and resveratrol molecules were discarded from the protein co-ordinate file. Hydrogen atoms and charges were added to both protein and ligand using Sybyl7.2 (Tripos) before converting their files into mol2 format. MCA-NAT was also energy-minimized via 1000 cycles of conjugate gradients. Automated docking was performed using the genetic algorithm GOLD (Genetic Optimization for Ligand Docking, version 3.1; Cambridge Crystallographic Data Centre, Cambridge, U.K.) [33]. The implementation of the program for QR2 was first validated by obtaining docking orientations for resveratrol, which were nearly identical with the orientation observed in the X-ray structure. The binding site was defined by using only the amino acid residues within 10 Å of resveratrol in the X-ray structure. The GOLD chemical score was chosen as the fitness function, and the default settings were used in the calculations. Default cut-off values of 2.5 Å for hydrogen bonds and 4 Å for van der Waals interactions were employed. The two metal ions, one in each monomer of the asymmetric unit, were set to allow tetravalent co-ordination according to a Zn2+ ion type. Early termination was allowed for results differing by less than 1.5 Å in ligand all-atom RMSD (root mean square deviation). After completion of the docking computational runs, visual analysis of the results was performed using Sybyl7.2 in order to determine any hydrogen bonds and van der Waals interactions formed between the ligand and QR2.
RESULTS
Expression and purification of human QR2
QR2 is a cytosolic protein that can be easily expressed and purified from bacteria. The purification procedure involved three chromatographic steps including a DEAE-Sepharose weak anion-exchange step, a Superdex 75 gel-filtration step and a Mono-Q strong anion-exchange step. In the end, approx. 30 mg of pure QR2 was obtained per litre of cell culture (Supplementary Figure S1 at http://www.BiochemJ.org/bj/413/bj4130081add.htm). The final specific activity of pure QR2 (5 nM) with the substrate menadione (30 μM) and the co-substrate NMeH (100 μM) was approx. 12 300 units/mg which corresponds to a turnover number (kcat value) of 205 s−1.
Determination of IC50 values for melatonin and analogues
Since melatonin and a variety of its analogues are known to bind to MT3, and since MT3 and QR2 are considered the same proteins, we determined the IC50 values of melatonin and a series of structurally related analogues and compared these values with the IC50 of resveratrol, a known QR2 inhibitor [12] (Table 1). Using menadione and NMeH as the substrates, we determined an IC50 of 11 μM for melatonin and 1 μM for 2-iodomelatonin. The IC50 value for resveratrol is 1 μM under these assay conditions, indicating that it is a better inhibitor than melatonin by one order of magnitude. By adding a single iodine atom to the 2-position of the indole ring, 2-idodomelatonin showed an affinity for QR2 that was almost one order of magnitude stronger than melatonin. Among the compounds studied, 5-hydroxytryptamine displayed the weakest inhibitory activity towards the QR2 enzyme, with an IC50 value of 54 μM, which is approx. 5- and 50-fold higher than that of melatonin and 2-iodomelatonin.
Table 1.
Comparison of the IC50 values for compounds binding to either recombinant human QR2 or the cytosolic fraction of mammalian cell lines overexpressing human QR2
| Compound | Structure | IC50 value |
|
|---|---|---|---|
| Recombinant (μM)* | Mammalian cells (μM)† | ||
| Melatonin | 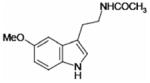 |
11.3 ± 2.1 | 130 ± 35 |
| 2-Iodomelatonin | 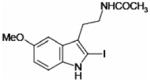 |
1.1 ± 0.2 | 16 ± 2 |
| N-Acetyl-5-hydroxytryptamine | 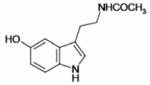 |
9.9 ± 1.8 | 99 ± 14 |
| 5-Hydroxytryptamine | 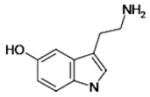 |
53.9 ± 19.6 | ND |
| MCA-NAT | 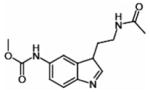 |
ND | 295 ± 19 |
| Resveratrol | 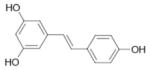 |
0.96 ± 0.13 | 9.1‡ |
Results are means ± S.D. for triplicate experiments. ND, not determined.
The co-substrate used in the reaction was NMeH.
Data taken from [37] using the cytosolic fraction of mammalian cell lines overexpressing human QR2 in the presence of dihydrobenzylnicotinamide as co-substrate.
J.A. Boutin and P. Delagrange, unpublished work.
Binding thermodynamics of melatonin and 2-iodomelatonin to QR2
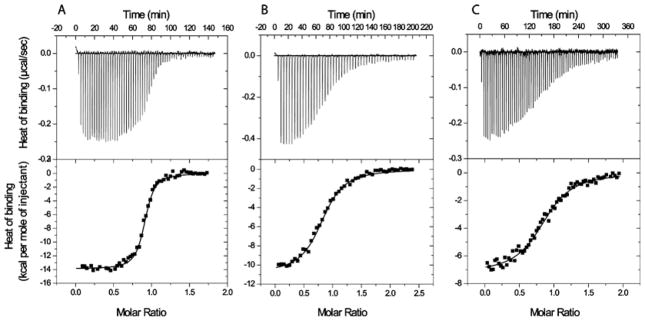
Since the IC50 values are derived from kinetic data on the enzyme in the presence of substrates, they are not bona fide thermodynamic constants. Therefore we determined the dissociation constant (Kd) values for melatonin, 2-iodomelatonin and resveratrol from the free QR2 enzyme using ITC. The Kd value for resveratrol served as a control for the ITC experiments since it has been reported to bind tightly to the enzyme and since its structure in complex with QR2 has been determined [12]. The binding isotherm for the interaction of resveratrol with QR2 is shown in Figure 1(A). Resveratrol was found to interact strongly with QR2 with a Kd of 39 nM. This value is in good agreement with the Kd value of 35 nM determined by Buryanovskyy et al. [12], who titrated QR2 with increasing concentrations of resveratrol, while monitoring the change in intrinsic tryptophan fluorescence [12]. After validating the ITC methodology using resveratrol, we next performed ITC titrations of QR2 with melatonin (Figure 1B) and 2-iodomelatonin (Figure 1C). The Kd values for melatonin and 2-iodomelatonin together with their thermodynamic parameters are reported in Table 2.
Figure 1. ITC of human QR2 with various ligands.
QR2 was titrated with (A) resveratrol, (B) melatonin and (C) 2-iodomelatonin. Each titration experiment was carried out in duplicate or triplicate. ITC traces (upper panels) and binding isotherms (lower panels) are shown for each ligand. The continuous line in the lower plot represents a best fit of the data to a single-binding-site model which was used to derive the values for the fitted parameters ΔG, ΔH, ΔS and the stoichiometry of binding. The Kd values for each titration experiment were calculated from the ΔG values, and mean values are shown. The resulting K d values were as follows: (A) resveratrol, 39 ± 0.011 nM; (B) melatonin, 1.4 ± 0.43 μM; and (C) 2-iodomelatonin, 0.7 ± 0.4 μM. The mean values of the thermodynamic parameters ΔG, ΔH and ΔS, together with the stoichiometry of binding (h), are shown in Table 2.
Table 2.
Thermodynamic parameters of resveratrol, melatonin and 2-iodomelatonin for binding to QR2
| Compound | ΔH (kcal · mol−1) | −TΔS (kcal · mol−1) | ΔG (kcal · mol−1) | Kd (μM) | h |
|---|---|---|---|---|---|
| Resveratrol | −14.6 ± 0.15 | 5.01 ± 0.61 | −9.8 ± 0.50 | 0.039 ± 0.011 | 0.9 |
| Melatonin | −10.45 ± 0.4 | 2.38 ± 0.11 | −8.07 ± 0.3 | 1.4 ± 0.43 | 0.8 |
| 2-Iodomelatonin | −7.85 ± 0.62 | −0.65 ± 0.24 | −8.5 ± 0.38 | 0.70 ± 0.4 | 0.8 |
Results are means ± S.D. for duplicate or triplicate experiments. h is the Hill coefficient, a measure of the stoichiometry of binding.
The binding of resveratrol, melatonin and 2-iodomelatonin to QR2 are almost entirely driven by enthalpy (ΔH) as observed from their large negative, i.e. favourable, enthalpy values ranging from −7.9 to −14.6 kcal/mol (1 kcal=4.184 kJ) (Table 2). A comparison of the ΔG values for melatonin and 2-iodomelatonin indicate that they are nearly identical (−8.1 compared with −8.5 kcal/mol). The ΔH value for melatonin (−10.45 kcal/mol), on the other hand, is approx. 2.6 kcal/mol more favourable than the ΔH value for 2-iodomelatonin (−7.85 kcal/mol). The near equivalent ΔG values, despite the different ΔH values, stems from the differences in the ΔS values where TΔS is 2.4 kcal/mol for melatonin and −0.65 kcal/mol for iodomelatonin. Such a trend is characteristic of a process known as enthalpy/entropy compensation. Such compensation occurs upon binding of melatonin and 2-iodomelatonin to QR2: the binding entropy increases when the binding enthalpy decreases, resulting in a constant value of free energy of binding. As a result, a favourable enzyme–ligand complex is formed [34]. Although the ΔH value for resveratrol is approx. 4.2 kcal/mol more favourable (more negative) than the value for melatonin, and also 6.8 kcal/mol more favourable than the value for 2-iodomelatonin, the differences between the ΔG values of resveratrol, melatonin and 2-iodomelatonin are only approx. 1.7 kcal/mol. The smaller difference is due to a more unfavourable value of TΔS of 5 kcal/mol for resveratrol. Thus, despite a very strong enthalpic driving force for the interaction between resveratrol and QR2, the strength of interaction is weakened by the TΔS term.
Mechanism of inhibition of QR2 by melatonin
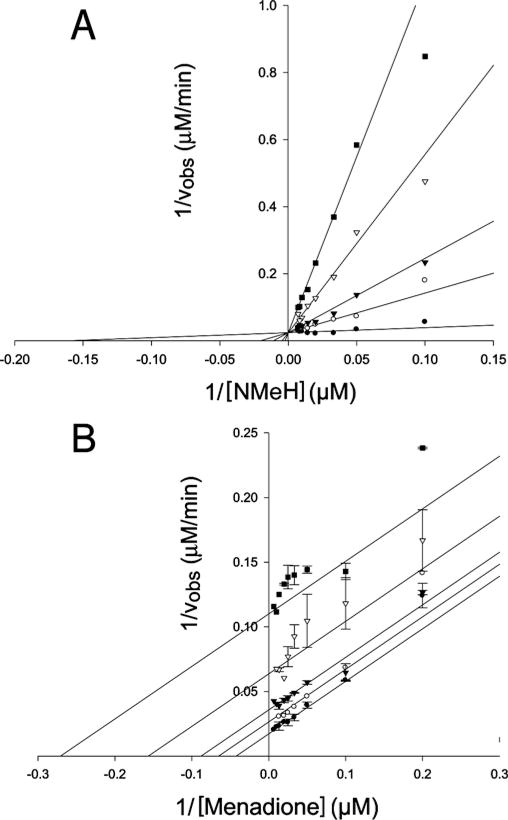
To determine the mechanism of inhibition of QR2 activity by melatonin, steady-state kinetic studies were performed by varying the concentrations of either the substrate (menadione) or the co-substrate (NMeH). The steady-state kinetic data are plotted in double-reciprocal form in Figure 2. The data were fitted to equations describing competitive, non-competitive and uncompetitive inhibition models using non-linear regression, and the best-fit lines are shown. From the various fits and the patterns of lines observed in the double-reciprocal plots, it was determined that melatonin is a competitive inhibitor towards the co-substrate NMeH (Ki = 7.2 μM) and an uncompetitive inhibitor towards the substrate menadione (Ki = 92 μM). These results indicate that melatonin binds to the free non-reduced (i.e. FAD) form of QR2. The uncompetitive inhibition with respect to menadione would be expected for a kinetic reaction mechanism that is Ping Pong. In such a mechanism, the inhibitor, melatonin, is directly dependent upon the substrate, menadione, for the conversion of the reduced enzyme form, E*–FADH2, back to the oxidized enzyme form, E–FAD. In other words, melatonin can only bind to the E–FAD form of QR2 and not to the E*–FADH2 form. In the latter case, E* represents a different conformationally distinct enzyme form that is commonly designated as ‘F’ in Ping Pong mechanisms.
Figure 2. Steady-state inhibition of QR2 by melatonin.
(A) Kinetic data were recorded at a fixed concentration of menadione (30 μM) and at various concentrations of NMeH as indicated. The resulting kinetic parameters are Vmax = 41.8 ± 2.2 μM/min, Km = 6.2 ± 2.3 μM, K i = 7.2 ± 2.4 μM; r2 =0.93. (B) Kinetic data were recorded at a fixed concentration of NMeH (100 μM) and at various concentrations of menadione as indicated. The resulting kinetic parameters are Vmax = 57.5 ± 1.6 μM/min, Km = 23.2 ± 1.9 μM, Ki = 91.6 ± 4.6 μM; r2 = 0.97. The data are plotted as a Lineweaver–Burk plot of QR2 activity with 0 (●), 50 (○), 100 (▼), 250 (▽) and 500 (■) μM melatonin. Data points are the means of three replicates. Continuous lines represent best-fit lines calculated using the kinetic module in SigmaPlot 9.0.
X-ray structure of QR2 in complex with melatonin
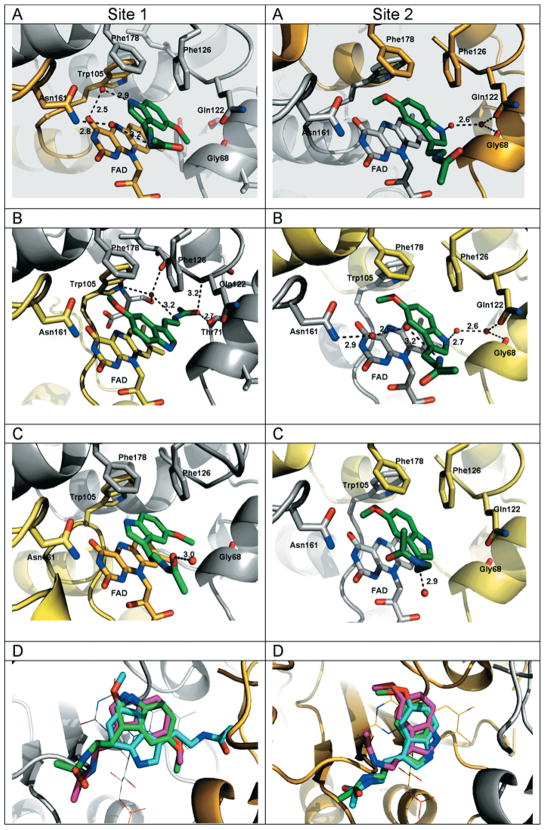
To gain insight into the binding mode of melatonin to the free FAD form of QR2, we determined the crystal structure of QR2 in complex with melatonin. We collected, processed and refined complete X-ray datasets on three different crystals of the same QR2–melatonin complex in order to probe for any stochastic conformational differences. The three crystals diffracted to about the same resolution (1.5, 1.7 and 1.8 Å) (Supplementary Table S1). QR2 is a dimer in solution and crystallizes as a dimer in the asymmetric unit (biological dimer) with two nearly equivalent catalytic sites that we will define here as site 1 (chain A) and site 2 (chain B) (Supplementary Figure S2A at http://www.BiochemJ.org/bj/413/bj4130081add.htm). Each site consists of a deep hydrophobic cavity with one face exposed to the solvent (Supplementary Figure S2B). Electron density for melatonin was clearly visible in both active sites of the dimer and in all three of the structures from different crystals (Supplementary Figure S3 at http://www.BiochemJ.org/bj/413/bj4130081add.htm). Surprisingly, the orientations of melatonin differ between sites 1 and 2 of a single dimer (Figure 3). Moreover, the orientations differ between site 1 in the three crystal structures. Thus melatonin binds to the enzyme in three major orientations: in site 1, melatonin adopts two different binding modes (Figure 3D, Site 1), whereas in site 2, melatonin adopts essentially the same orientation (Figure 3D, Site 2), which is different from the orientations adopted in site 1.
Figure 3. Structures of QR2 complexed to melatonin.
(A) Sites 1 and 2 for the complex at 1.5 Å resolution. (B) Sites 1 and 2 for the complex at 1.7 Å resolution. (C) Sites 1 and 2 for the complex at 1.8 Å resolution. (D) A superposition of the three sites of each monomer is shown to summarize the three different orientations adopted by melatonin. The protein is shown as a ribbon representation. The side chains of the residues in the active site are shown as stick representations, coloured according to subunit type (chain A in yellow, chain B in grey). FAD and melatonin are shown as stick representations coloured according to atom type. FAD is coloured according to which monomer it belongs. Hydrogen bonds are shown as broken lines, water molecules as red spheres. Figure prepared using PyMOL (DeLano Scientific; http://www.pymol.org).
In the first orientation, common to site 1 of two crystal structures (Figures 3A and 3C, Site 1), the indole moiety of melatonin stacks in parallel and on top of the FAD isoalloxazine ring. Hydrophobic interactions appear to be the strongest determinants in melatonin binding. Hydrophobic interactions (distance < 4 Å) with Trp105, Gly68′, Phe126′ and Phe178′ help to stabilize melatonin binding via interactions with the indole ring and carboxymethyl group. The pyrrole nitrogen is able to interact with the carbonyl oxygen in the 2-position of the FAD ring through a water molecule. In one structure, the amide nitrogen on the melatonin side chain makes a hydrogen bond with Asn161 via a relay through a water molecule. In contrast, in the structure of another complex, the side chain of melatonin is oriented more toward the solvent-exposed surface, and the carbonyl oxygen is hydrogen-bonded to a water molecule.
In the second orientation (Figure 3B, Site 1), melatonin is positioned so that the indole ring only partially stacks on top of the flavin cofactor ring. The long side chain of melatonin is pointed towards the interior of the active site, which allows the carbonyl oxygen to bind directly to both the OH group of Thr71 and the backbone nitrogen of Gln122. Once again, the melatonin ring and carboxymethyl groups make several hydrophobic interactions with Phe106, Trp105, Tyr67′ and Cys121′. Finally, in the third orientation, analogous in site 2 of each structure (Figures 3A–3C, Site 2), melatonin is oriented so that only its benzene ring stacks on top of the FAD piperazine-like moiety. The melatonin side chain points only towards the solvent-exposed surface in this third orientation.
Although the general orientations of melatonin in site 2 of all three structures are very similar, the neurohormone does not make the same interactions with residues within the active sites. In the two highest-resolution structures (1.5 and 1.7 Å), the pyrrole nitrogen of melatonin interacts with water molecules that are bridged to Gly68, Gln122 and Asn66. In the 1.8 Å structure, only the side-chain nitrogen of melatonin forms a hydrogen bond with a water molecule. Interactions of the melatonin indole ring and carboxymethyl group with several hydrophobic residues (Phe126, Phe178, Phe106′, Trp105′ and Met154′) are also observed.
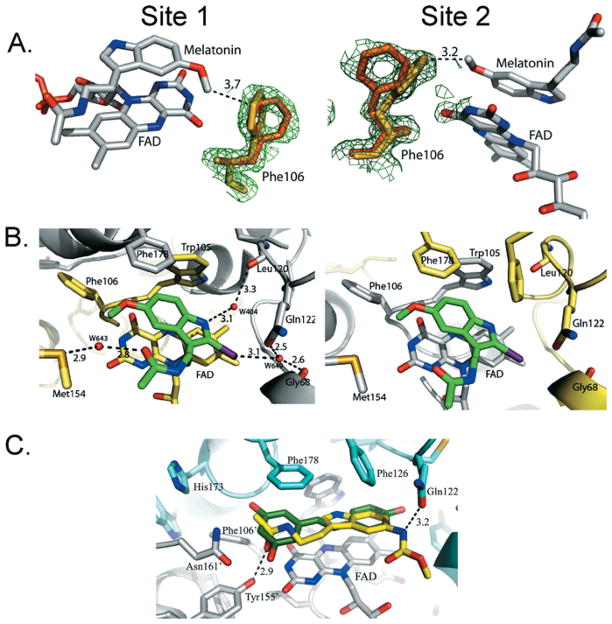
Interestingly, for the two higher-resolution structures, we observed differences in the position of the Phe106 side chain. The phenyl ring of Phe106 is disordered over two different conformations, which are clearly visible in the electron-density map (Figure 4A). This motion has not been observed or noted in any of the structures of QR2 published previously. In one orientation, Phe106 occupies the same position observed in the other published QR2 structures [12,35,36], whereas, in the second new orientation, Phe106 is able to come closer to melatonin, i.e. within a distance of 4 Å, and to make van der Waals contacts with melatonin’s methoxy side chain. In the structure of QR2 in complex with resveratrol [12], the resorcinol moiety of resveratrol occupies the portion of the active site in close proximity to Phe106, which forces the phenyl side chain of Phe106 to the back of the pocket, thereby restraining its movement.
Figure 4. QR2 active-site structural features with bound inhibitors.
(A) Stick representation of Phe106 in the two orientations adopted in the active sites of two complexes. The two panels represent the view of the two active sites in the QR2–melatonin complex at 1.7 Å resolution. One orientation is shown in yellow, and the other is in orange. The 2Fo − Fc electron-density map at 1σ (green mesh) is shown only around the aminoacidic residue for clarity. FAD and melatonin are shown in stick representations and are coloured according to residue type. (B) Images of the two QR2 active sites with bound 2-iodomelatonin showing the side chains of the residues as stick representations, coloured according to subunit type. 2-Iodomelatonin adopts a single orientation in the QR2 active sites. FAD and melatonin are shown as stick representations coloured according to atom type. Hydrogen bonds (broken lines) and water molecules (red spheres) are shown only for one site (left). (C) Result of MCA-NAT docking in QR2 active site and its superposition on resveratrol. The enzyme is coloured according to subunit type. FAD is grey, MCA-NAT is yellow, and resveratrol is green. FAD, MCA-NAT and some of the active-site residues are in stick representation. The hydrogen-bond network between MCA-NAT and QR2 residues is depicted as broken lines. Prepared using PyMOL (DeLano Scientific; http://www.pymol.org).
X-ray structure of QR2 in complex with 2-iodomelatonin
We next determined the X-ray structure of QR2 bound to 2-iodo-melatonin. We collected full X-ray datasets from two different crystals to resolutions of 1.6 and 2.3 Å, and we refined and determined each structure independently (Supplementary Table S1). The high quality of the density maps allowed an unbiased positioning of 2-iodomelatonin within the active sites. In contrast with the multiple orientations observed for melatonin, 2-iodomelatonin adopts a single orientation in the QR2 active site of both X-ray structures (Figure 4B). This orientation is analogous to the binding mode adopted by melatonin in site 2 of all the structures. 2-Iodomelatonin is positioned so that its indole moiety is stacked on top of the FAD cofactor ring. The iodine atom forms a hydrogen bond with a water molecule that is also hydrogen-bonded to Gly68′ and Gln122′. The side-chain carbonyl group and the pyrrole nitrogen of 2-iodomelatonin interact with Met154 and Leu120′ (Figure 4B). As is the case for melatonin, a hydrogen-bond network between 2-iodomelatonin and QR2 is established by a network of bridging water molecules.
Docking of MCA-NAT in the QR2-binding pocket
In order to gain further insights into the selectivity of QR2 towards different melatonin analogues, we explored the binding mode of MCA-NAT, a selective MT3 ligand, using computational docking methods. Docking of MCA-NAT in the QR2 active site was performed using the program GOLD [33]. The GOLD fitness function was used to rank the compound according to its ability to bind favourably within the QR2 active site. First, we used the X-ray structure of the QR2–resveratrol complex as a structural guide to define parameters and validate our computational strategy. The binding orientations and geometries for resveratrol in the QR2 active site are superimposable and consistent with the published X-ray structure (Supplementary Figure S4A at http://www.BiochemJ.org/bj/413/bj4130081add.htm).
Since our computational approach was validated with resveratrol, we next docked MCA-NAT to QR2. Similarly to resveratrol, the solutions generated via the GOLD program all superimposed on each other (Supplementary Figure S4B). Since the solutions resulted in an almost identical docked pose, the docked solution with the highest fitness score was selected as the predicted ligand-binding mode. MCA-NAT is positioned within the active site of QR2 so that its indole ring is stacked on top of the isoalloxazine moiety of the FAD cofactor (Figure 4C). The orientation resembles that adopted by melatonin in site 1 of two structures, with the nitrogen atom of the pyrrole ring pointing towards the core of the cavity. However, in contrast with melatonin, MCA-NAT is able to make specific interactions with the active-site residues. The carbonyl oxygen on the methoxycarbonylamino side chain makes a hydrogen bond with Tyr155, whereas the amidic nitrogen on the other side chain of the ligand interacts with the carboxyl oxygen of Gln122. Multiple hydrophobic contacts are formed between MCA-NAT and the side chains of Phe106′, Phe126, Phe178 and Gly68.
DISCUSSION
We have determined the crystal structures of melatonin and 2-iodomelatonin in complex with the human QR2 enzyme, which are the first X-ray structures of melatonin or an analogue of melatonin to be determined in complex with a protein. We also showed that melatonin and its iodinate analogue bind directly to the QR2 active site and not to an allosteric or ancillary MT3 binding site as envisioned previously [11,37].
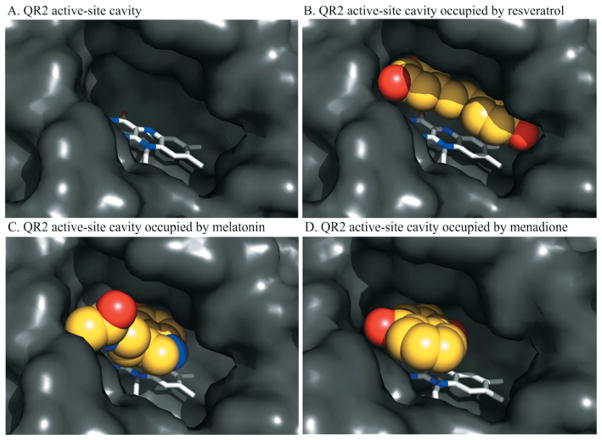
The X-ray structures of QR2 in complex with melatonin and 2-iodomelatonin, together with the structure of QR2 bound to resveratrol published previously [12], provide a precise molecular topography of these molecules binding within the active site of the enzyme. A common feature present in each of the structures of QR2–inhibitor complexes is the presence of a central flat hydrophobic moiety surrounded by polar side groups. The differences observed in the inhibitory potencies of ligands probably depend on both the size of the molecules and the subsequent interactions that result. In fact, potent and selective QR2 inhibitors, such as resveratrol, can fill the QR2 cavity almost completely (Figure 5). Moreover, these inhibitors can extend to reach different active-site residues with which they make specific polar interactions [12]. In contrast, weaker and smaller inhibitors and substrates, such as melatonin and menadione [38] (Figure 5), partially occupy the large active-site cavity, consequently making fewer hydrogen bonds and hydrophobic contacts with QR2.
Figure 5. Comparison of the binding mode between resveratrol, melatonin and menadione in the QR2-binding site.
(A) The empty active site of human QR2 apoenzyme. Binding mode of (B) resveratrol, (C) melatonin and (D) menadione in the QR2 active-site cavity. The QR2 active site in each panel is shown as a surface representation. The FAD cofactor is represented as a stick model and is coloured grey, and the ligands (yellow) are represented as space-filling models.
The results of the inhibition assays, i.e. the IC50 values, indicate the following rank order of affinities: resveratrol≥2-iodomel-atonin>N-acetyl-5-hydroxytyptamine ≥ melatonin>5-hydroxytryptamine (Table 1), which are in relative agreement with IC50 values reported previously using mammalian cells or hamster membrane homogenates [3,4,37]. The differences in our IC50 values compared with those reported in the literature [4,37] (Table 1) probably stem from the different assay conditions used, including different substrate–co-substrate pairs and concentrations. Since the kinetic mechanism for QR2 is Ping Pong, the degree of inhibition, i.e. IC50 values, for compounds will be strongly influenced by the substrates and their concentrations [39]. To address this problem, we determined the dissociation constants for melatonin, 2-iodomelatonin and resveratrol by ITC. In contrast with the rank order of affinities determined by IC50 values, the rank order of affinity based on the Kd values is resveratrol ≫ 2-iodomelatonin≥melatonin. Interestingly, this trend is reflected in the crystal structure of the complexes in that resveratrol and 2-iodomelatonin have well-ordered electron-density maps and only one orientation for the inhibitor, whereas melatonin binds with multiple orientations.
The higher entropy associated with the binding of melatonin to QR2 compared with resveratrol implies increased rotational and conformational freedom of melatonin which is consistent with the electron density and multiple orientations observed. The consequence of the increase in flexibility is a decrease in the number of stable bonds within the ligand–protein complex. As a result, the enthalpy associated with melatonin binding is approx. 4 kcal/mol less than that of resveratrol, suggesting the formation of weaker interactions between the ligand and the enzyme. The thermodynamic parameters for 2-iodomelatonin are analogous to that of melatonin: higher entropy and lower enthalpy of binding compared with resveratrol. In the case of 2-iodomelatonin, however, the molecule adopts the same orientation in both of the active sites of the dimer and in the two structures determined. The favourable ΔS value obtained for 2-iodomelatonin is likely to be due to the presence of the iodine atom and the desolvation of this group. According to the hydrophobic effect, water molecules around a non-polar atom are more ordered and organized such that they tend to form relatively unstable cages with low entropy. The transfer of the structured waters into the more disordered bulk solvent upon binding of the ligand to the enzyme active site would result in increased entropy [40].
Based on the relatively weak affinity, i.e. micromolar IC50 and Kd values, for melatonin and the fact that, over a 24 h period, plasma concentrations of melatonin are only in the low-nanomolar range [37,41], we believe that a physiological role for QR2 as a functionally important binding site for endogenous melatonin is not likely. MT3 was identified by the utilization of 2-iodomelatonin in competitive binding assays. 2-Iodomelatonin has a higher specific activity compared with tritiated melatonin, and therefore has led to the detection of moderate non-physiological melatonin-affinity sites in animal tissue homogenates. Therefore QR2, i.e. MT3, is not likely to be a physiologically relevant binding site of melatonin in vivo.
On the other hand, it is possible that QR2 may function to bind exogenous melatonin, e.g. from the diet. A plethora of data has been published using pharmacological concentrations of melatonin (1 μM and beyond). Those experiments described various effects for this neurohormone. At least some of these effects could be mediated by MT3, thereby shedding light on the mechanism by which melatonin can have these effects, particularly these antioxidant effects. Furthermore, we cannot disregard the possibility that dietary melatonin from plant sources could directly elevate the circulating levels of melatonin within the body to levels required to inhibit QR2 activity [16,42]. It could be possible to achieve melatonin concentrations in the body to the levels required to inhibit QR2 activity, but such studies have not yet been conducted.
We also cannot disregard the possibility that the IC50 values for melatonin measured in vitro do not reflect the real inhibitory potency of the hormone in vivo. The identity and concentration of both the reduced co-substrate and the substrate that ultimately is reduced are uncertain. As described above, since the degree of inhibition of an enzyme with a Ping Pong kinetic mechanism depends upon the nature of the substrate and cofactor utilized, and since the ‘real’ substrate and co-substrate utilized by QR2 under physiological conditions are not known, it is possible that, in vivo, the concentration of melatonin required to inhibit the enzyme could be lower than that found in vitro. It is also possible that, in certain areas, particularly in brain, local concentrations of melatonin could be elevated, particularly during the night, to levels that would cause marked inhibition of QR2. These possibilities need to be addressed via in depth in vivo studies.
In summary, the crystal structure of melatonin and 2-iodomel-atonin in complex with QR2 provide a detailed description of the enzyme active site that can now be utilized in the design of new and potent inhibitors of the enzyme. These inhibitors might be instrumental in discriminating the role of MT1, MT2 and QR2/MT3 in mediating the physiological and/or possible therapeutic effects attributed to the neurohormone.
Supplementary Material
Acknowledgments
We thank Dr Philippe Delagrange (Institut de Recherches Servier) for his input into this work. Special thanks also go to Dr Klavs Dolmer (University of Illinois at Chicago) for helpful comments on the ITC experiments. We are grateful to the BioCARS (14-BM-C) beamline personnel at the Advanced Photon Source, Argonne National Laboratory, for their assistance in beamline operations. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract no. W-31-109-Eng-38. Use of the BioCARS Sector 14 was supported by the National Institutes of Health, National Center for Research Resources, under grant number RR007707. This publication was made possible by grant numbers CA92744 and CA48112 from the National Cancer Institute (NCI) at the National Institutes of Health. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NCI.
Abbreviations used
- DTT
dithiothreitol
- GOLD
Genetic Optimization for Ligand Docking
- IPTG
isopropyl β-D-thiogalactoside
- ITC
isothermal titration calorimetry
- MCA-NAT
5-methoxycarbonylamino-N-acetyltryptamine
- MT3
melatonin-binding site 3
- NMeH
N-methyldihydronicotinamide
- NQO2
NNRH (-ribosyldihydronicotinamide):quinone oxidoreductase 2
- QR2
quinone reductase 2
References
- 1.Arendt J. Melatonin. Clin Endocrinol. 1988;29:205–229. doi: 10.1111/j.1365-2265.1988.tb00263.x. [DOI] [PubMed] [Google Scholar]
- 2.Boutin JA, Audinot V, Ferry G, Delagrange P. Molecular tools to study melatonin pathways and actions. Trends Pharmacol Sci. 2005;26:412–419. doi: 10.1016/j.tips.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 3.Dubocovich ML. Pharmacology and function of melatonin receptors. FASEB J. 1988;2:2765–2773. doi: 10.1096/fasebj.2.12.2842214. [DOI] [PubMed] [Google Scholar]
- 4.Molinari EJ, North PC, Dubocovich ML. 2-[125I]iodo-5-methoxycarbonylamino-N-acetyltryptamine: a selective radioligand for the characterization of melatonin ML2 binding sites. Eur J Pharmacol. 1996;301:159–168. doi: 10.1016/0014-2999(95)00870-5. [DOI] [PubMed] [Google Scholar]
- 5.Nosjean O, Ferro M, Coge F, Beauverger P, Henlin JM, Lefoulon F, Fauchere JL, Delagrange P, Canet E, Boutin JA. Identification of the melatonin-binding site MT3 as the quinone reductase 2. J Biol Chem. 2000;275:31311–31317. doi: 10.1074/jbc.M005141200. [DOI] [PubMed] [Google Scholar]
- 6.Liao S, Dulaney JT, Williams-Ashman HG. Purification and properties of a flavoprotein catalyzing the oxidation of reduced ribosyl nicotinamide. J Biol Chem. 1962;237:2981–2987. [PubMed] [Google Scholar]
- 7.Liao S, Williams-Ashman HG. Enzymatic oxidation of some non-phosphorylated derivatives of dihydronicotinamide. Biochem Biophys Res Commun. 1961;4:208–213. doi: 10.1016/0006-291x(61)90272-8. [DOI] [PubMed] [Google Scholar]
- 8.Zhao Q, Yang XL, Holtzclaw WD, Talalay P. Unexpected genetic and structural relationships of a long-forgotten flavoenzyme to NAD(P)H:quinone reductase (DT-diaphorase) Proc Natl Acad Sci USA. 1997;94:1669–1674. doi: 10.1073/pnas.94.5.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liao S, Williams-Ashman HG. Biochem Pharmacol. 1961;6 [Google Scholar]
- 10.Ernster L. DT diaphorase: a historical review. Chem Scripta. 1987;27A:1–13. [Google Scholar]
- 11.Vella F, Ferry G, Delagrange P, Boutin JA. NRH:quinone reductase 2: an enzyme of surprises and mysteries. Biochem Pharmacol. 2005;71:1–12. doi: 10.1016/j.bcp.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 12.Buryanovskyy L, Fu Y, Boyd M, Ma Y, Hsieh TC, Wu JM, Zhang Z. Crystal structure of quinone reductase 2 in complex with resveratrol. Biochemistry. 2004;43:11417–11426. doi: 10.1021/bi049162o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Long DJ, 2nd, Iskander K, Gaikwad A, Arin M, Roop DR, Knox R, Barrios R, Jaiswal AK. Disruption of dihydronicotinamide riboside:quinone oxidoreductase 2 (NQO2) leads to myeloid hyperplasia of bone marrow and decreased sensitivity to menadione toxicity. J Biol Chem. 2002;277:46131–46139. doi: 10.1074/jbc.M208675200. [DOI] [PubMed] [Google Scholar]
- 14.Chomarat P, Coge F, Guenin SP, Mailliet F, Vella F, Mallet C, Giraudet S, Nagel N, Leonce S, Ferry G, et al. Cellular knock-down of quinone reductase 2: a laborious road to successful inhibition by RNA interference. Biochimie. 2007;89:1264–1275. doi: 10.1016/j.biochi.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 15.Mailliet F, Ferry G, Vella F, Thiam K, Delagrange P, Boutin JA. Organs from mice deleted for NRH:quinone oxidoreductase 2 are deprived of the melatonin binding site MT3. FEBS Lett. 2004;578:116–120. doi: 10.1016/j.febslet.2004.10.083. [DOI] [PubMed] [Google Scholar]
- 16.Tan DX, Reiter RJ, Manchester LC, Yan MT, El-Sawi M, Sainz RM, Mayo JC, Kohen R, Allegra M, Hardeland R. Chemical and physical properties and potential mechanisms: melatonin as a broad spectrum antioxidant and free radical scavenger. Curr Top Med Chem. 2002;2:181–197. doi: 10.2174/1568026023394443. [DOI] [PubMed] [Google Scholar]
- 17.James AM, Smith RA, Murphy MP. Antioxidant and prooxidant properties of mitochondrial coenzyme Q. Arch Biochem Biophys. 2004;423:47–56. doi: 10.1016/j.abb.2003.12.025. [DOI] [PubMed] [Google Scholar]
- 18.Galinier A, Carriere A, Fernandez Y, Caspar-Bauguil S, Periquet B, Periquet A, Penicaud L, Casteilla L. Site specific changes of redox metabolism in adipose tissue of obese Zucker rats. FEBS Lett. 2006;580:6391–6398. doi: 10.1016/j.febslet.2006.10.052. [DOI] [PubMed] [Google Scholar]
- 19.Wang Z, Hsieh TC, Zhang Z, Ma Y, Wu JM. Identification and purification of resveratrol targeting proteins using immobilized resveratrol affinity chromatography. Biochem Biophys Res Commun. 2004;323:743–749. doi: 10.1016/j.bbrc.2004.08.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr. 1993;26:795–800. [Google Scholar]
- 21.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 22.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr Sect D Biol Crystallogr. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 23.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr Sect A Found Crystallogr. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 24.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr Sect D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 25.Kleywegt GJ, Jones TA. Databases in protein crystallography. Acta Crystallogr Sect D Biol Crystallogr. 1998;54:1119–1131. doi: 10.1107/s0907444998007100. [DOI] [PubMed] [Google Scholar]
- 26.Kleywegt GJ, Henrick K, Dodson EJ, van Aalten DMF. Pound-wise but penny-foolish: how well do micromolecules fare in macromolecular refinement? Structure. 2003;11:1051–1059. doi: 10.1016/s0969-2126(03)00186-2. [DOI] [PubMed] [Google Scholar]
- 27.Kleywegt GJ, Jones TA. Model-building and refinement practice. Methods Enzymol. 1997;277:208–230. doi: 10.1016/s0076-6879(97)77013-7. [DOI] [PubMed] [Google Scholar]
- 28.Kleywegt GJ. Making the most of your search model. Newsl Protein Crystallogr. 1996;32:32–36. [Google Scholar]
- 29.Morris AL, MacArthur MW, Hutchinson EG, Thornton JM. Stereochemical quality of protein structure coordinates. Proteins. 1992;12:345–364. doi: 10.1002/prot.340120407. [DOI] [PubMed] [Google Scholar]
- 30.Lovell S, Davis IW, Arendall WB, III, de Bakker PIW, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Cα geometry:ϕ, ψ, and Cβ deviation. Proteins Struct Funct Genet. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 31.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ortiz-Maldonado M, Gatti D, Ballou DP, Massey V. Structure–function correlations of the reaction of reduced nicotinamide analogues with p-hydroxybenzoate hydroxylase substituted with a series of 8-substituted flavins. Biochemistry. 1999;38:16636–16647. doi: 10.1021/bi991603u. [DOI] [PubMed] [Google Scholar]
- 33.Jones G, Willett P, Glen RC, Leach AR, Taylor R. Development and validation of a genetic algorithm for flexible docking. J Mol Biol. 1997;267:727–748. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- 34.Lumry R, Rajender S. Enthalpy–entropy compensation phenomena in water solutions of proteins and small molecules: a ubiquitous property of water. Biopolymers. 1970;9:1125–1227. doi: 10.1002/bip.1970.360091002. [DOI] [PubMed] [Google Scholar]
- 35.AbuKhader M, Heap J, De Matteis C, Kellam B, Doughty SW, Minton N, Paoli M. Binding of the anticancer prodrug CB1954 to the activating enzyme NQO2 revealed by the crystal structure of their complex. J Med Chem. 2005;48:7714–7719. doi: 10.1021/jm050730n. [DOI] [PubMed] [Google Scholar]
- 36.Foster CE, Bianchet MA, Talalay P, Zhao Q, Amzel LM. Crystal structure of human quinone reductase type 2, a metalloflavoprotein. Biochemistry. 1999;38:9881–9886. doi: 10.1021/bi990799v. [DOI] [PubMed] [Google Scholar]
- 37.Mailliet F, Ferry G, Vella F, Berger S, Coge F, Chomarat P, Mallet C, Guenin SP, Guillaumet G, Viaud-Massuard MC, et al. Characterization of the melatoninergic MT3 binding site on the NRH:quinone oxidoreductase 2 enzyme. Biochem Pharmacol. 2005;71:74–88. doi: 10.1016/j.bcp.2005.09.030. [DOI] [PubMed] [Google Scholar]
- 38.Kwiek JJ, Haystead TA, Rudolph J. Kinetic mechanism of quinone oxidoreductase 2 and its inhibition by the antimalarial quinolines. Biochemistry. 2004;43:4538–4547. doi: 10.1021/bi035923w. [DOI] [PubMed] [Google Scholar]
- 39.Preusch PC, Siegel D, Gibson NW, Ross D. A note on the inhibition of DT–diaphorase by dicoumarol. Free Radical Biol Med. 1991;11:77–80. doi: 10.1016/0891-5849(91)90191-5. [DOI] [PubMed] [Google Scholar]
- 40.Sturtevant JM. Heat capacity and entropy changes in processes involving proteins. Proc Natl Acad Sci USA. 1977;74:2236–2240. doi: 10.1073/pnas.74.6.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pandi-Perumal SR, Srinivasan V, Maestroni GJ, Cardinali DP, Poeggeler B, Hardeland R. Melatonin: Nature’s most versatile biological signal? FEBS J. 2006;273:2813–2838. doi: 10.1111/j.1742-4658.2006.05322.x. [DOI] [PubMed] [Google Scholar]
- 42.Reiter RJ, Tan DX, Manchester LC, Simopoulos AP, Maldonado MD, Flores LJ, Terron MP. Melatonin in edible plants (phytomelatonin): identification, concentrations, bioavailability and proposed functions. World Rev Nutr Diet. 2007;97:211–230. doi: 10.1159/000097917. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.