Summary
Regulated relocalization of signaling and trafficking proteins is crucial for the control of many cellular processes, and is driven by a series of domains that respond to alterations at membrane surfaces. The first examples of these domains – conditional peripheral membrane proteins – included C1, C2, PH, PX, and FYVE domains, which specifically recognize single tightly regulated membrane components such as diacylglycerol or phosphoinositides. The structural basis for this recognition is now well understood. Efforts to identify additional domains with similar functions that bind other targets (or participate in unexplained cellular processes) have not yielded many more examples of specific phospholipid-binding domains. Instead, most of the recently discovered conditional peripheral membrane proteins bind multiple targets (each with limited specificity), relying on coincidence detection, and/or recognizing broader physical properties of the membrane such as charge or curvature. This broader range of recognition modes presents significant methodological challenges for a full structural understanding.
Introduction
Numerous protein domains or modules drive functionally important membrane recruitment of their ‘host’ proteins by recognizing key features of the membrane surface (Lemmon, 2008). These domains are peripheral membrane proteins. Some are constitutively membrane associated. Others are highly regulated, and may be thought of as “conditional peripheral membrane proteins”. It could be argued that this class of membrane proteins is one of the most straightforward for structural study. This is true for the domains themselves, but understanding how they bind membrane surfaces remains a major challenge. In this article we will review what is known about defined globular membrane-association domains for which there is clear structural understanding. We then discuss ongoing efforts to identify new conditional peripheral membrane proteins/domains. Although initially described examples recognize single well-defined binding targets, several more recently identified domains must engage multiple binding targets simultaneously to drive membrane association or recognize poorly defined physical characteristics of membranes. As the field progresses, it is increasingly clear that these domains have evolved to bind biological membranes themselves, and not their constituent lipids or other components. This creates significant challenges for studying the structural basis of subcellular targeting by these domains, which will require the development of new methods.
Domains that bind stereospecifically to membrane lipids
Discovery of stereospecific phosphoinositide-binding domains
During the 1990s, two overlapping classes of conditional peripheral membrane proteins with specific lipid recognition properties were identified:
Those that are spatially restricted – selectively binding lipids found only in specific subcellular compartments.
Those that are temporally restricted – specifically recognizing lipids that are generated only transiently (in cell signaling) or requiring the presence of another signaling molecule (such as Ca2+) for membrane binding.
The first conditional peripheral membrane proteins described were the conserved C1 and C2 regulatory regions from protein kinase C (PKC), which together ‘decode’ diacylglycerol (DAG) and calcium signals to recruit PKC to the plasma membrane and promote its activation (Oancea and Meyer, 1998).
Phosphoinositides became known as binding targets for conditional peripheral membrane proteins in the 1990s (Janmey, 1995). Phosphatidylinositol-4,5-bisphosphate (PtdIns(4,5) P2), which accounts for ~1% of inner leaflet phospholipid in mammalian cell plasma membranes, is well known to regulate cytoskeletal organization and is rapidly turned over in response to numerous different signals (Di Paolo and De Camilli, 2006). PtdIns(4,5)P2 associates with the actin regulatory proteins gelsolin (Janmey and Stossel, 1987) and profilin (Lassing and Lindberg, 1985), reducing the affinity of both proteins for monomeric actin and thus promoting nucleated actin polymerization. In 1994, certain pleckstrin homology (PH) domains were found to bind PtdIns(4,5)P2 (Harlan et al., 1994), and this was soon extended to other phosphoinositides (Di Paolo and De Camilli, 2006). Subsequently, two distinct domains were identified that bind the endosomal phosphoinositide PtdIns3P. These were the FYVE (for Fab1, YOTB, Vac1, EEA1) domain (Kutateladze, 2006) and the PX (for Phox homology) domain (Seet and Hong, 2006), with the first reports appearing respectively in 1998 and 2001.
Thus, between 1995 and 2001, successive examples were rapidly identified of domains that specifically recognize PtdIns(4,5)P2, PtdIns(3,4,5)P3/PtdIns(3,4)P2, and PtdIns3P. This cemented the function of phosphoinositides as signaling molecules, recognized by conditional peripheral membrane proteins that serve as phosphoinositide ‘effectors’. Efforts were also spurred to identify new effector domains – with the appealing notion that they translate a cellular ‘phosphoinositide code’ (Kutateladze, 2010), including the ‘orphan’ phosphoinositides PtdIns(3,5)P2 (Dove et al., 1997) and PtdIns5P (Rameh et al., 1997), as well as PtdIns4P. The additional anticipated domains have been notoriously difficult to identify. Indeed, it remains unclear whether PtdIns(3,5)P2 and PtdIns5P have bona fide effectors (Lemmon, 2008). Domains that bind PtdIns4P have been identified, but most are not selective (Levine and Munro, 2002). In fact, very few new specific phosphoinositide binding domains have been uncovered since 2000. Instead, a range of modules has been uncovered that recognize broader features of membrane surfaces, and combinations of potential targets. How this recognition is achieved, and what its consequences are for membrane structure and function, remains unclear in many cases, and will require challenging structural analysis of membrane/protein complexes.
Structural views of intracellular conditional peripheral membrane proteins that specifically recognize lipid headgroups
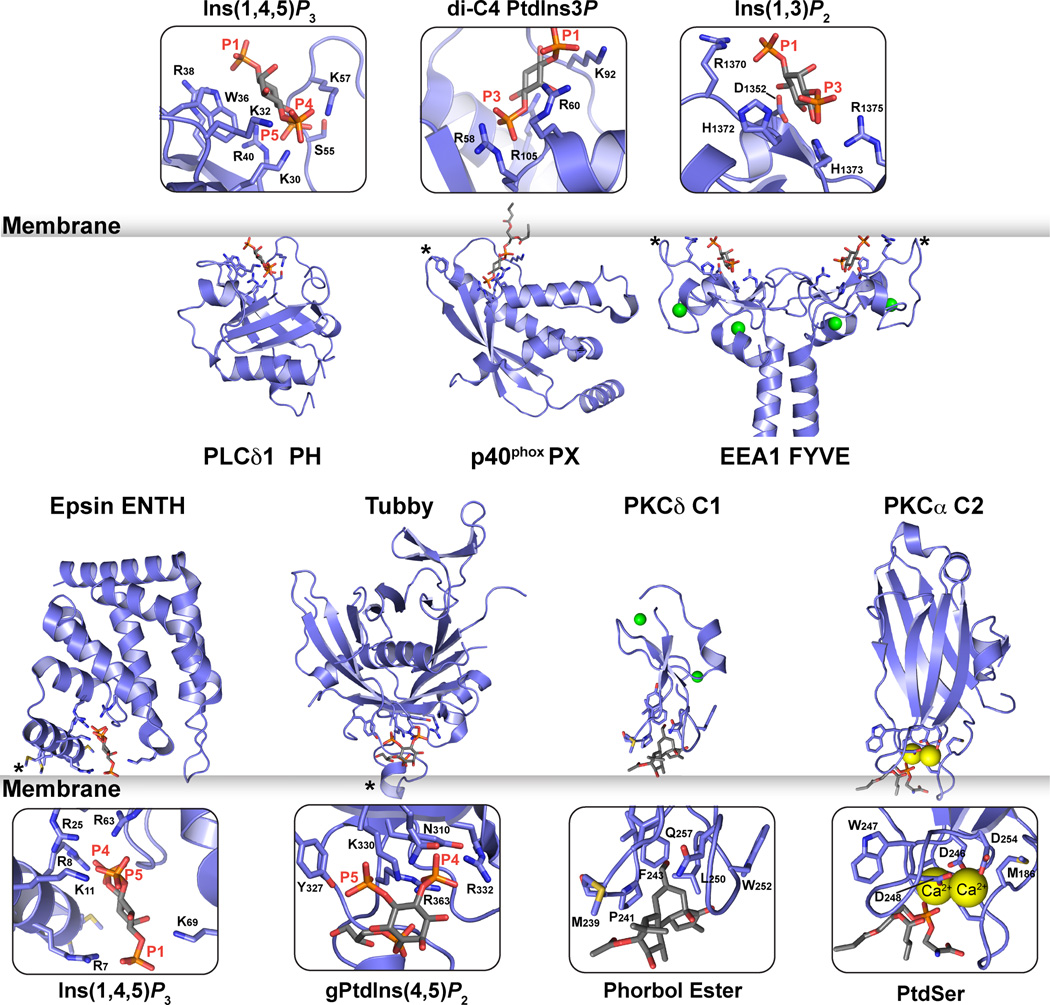
Figure 1 shows crystal structures of seven classes of domain that stereospecifically recognize the headgroup of their lipid binding partners. In each case it was possible to crystallize a complex between the domain and a short-chain variant of the target lipid or its isolated headgroup.
Figure 1. Domains that stereospecifically recognize phospholipid headgroups.
Cartoon representations of seven domains that stereospecifically recognize their lipid targets. Top row: the PLC-δ1 PH domain bound to Ins(1,4,5)P3 (Ferguson et al., 1995); the PX domain of p40phox bound to dibutanoyl PtdIns3P (Bravo et al., 2001); and a coiled coil-mediated dimer of the EEA1 FYVE domain bound to Ins(1,3)P2 (Dumas et al., 2001). PDB codes are 1MAI, 1H6H and 1JOC respectively. Bottom row: epsin ENTH domain bound to Ins(1,4,5)P3 (Ford et al., 2002); tubby C-terminal domain bound to 1-glycerophosphoryl-Ins(4,5)P2 (Santagata et al., 2001); PKCδ C1 domain bound to phorbol-13-myristate (Zhang et al., 1995), and the PKCα C2 domain bound to dicaproyl-phosphatidylserine and calcium ions (Verdaguer et al., 1999). PDB codes are 1H0A, 1I7E, 1PTR and 1DSY respectively. Ligands and residues involved in stereospecific recognition are shown in stick representation in the zoomed views. Asterisks mark regions proposed to penetrate the membrane surface. Zn2+ ions are shown as green spheres, and Ca2+ ions as yellow spheres.
PH Domains
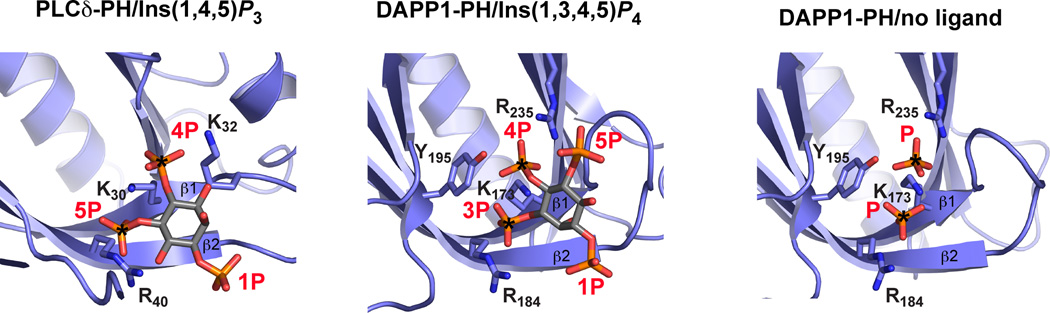
The PH domain of PLCδ1 (Ferguson et al., 1995) provided the first structural view of how a conditional peripheral membrane protein can associate with membrane surfaces by binding specifically to a phosphoinositide headgroup. The PLCδ1 PH domain binds ~10-fold more strongly to the free PtdIns(4,5)P2 headgroup (Ins(1,4,5)P3) than to the lipid in a membrane context (Lemmon et al., 1995). As shown in Figure 1, the PLCδ1 PH domain – like all PH domains – adopts a 7-stranded β-sandwich structure, with one of its splayed corners capped by a C-terminal α-helix. The headgroup binding site lies at the other splayed corner, defined primarily by the loop that connects strands β1 and β2 (see Figure 2). A basic sequence ‘motif’ in this loop forms a ‘platform’ for headgroup binding: KXn (K/R) XR. The first lysine projects from strand β1 (K30 in PLCδ1-PH), and the last arginine projects from strand β2 (R40 in PLCδ1-PH) to create binding sites for two phosphate groups as shown in Figure 2. The distance between these sites precisely matches that between two vicinal phosphates on an inositol ring (the 4- and 5-phosphates of Ins(1,4,5)P3 in PLCδ1 or the 3- and 4-phosphates of Ins(1,3,4,5)P4 bound to DAPP1-PH) – suggesting that PH domain ligands must have two vicinal phosphates. Indeed, this is consistent with the fact that no PH domain has been identified that specifically recognizes PtdIns3P, PtdIns(3,5)P2 or PtdIns4P – which all lack a vicinal phosphate pair. All PH domains that contain the KXn (K/R) XR motif bind phosphoinositides (Yu et al., 2004). The motif is also retained in more complete motifs or algorithms that predict PH domain binding to PtdIns(3,4,5)P3 and PtdIns(3,4)P2 (Isakoff et al., 1998; Lietzke et al., 2000; Park et al., 2008), and can be rationalized structurally (Ferguson et al., 2000; Lietzke et al., 2000).
Figure 2. PH domains recognize vicinal phosphate pairs.
From left to right are shown Ins(1,4,5)P3 bound to PLCδ-PH (Ferguson et al., 1995); Ins(1,3,4,5)P4 bound to the DAPP1 PH domain (Ferguson et al., 2000); and two phosphate groups bound to DAPP1-PH (Ferguson et al., 2000), from PDB entries 1MAI, 1FAO, and 1FB8. Each PH domain is identically orientated, and the view centered on the inositol phosphate-binding site. Strands β1 and β2 of the PH domain are marked, as are phosphate groups and key basic side-chains that interact with them. The two vicinal phosphates that lie in a common location in these and all other high-affinity PH domains are marked with asterisks.
The SMART database (Letunic et al., 2008) lists 329 different PH domains in 284 human proteins. Among those with the KXn (K/R) XR motif, ~40 were predicted or shown to bind PtdIns(3,4,5)P3/PtdIns(3,4)P2 (Park et al., 2008). In humans, only PH domains from PLCδ relatives specifically bind PtdIns(4,5)P2 (Lemmon, 2008). A few additional PH domains with the KXn (K/R) XR motif bind phosphoinositides with little or no stereospecificity – although the structural basis for this is not yet clear. There is an additional set of PH domains that bind ‘non-canonically’ to phosphoinositides and lack the KXn (K/R) XR motif. These include the ArhGAP9, Tiam1 and spectrin PH domains (Ceccarelli et al., 2007; Hyvönen et al., 1995) as well as the GLUE domain from yeast Vps36p (Teo et al., 2006), which has a PH domain fold ‘split’ by a large insertion between strands β6 and β7. Each of these PH domains binds phosphoinositide headgroups (with limited stereospecificity) on the opposite face of the β1/β2 loop from that seen for PLCδ1-PH and DAPP1-PH in Figure 2 (Lemmon, 2008). A recent description of phosphoserine bound to the evectin-2 PH domain also showed a different location for an anion binding site on a PH domain (Uchida et al., 2011) – although neither the affinity nor specificity of this PH/phosphoserine interaction is clear. It is important to note that data from S. cerevisiae suggest that the majority of PH domains may not bind phosphoinositides (Gallego et al., 2010; Yu et al., 2004), arguing that other lipid or protein targets should be sought.
PX domains
PX domains contain an N-terminal three-stranded β-meander that abuts a C-terminal α-helical subdomain (Seet and Hong, 2006). In the crystal structure of the p40phox PX domain bound to dibutanoyl-PtdIns3P (Bravo et al., 2001), the phosphoinositide monomer lies in a positively charged pocket between these two structural elements (Figure 1). A limited number of conserved basic residues form hydrogen bonds with the headgroup, consistent with a modest affinity (KD ~5µM) for the monomeric dibutanoyl PtdIns3P. In addition, two tyrosines contact the inositol ring and glycerol backbone respectively. Association of the PX domain with PtdIns3P-containing membranes appears to be strengthened substantially by involvement of the membrane interaction loop marked with an asterisk in Figure 1. Residues in this loop show substantial NMR chemical shift changes upon binding to PtdIns3P-containing micelles (Cheever et al., 2001), and monolayer or micelle insertion studies of PX domains indicate that this loop promotes binding by penetrating the membrane (Seet and Hong, 2006).
PX domains are found in 42 different human proteins (Letunic et al., 2008), the majority being sorting nexins (SNXs) involved in the retromer complex that directs retrograde trafficking of Golgi resident proteins from endosomes to the trans Golgi. Although all S. cerevisiae PX domains appear to prefer PtdIns3P, there are reports of distinct phosphoinositide specificities for several mammalian examples that can be rationalized structurally based on the arrangement of basic side-chains in the binding site (Seet and Hong, 2006). Interestingly only 4 of the 15 S. cerevisiae PX domains bind to PtdIns3P with high (micromolar range KD) affinity. The ‘weak’ PtdIns3P binders appear to rely on protein oligomerization to increase avidity of membrane binding, or cooperation with adjacent BAR domains (see below) – both being important mechanisms for membrane assembly of the retromer complex.
FYVE domains
FYVE domains are zinc fingers of ~70 amino acids that also bind specifically to PtdIns3P (Kutateladze, 2006), and occur 30 times across 29 human proteins (Letunic et al., 2008). As shown in Figure 1 (top right), each FYVE finger comprises two β-hairpins plus a small C-terminal α-helix, and is held together by two tetrahedrally coordinated Zn2+ ions (Misra and Hurley, 1999). A conserved basic motif (RR/KHHCR) in the first β-strand defines a shallow positively charged pocket for PtdIns3P binding, and all but two of the direct hydrogen bonds seen between the PtdIns3P headgroup and the FYVE domain involve residues in this motif (Dumas et al., 2001; Kutateladze, 2006). Although FYVE domains bind specifically to PtdIns3P, they bind the monomeric lipid (or headgroup) rather weakly (Dumas et al., 2001). High affinity binding to PtdIns3P-containing membranes requires insertion of hydrophobic side-chains and/or dimerization of the FYVE domain-containing protein to enhance avidity (Kutateladze, 2006). A long N-terminal helical extension from the early endosome antigen-1 (EEA1) FYVE domain, for example, drives coiled coil-mediated dimerization of this domain, allowing high-avidity multivalent FYVE domain-mediated binding to the membrane surface (Dumas et al., 2001).
ENTH domain
The ~140 amino acid epsin NH2-terminal homology (ENTH) domain – found in 9 different human proteins (Letunic et al., 2008) – consists of a superhelical solenoid of α-helices (Figure 1). The epsin ENTH domain binds the PtdIns(4,5)P2 headgroup in a well-defined pocket that is only seen following ligand binding (Ford et al., 2002). The amphipathic ‘helix 0’ (marked with an asterisk in Figure 1), adjacent to the inositol phosphate headgroup, is unstructured in the absence of bound ligand (Hyman et al., 2000). It becomes ordered upon PtdIns(4,5)P2 binding and penetrates the membrane to promote membrane curvature (Ford et al., 2002).
Tubby domains
The 260 amino acid C-terminal domain from the transcription factor tubby has shown promise alongside PLCδ-PH as a probe for monitoring cellular PtdIns(4,5)P2 (Szentpetery et al., 2009). Tubby displays PtdIns(4,5)P2-dependent plasma membrane association, and its C-terminal domain binds PtdIns(4,5)P2-containing membranes in vitro (Santagata et al., 2001; Szentpetery et al., 2009) – greatly preferring membrane-embedded PtdIns(4,5)P2 over free Ins(1,4,5)P3, by contrast with PLCδ-PH. Tubby also fails to distinguish PtdIns(4,5)P2 from PtdIns(3,4)P2 or PtdIns(3,4,5)P3 (Santagata et al., 2001). A crystal structure of the tubby C-terminal domain bound to a PtdIns(4,5)P2 headgroup fragment (Santagata et al., 2001) revealed a relatively surface-exposed binding site for the headgroup fragment (Figure 1), consistent with the lack of stereospecificity. The requirement for the membrane surface to achieve high affinity binding may result from an adjacent loop (marked with an asterisk in Figure 1) that could insert into the membrane and/or associate with the apolar/polar interfacial region. Other basic patches on the protein may provide additional membrane interaction sites required for high affinity binding.
C1 Domains
Protein kinase C conserved region 1 (C1) domains, were the ‘founder’ conditional peripheral membrane protein modules. They are zinc finger-like domains of ~50 amino acids (Colon-Gonzalez and Kazanietz, 2006) with the signature motif: HX12CX2CX13–14CX2CX4HX2CX7C (H is histidine, C is cysteine, and X is any other amino acid). Some 87 C1 domains have been identified in 66 human proteins (Letunic et al., 2008). ‘Typical’ C1 domains bind DAG and phorbol esters, whereas so called atypical C1 domains share the same structure but do not bind these targets. The crystal structure of the second (of 2) C1 domains from PKCδ (C1B) bound to phorbol-13-myristate (Zhang et al., 1995) provided important insights into how this domain binds membranes (Figure 1). In addition to binding the phorbol ester, the C1 domain inserts a band of hydrophobic side-chains into the apolar milieu of the membrane, as seen in monolayer insertion studies (Medkova and Cho, 1999). This mode of binding also places basic residues in the C1 domain against the membrane surface, likely explaining (at least for PKCδ C1B) why targets are bound most strongly when present in negatively charged membranes (Colon-Gonzalez and Kazanietz, 2006).
C2 Domains
PKC C2 (for conserved region 2) domains are ~130 amino acid 8-stranded antiparallel β sandwich modules (Cho and Stahelin, 2006), not to be confused with the unrelated extracellular discoidin C2 domains (Lemmon, 2008). Some 238 PKC C2 domains are found across 140 different human proteins (Letunic et al., 2008). Canonical C2 domains, including the PKCα C2 domain depicted in Figure 1 (Verdaguer et al., 1999), are Ca2+-dependent phospholipid-binding domains that bind phosphatidylserine. The binding site involves three key inter-strand loops, and is acidic in character rather than basic as in PH, PX and FYVE domains. Two Ca2+ ions bind to this site and effectively form a ‘bridge’ between the C2 domain and phosphatidylserine in the membrane (Cho and Stahelin, 2006). The binding characteristics and specificities of C2 domains vary widely. Some have also been reported to bind selectively to phosphoinositides (Sánchez-Bautista et al., 2006), through a basic site that is adjacent to the region that binds Ca2+ and phosphatidylserine (see below).
Searching for new phospholipid-binding domains and new specificities
As our understanding of the domains in Figure 1 has developed, many groups have sought additional domains that might function as effectors for lipids with no current binding partner – or which might explain other lipid-regulated cellular phenomena. A variety of approaches have been employed – from screening yeast proteome microarrays for phosphoinositide binders (Zhu et al., 2001) to mass spectrometric identification of proteins that bind immobilized phosphoinositides (Catimel et al., 2008; Catimel et al., 2009; Dixon et al., 2011), screening candidate proteins with lipid overlay assays (Dippold et al., 2009; Gallego et al., 2010), and others (Lewis et al., 2011). In parallel, genetic approaches have sought effectors for certain lipids, and functionally-driven investigations have uncovered additional potential phosphoinositide or phospholipid binding domains, examples of which are summarized here.
Contrary to most expectations, few examples of new phosphoinositide specificities have been found that are not included in the domain descriptions above. Moreover, it is interesting that rather few clear new classes of globular phospholipid binding domain have emerged from these studies. Analyses of PtdIns(4,5)P2 and PtdIns(3,5)P2 interactomes (Catimel et al., 2008) identified proteins with HEAT repeats as the only potential new class of phosphoinositide-binding domains – although these are structurally related to the ENTH domain shown in Figure 1 (De Camilli et al., 2002). Other interactors included β-arrestin, already known to bind inositol phosphates (Milano et al., 2006), actin regulatory proteins, and several small GTPases.
Domains with selectivity for PtdIns5P, PtdIns4P and PtdIns(3,5)P2
Numerous studies have sought domains that selectively recognize PtdIns4P, PtdIns5P or PtdIns(3,5)P2 – which all lack the required pair of vicinal phosphates recognized by ‘canonical’ specific PH domains (Figure 2). Specific recognition of PtdIns3P, which also lacks this vicinal phosphate pair, involves distinct sets of modules (PX and FYVE domains). With the caveat that potential ‘leads’ in the reported interactomes have not yet been fully analyzed, we summarize below the status of specific binders or effectors for PtdIns4P, PtdIns5P and PtdIns(3,5)P2.
PtdIns4P
There has been some confusion over the existence of PtdIns4P-specific PH domains. The PH domain from FAPP1 (phosphatidylinositol-four-phosphate adaptor protein-1) was reported to bind PtdIns4P selectively in lipid-overlay studies (Dowler et al., 2000). However, PtdIns4P specificity is not evident in studies of lipid vesicle binding for FAPP1-PH or the related PH domains from oxysterol binding protein (OSBP), Osh1p, and Osh2p (Lenoir et al., 2010; Levine and Munro, 2002; Stahelin et al., 2007; Yu et al., 2004) – suggesting that the apparent specificity was an artifact of the lipid-overlay method. It is clear that all of these PH domains do bind PtdIns4P, but they also bind PtdIns(4,5)P2, PtdIns3P, and other phosphoinositides with the same affinity. Nonetheless, these PH domains all show clear Golgi localization that depends on PtdIns4P production (Godi et al., 2004; Levine and Munro, 2002; Stefan et al., 2002). It is now clear that Golgi localization does not result from the inherent PtdIns4P-binding selectivity of these PH domains per se: they are not useful as specific intracellular ‘probes’ for PtdIns4P. Rather – as discussed below for other examples – these PH domains simultaneously recognize two targets, a phosphoinositide and the small G-protein Arf1 (Godi et al., 2004; Levine and Munro, 2002). The dependence of FAPP1/OSBP/Osh PH domain Golgi localization on PtdIns4P production (rather than other phosphoinositides) reflects the predominance of this lipid in the Golgi (alongside Arf1) rather than its selective recognition by the PH domains.
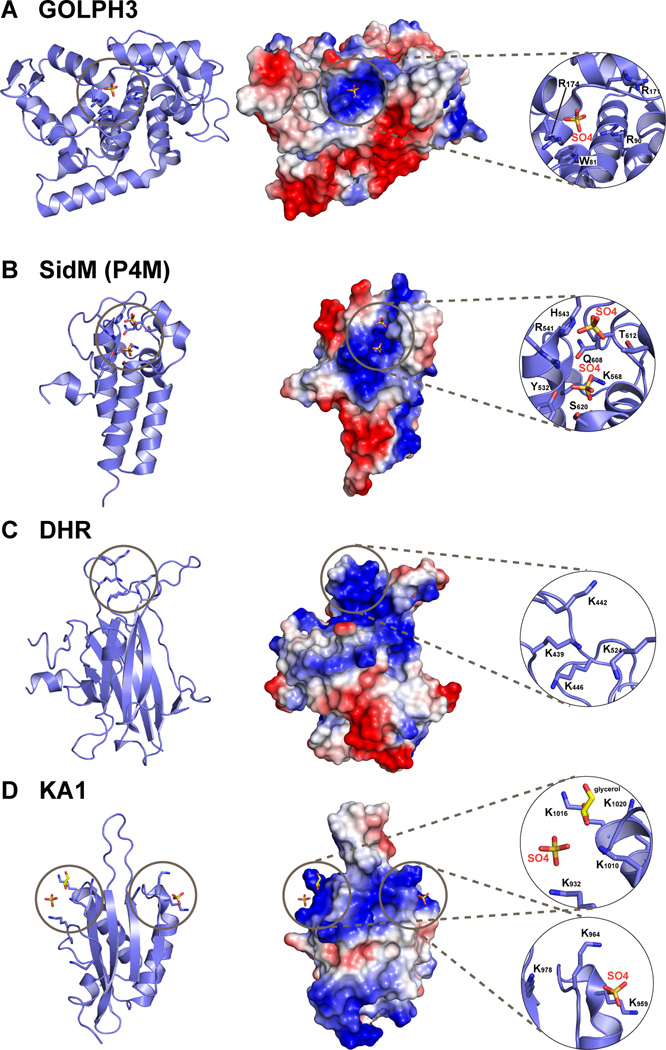
A novel PtdIns4P-specific protein with no previously recognized phosphoinositide-binding domains was recently identified in a lipid overlay-based screen of proteins from D. melanogaster (Dippold et al., 2009). This was the Golgi-localized protein GOLPH3/GMx33/GPP34/MIDAS, for which the precise functions are not yet clear. Unlike the PH domains mentioned above, PtdIns4P specificity of GOLPH3 is retained in membrane binding studies. Its S. cerevisiae homolog Vps74p shows clearer selectivity, binding modestly to PtdIns4P-containing membranes (KD = 8.9µM) but not to those containing equivalent amounts of other phosphoinositides (Wood et al., 2009). Crystal structures of Vps74p and GOLPH3 (Schmitz et al., 2008; Wood et al., 2009) revealed a globular domain with a 4-helix bundle at its core, surrounded by a series of amphipathic helices that are almost perpendicular to this bundle (Figure 3A). Although no PtdIns4P was included in the crystals, a bound sulfate ion was seen in a conserved basic site in GOLPH3, adjacent to a β-hairpin that may insert into the membrane (Wood et al., 2009). The importance of this basic ‘pocket’ for phosphoinositide binding was established by mutational analysis. Intriguingly, these studies suggest that specific PtdIns4P recognition is associated with a completely different structural scaffold from those described above: one that has presumably emerged independently of domains that specifically recognize PtdIns3P (PX and FYVE domains) or phosphoinositides with vicinal phosphates (PH domains).
Figure 3. Recent additions to the list of phospholipid binding domains.
Structures of newly reported phospholipid-binding domains are shown in cartoon representation (left); in surface representation (middle) colored according to electrostatic potential (blue is positive, red is negative); and with a close-up view of the proposed lipid or anion binding sites (right). A. The helical GOLPH3 protein (Wood et al., 2009), from PDB entry 3KN1. A sulfate ion lies in the basic patch implicated in PtdIns4P binding. B. The P4M PtdIns4P-binding domain of the Legionella pneumophila SidM protein, from PDB entry 3N6O, which also includes an adjacent GEF domain that is not shown (Schoebel et al., 2010). Two bound sulfate ions are marked. C. The uncomplexed Dock Homology Region-1 (DHR-1) domain (Premkumar et al., 2010) from PDB entry 3L4C. D. The KA1 domain from the S. cerevisiae Kcc4p kinase (Moravcevic et al., 2010) from PDB entry 3OST, which has two primary regions of positive charge on its surface that contribute to non-specific association with negatively-charged membrane surfaces, each with a bound sulfate ion.
Another domain with specificity for PtdIns4P is the P4M domain from the Legionella pneumophila DrrA/SidM protein, involved in redirecting membrane trafficking within infected host cells (Schoebel et al., 2010). A crystal structure of a DrrA/SidM fragment containing the GEF and P4M domains revealed that the ~100 amino acid P4M domain is a helical bundle (Figure 3B) with two sulfate ions bound close to the tips of its three central helices – in a positively-charged pocket (Schoebel et al., 2010). The distance between these two bound sulfates is the same as that between the 1- and 4-phosphates in PtdIns4P. Thus, just as the distance between two anion-binding sites in PH domains (Figure 2) appears to define selectivity for phosphoinositides with a pair of vicinal phosphates, the precise disposition of two anion binding sites in the DrrA/SidM P4M domain may define its distinct selectivity (and high affinity) for PtdIns4P.
PtdIns5P
The functions of PtdIns5P remain unclear. There is evidence that it plays a role in regulating intracellular trafficking, and infection by several intracellular pathogens causes elevation of host PtdIns5P levels and altered endosomal trafficking (Ramel et al., 2011). The C-terminal region of the tumor suppressor ING2, which contains a PHD (plant homeodomain) zinc finger, was reported to bind nuclear PtdIns5P (Gozani et al., 2003). However, PHD fingers have since been shown to function as ‘readers’ of the methylation state of lysines and arginines in the histone H3 amino terminus (Sanchez and Zhou, 2011). The only structural data on PtdIns5P binding by a PHD finger (Huang et al., 2007) indicate very weak binding (KD in the millimolar range by NMR), leaving their status as phosphoinositide-binding domains unconvincing.
PtdIns(3,5)P2
Efforts to identify PtdIns(3,5)P2-specific binding domains have also yielded fewer potential targets than initially expected (Dove et al., 2009). The PROPPINs represent the most convincing candidates (Michell et al., 2006), predicted β-propeller proteins that include Atg18p from S. cerevisiae and mammalian WIPI proteins. Atg18p shows clear selectivity for PtdIns(3,5)P2-containing membranes in vitro, whereas some of its homologues in yeast and mammals also appear to bind PtdIns3P. No structural characterization of PtdIns(3,5)P2 binding to any of the PROPPINs has yet been reported, but mutational analyses suggest that the PtdIns(3,5)P2 headgroup binds to the center of the β-propeller. Interestingly, a general repressor of transcription in S. cerevisiae was also reported to bind PtdIns(3,5)P2 through its β-propeller region (Han and Emr, 2011), which is among the closest sequence relatives of Atg18p. This study also reported lipid overlay studies suggesting that the PHD finger-containing Cti6p protein can bind PtdIns(3,5)P2. It is intriguing that the most convincing PtdIns(3,5)P2 effector (Atg18p) has yet another structural scaffold – distinct from domains that recognize PtdIns3P (PX and FYVE), PtdIns4P (the helical Vps74p) or phosphoinositides with vicinal phosphates (PH domains).
Newly identified domains that bind PI3K products
Since the discovery that several PH domains bind lipid second messenger products of PI3K, a great deal of effort has been expended to identify the whole complement of ‘effector domains’ for these lipids. Several have been discussed above. These studies have defined sophisticated sequence profiles for identifying PH domains that bind PI3K products (Lietzke et al., 2000; Park et al., 2008), new potential effectors such as IQGAP (Dixon et al., 2011), and many other potential effectors that have still to be evaluated. In parallel, numerous studies have focused on proteins known to be likely PtdIns(3,4,5)P2/PtdIns(3,4)P2 effectors, and have asked which of their constituent domains are responsible for phosphoinositide binding.
DHR-1 domain
One example is the Dock Homology Region-1, or DHR-1 domain, required for targeting Dock1 family Rho family guanine nucleotide exchange factors to PtdIns(3,4,5)P3-containing membranes. A crystal structure of the Dock180 DHR-1 domain (Premkumar et al., 2010) revealed that it is a C2 domain with several additional large insertions (Figure 3C). DHR1 binding to PtdIns(3,4,5)P3 is Ca2+-independent, of modest affinity (KD ~3µM), but is not substantially stronger than binding to PtdIns(4,5)P2 in vitro. A combination of modeling and mutational studies suggested that the phosphoinositide binding site lies in the same region of the C2 domain as the phosphatidylserine binding site in classical C2 domains (Premkumar et al., 2010), but is defined by a series of basic side-chains (as in PH domains) by contrast with the Ca2+ binding site in classical C2 domains.
A phosphoinositide-binding site in inositol pyrophosphate kinases
Studies of inositol pyrophosphate kinases called PPIP5Ks have identified another interesting phosphoinositide binding site in which the same β1/β2 loop sequence motif from PH domains (Figure 2) that predicts PI3K product binding appears to have been ‘spliced’ into an acid phosphatase-like domain within the protein (Gokhale et al., 2011). Although this phosphoinositide-binding domain has not yet been characterized structurally, specificity studies indicate that it recognizes vicinal phosphate pairs – perhaps in a manner similar to that seen for PH domains in Figure 2.
The SYLF domain
Although not yet structurally characterized, the so-called SYLF domain (for the names of the proteins that contain it: SH3YL1, Ysc84p/Lsb4p, Lsb3p and plant FYVE proteins) has also been proposed as a novel phosphoinositide-binding domain (Hasegawa et al., 2011). This ~230 amino acid domain appears to bind equally well to liposomes containing PtdIns(3,4,5)P3, PtdIns(4,5)P2 or PtdIns(3,5)P2, and requires an amphipathic N-terminal helical region to do so.
The BATS domain
The ~80 amino acid BATS domain (for Barkor/Atg14(L) autophagosome targeting sequence) is found at the C-terminus of the autophagic adaptor protein Barkor/Atg14. Although not well characterized, the BATS domain appears to bind both PtdIns(4,5)P2 and PtdIns3P, specifically preferring high-curvature membranes containing PtdIns3P utilizing a C-terminal amphipathic helix (Fan et al., 2011).
New PtdIns(4,5)P2-binding proteins: Headgroup recognition or polyanion binding?
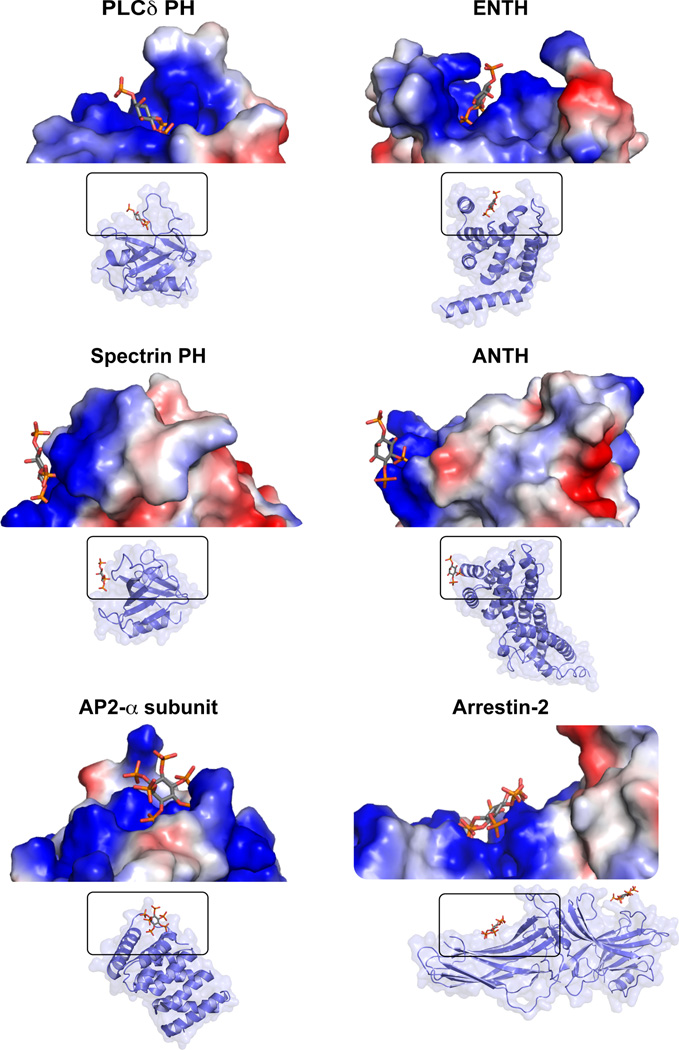
Since PtdIns(4,5)P2 is more abundant than other phosphoinositides and is among the most highly charged phospholipids, specificity constraints on its effector proteins are less stringent than for domains that must specifically recognize the much less abundant PI3K products. Accordingly, many PtdIns(4,5)P2-binding domains and proteins appear quite promiscuous when their phosphoinositide-binding specificity is interrogated. Many are unstructured clusters of basic residues, to which readers are directed elsewhere for excellent reviews (McLaughlin and Murray, 2005). Several others are globular domains that also have protein binding partners, and have been found to bind PtdIns(4,5)P2 in the course of functional studies. Examples include FERM domains (Frame et al., 2010), PDZ domains (Gallardo et al., 2010), PTB domains (DiNitto and Lambright, 2006), the AP180 N-terminal homology (or ANTH) domain (Ford et al., 2001), the amino-terminal part of the AP2 α subunit (Collins et al., 2002), and arrestin (Milano et al., 2006). In several cases, crystal structures have been determined in complex with short-chain phosphoinositides or headgroups. A characteristic shared by many of these proteins is that their phosphoinositide-binding site simply comprises a set of basic side-chains that create a positively charged surface ‘patch’ – contrasting with the well-defined basic pockets seen for the binding sites of domains that specifically recognize particular phosphoinositide isomers. Figure 4 illustrates this contrast. Whereas the PtdIns(4,5)P2 headgroup projects into a well-defined pocket in the specific PLCδ1 PH domain, it simply abuts the surface of the spectrin PH domain (which binds all PtdInsP2 isomers with similar affinities). Similarly, whereas the relatively specific ENTH domain has a well-defined PtdIns(4,5)P2-binding pocket, the structurally analogous ANTH domain uses a surface-lying basic patch for its promiscuous phosphoinositide binding (Ford et al., 2001). Non-specific, electrostatically driven, binding of inositol phosphates to similar basic patches is also seen in the α-subunit of AP2 (Collins et al., 2002) and arrestin-2 (Milano et al., 2006) as illustrated in Figure 4.
Figure 4. Comparison of specific and non-specific phosphoinositide binding domains.
The well defined specific pockets in which Ins(1,4,5)P3 binds to PLCδ-PH (Ferguson et al., 1995) and the epsin ENTH domain (Ford et al., 2002) are contrasted with the surface-lying and solvent-exposed locations of the promiscuous inositol phosphate binding sites on the spectrin PH domain (Hyvönen et al., 1995), the AP180 ANTH domain (Ford et al., 2001), the α subunit of the endocytic AP2 adaptor (residues 9–185 of PDB entry 2VGL are shown) (Collins et al., 2002) and arrestin-2 (Milano et al., 2006). Cartoon representations of each domain are shown, from PDB entries 1MAI (PLCδ-PH/Ins(1,4,5)P3), 1H0A (ENTH/Ins(1,4,5)P3), 1BTN (spectrin-PH/Ins(1,4,5)P3), 1HFA (ANTH/PtdIns(4,5)P2 headgroup), 2VGL (AP2 α-subunit/InsP6), and 1ZSH (arrestin-2/InsP6). Above these representations is a close-up view in surface representation (colored by potential as in Figure 3) of the inositol phosphate-binding site.
Phospholipid-binding domains that recognize general features of the membrane
Membrane association driven by non-specific, delocalized electrostatic attraction appears to be relevant for a growing number of conditional peripheral membrane proteins. Domains in these proteins may act as sensors of membrane charge and/or curvature to fulfill their important biological roles. The cytoplasmic face of the plasma membrane is the most negatively charged membrane surface in mammalian cells, by virtue of its PtdIns(4,5)P2 and phosphatidylserine content. Studies employing cationic surface charge probes have shown that this surface charge is reduced to different extents in distinct endomembranes, depending on their phosphatidylserine content (Yeung et al., 2008). Membrane binding domains in cellular proteins that effectively sense surface charge will therefore interact most strongly with the plasma membrane, although some association will be seen with endosomes and lysosomes. The kinase-associated domain-1, or KA1 domain, recently identified as a phospholipid-binding domain by our laboratory (Moravcevic et al., 2010), epitomizes such membrane ‘charge sensors’. KA1 domains are found at the C-terminus of several kinases from yeast to mammals – including the human MARK kinases. Analysis of phospholipid binding specificity in vitro and in vivo revealed that KA1 domains do not distinguish between different anionic phospholipids. KA1 domains contain two interacting α-helices that lie on the concave surface of a four-stranded β-sheet (Figure 3D). One or two well-defined positively charged regions are found on the surface of KA1 domains with known structure, and were shown in mutational studies to be responsible for binding negatively-charged membranes (Moravcevic et al., 2010). In the case of yeast septin-associated kinases, the KA1 domain cooperates with adjacent domains to drive the protein to membranes that also contain a second binding partner. A combination of the KA1 domain and an adjacent low-affinity septin-binding domain, for example, can specifically target the kinase to septins only when they are assembled at the membrane surface – in a form of coincidence detection.
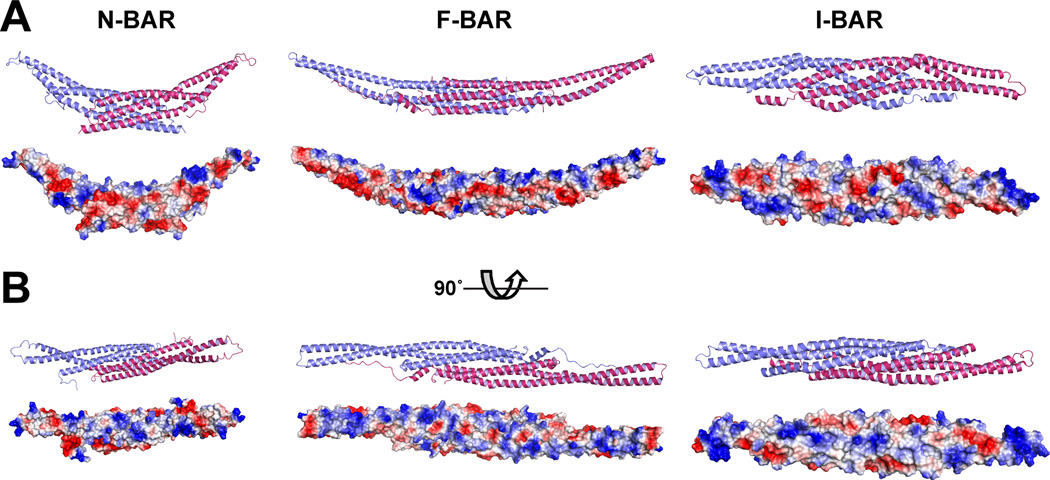
Members of an interesting group of conditional peripheral membrane proteins or domains – the BAR domain (Bin/Amphiphysin/Rvs) superfamily – are thought to promote or sense membrane curvature, another general membrane property. There are three main groups of domains in this superfamily: N-BAR, F-BAR and I-BAR domains (Figure 5). The groups share relatively little sequence identity with one another, but all form extended dimeric helical bundles that are characteristically ‘banana-shaped’ (Qualmann et al., 2011). The degree of curvature defined by the long axis of the dimer varies from group to group (and within groups) and is believed to determine the degree and direction of membrane curvature that is sensed or induced upon binding. The N-BAR domains promote a higher degree of curvatures than F-BAR domains as indicated in Figure 5A, and the I-BAR domain is thought to promote a small degree of negative curvature. The BAR superfamily has been recently well reviewed (Antonny, 2011; Qualmann et al., 2011), so functional details will not be discussed here. However, this is a family of proteins that combines delocalized electrostatic attraction with geometric definition. The membrane-binding face of each BAR family protein contains a series of positively charged patches (Figure 5B), and the basic residues within these patches have been shown by mutational studies to be important for membrane association. The multiple basic patches – which each represents a weak binding site – can only cooperate with one another in binding to the membrane surface if the geometry of the membrane conforms to that of the protein – i.e. becomes curved. In addition, some domains in the family insert amphipathic helices into the membrane to promote curvature (Qualmann et al., 2011). In isolation, BAR family domains can tubulate membranes (Frost et al., 2009), which may be important for their function. In cells, it has been suggested that the progressive recruitment of distinct BAR family proteins that induce different degrees of curvature may play an important role in driving budding and vesiculation in processes such as endocytosis (Qualmann et al., 2011); the different proteins selectively stabilizing intermediates in the process. The selectivity of successive recruitment could be defined in part by the adjacent PX, PH, SH3, or other domains that are frequently found adjacent to BAR family domains.
Figure 5. The BAR domain superfamily.
Structures are shown for representatives from the BAR superfamily: the amphiphysin N-BAR domain (Peter et al., 2004) from PDB entry 1URU; the Fbp17 F-BAR domain (Shimada et al., 2007) from PDB entry 2EFL; and the IRSp53/missing-in-metastasis I-BAR domain (Millard et al., 2005) from PDB entry 1Y2O. Two orientations are shown in both cartoon and surface representation (with electrostatic potential colored as in Figure 3). In A, the membrane-binding surface is open to the top of the page, revealing the ‘banana-shape’ and the different characteristic curvature of each domain. In B, the membrane-binding surface faces the reader, illustrating the clusters of basic residues thought to drive binding to the (curved) anionic membrane surface.
Emerging studies of the endosomal sorting complexes required for transport (or ESCRTs) suggest that similar principles may also define the ability of this complex machinery to drive membrane budding away from the cytosol (Hurley and Hanson, 2010). Like BAR family proteins, key components of ESCRT-III are helical proteins with extended basic surfaces, and undergo a tightly controlled assembly/disassembly cycle to drive processes such as the formation of multivesicular bodies, HIV budding, and cytokinesis (Henne et al., 2011).
Coincidence detection in binding to the membrane surface
The theme of coincidence detection has recurred several times during this review. Its occurrence may explain why fewer domains than expected stereospecifically recognize a single membrane component such as a phosphoinositide. An interesting study of S. cerevisiae protein-lipid interactions provides new food for thought in considering PH domains. An earlier proteome-wide study suggested that a surprisingly small fraction (~20%) of S. cerevisiae PH domains bind phosphoinositides (Yu et al., 2004), and suggested that most PH domains have alternative binding targets. In a more recent analysis of yeast phospholipid-binding domains, Gallego et al. (2010) identified numerous examples that bind sphingolipids – including BAR family proteins (amphiphysins), as well as ~50% of yeast PH domains. For one of these PH domains (from Slm1p or Yil105cp), biochemical and cellular studies showed that efficient membrane targeting specifically requires the presence of both PtdIns(4,5)P2 and dihydrosphingosine-1-phosphate (DHS-1P) in the membrane. Gallego et al. (2010) determined the Slm1p-PH crystal structure (Figure 6A) and docked PtdIns(4,5)P2 and DHS-1P respectively into the canonical PH domain binding site (green) and a second basic pocket (magenta). Mutations in the predicted DHS-1P binding site impaired membrane targeting, consistent with the proposed importance of this site. Intriguingly, other genetic data in yeast had already identified Slm1p as a molecular link between phosphoinositide and sphingolipid signaling (Tabuchi et al., 2006).
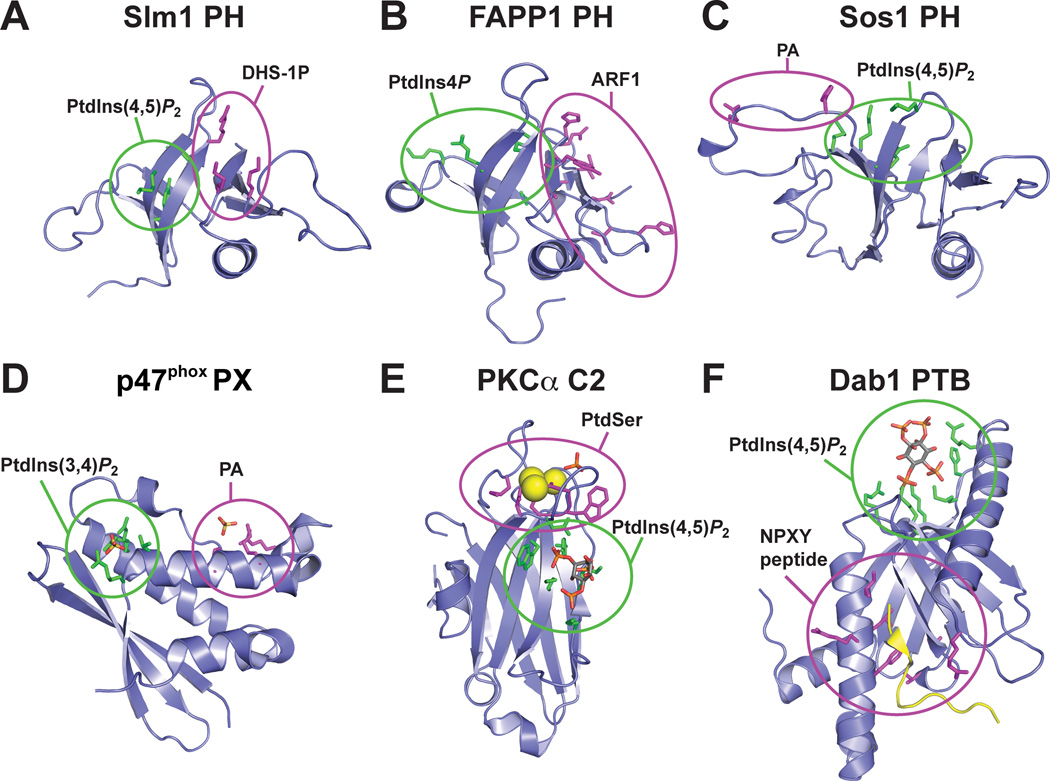
Figure 6. Phospholipid binding domains that bind dual targets.
Six phospholipid binding domains known to simultaneously bind two targets are shown in cartoon representation, with their phosphoinositide binding site highlighted in green and the ‘other’ binding site highlighted in magenta. A. The Slm1p PH domain (PDB code 3NSU) binds both PtdIns(4,5)P2 and dihydrophingosine-1-phosphate (DHS-1P) through adjacent sites on the domain (Gallego et al., 2010). B. The FAPP1 PH domain (PDB code 2KCJ) simultaneously binds phosphoinositides and Arf1 through the sites colored in the figure (He et al., 2011) to drive Golgi localization. C. NMR studies of the Sos1 PH domain (Zheng et al., 1997), shown from PDB entry 1AWE, confirmed that Ins(1,4,5)P3 bind to the same region as in other PH domains. An adjacent basic patch (magenta) was reported to bind phosphatidic acid (Zhao et al., 2006). D. The PX domain from p47phox (PTB code 1O7K) was also found to bind both to a phosphoinositide (PtdIns(3,4)P2, using the canonical PX domain binding site) and to PA, using a basic patch similar to that seen in Sos1-PH (Karathanassis et al., 2002). E. A structure of the PKCα C2 domain bound to a PtdIns(4,5)P2 headgroup (PDB code 3GPE) illustrates how this C2 domain can simultaneously bind phosphatidylserine (as in Figure 1) and phosphoinositides (Guerrero-Valero et al., 2009). F. The structure of the PTB domain from Dab1 (PDB code 1NU2) was solved in complex with both a 14-residue peptide (yellow) from the tail of apolipoprotein E receptor-2 (ApoER2) and Ins(1,4,5)P3 (Stolt et al., 2003), revealing how coincidence detection by this domain operates.
Similar modes of coincidence detection – in which a single domain binds more than one target – have been reported for other modules. As mentioned above, the FAPP1 PH domain and its relatives bind to both phosphoinositides (using the canonical PH domain binding site) and Arf1 (Godi et al., 2004; Levine and Munro, 2002). NMR-based mapping of the Arf1 binding site (He et al., 2011) places it adjacent to the phosphoinositide-binding site – on the outer surface of the β-sandwich (Figure 6B). The two binding sites are structurally independent, underlining the adaptability of PH domains as binding platforms. Simultaneous binding to phosphoinositides and Arf1 drives these PH domains to the membrane surface with high affinity. The PH domain from Sos1 and the PX domain from p47phox have been reported to use phosphatidic acid (PA) binding to augment their interactions with membrane phosphoinositides (PtdIns(4,5)P2 and PtdIns(3,4)P2 respectively). In both cases, the phosphoinositide headgroup binds to the canonical binding site for the respective domain class (Karathanassis et al., 2002; Zheng et al., 1997). The Sos1 PH domain and the p47 PX domain both have a motif of basic residues that has been inferred as a PA binding site (Karathanassis et al., 2002; Zhao et al., 2006), as shown in Figure 6C and D. Along similar lines, structural studies have revealed how the PKCα C2 domain can bind simultaneously to phosphatidylserine and PtdIns(4,5)P2 (Guerrero-Valero et al., 2009), and how the PTB domain from disabled-1 can accommodate an NPXY peptide and phosphoinositide headgroup in separate binding sites (Stolt et al., 2003). As shown in Figure 6E and F, in each case the two binding sites are disposed so that the single domain can associate with two targets in the same membrane – allowing coincidence detection. Coincidence detection of this sort can ensure that signaling molecules only bind to membrane receptors in the plasma membrane if PtdIns(4,5)P2 binding is required as part of a multivalent interaction. Alternatively, it can allow the integration of signaling responses – as in the p47phox case, where the PX domain will be recruited to the membrane only when PtdIns(3,4)P2 and PA are simultaneously produced. Many other possible combinations are possible, and there are many incidences (including the KA1 domain example mentioned above) where coincidence is detected by cooperation of two or more distinct domains in a protein.
Conclusions and Future Directions
Our understanding of membrane surface binding by conditional peripheral membrane proteins and domains has become quite sophisticated and extensive over the past two decades. Although the earliest examples were found to drive specific univalent recognition of individual membrane components, it is now clear that these are the exception rather than the norm. Most conditional peripheral membrane domains bind individual membrane components with relatively low affinity and specificity. Membrane-binding strength is enhanced through avidity effects involving multivalent interactions with several different membrane components that may be lipids, proteins, or both. Binding specificity is determined largely by the combination of components present in a particular membrane and by its physical properties (charge and curvature), greatly extending the number of possibilities beyond what would be possible with recognition of individual constituents alone. Thus, conditional peripheral membrane proteins can bind selectively to membrane protein targets in particular cellular membranes, and under specific signaling conditions – by recognizing them only in certain lipid contexts. Similarly, with domains that sense (or drive) curvature, specific phospholipids can be recognized only when present in membranes undergoing morphological changes (or their recognition can help drive the morphological changes).
The only aspect of membrane binding by conditional peripheral membrane proteins that is understood in precise structural detail is specific headgroup recognition. However, many conditional peripheral membrane proteins penetrate the membrane surface. Many of the domains also alter lateral distribution of lipid molecules, or change local membrane curvature. Methods for monitoring these changes structurally remain crude, and are largely limited to measurements of extent and depth of penetration. To fully understand the interplay between binding of conditional peripheral membrane proteins and alterations of the membranes that they bind, new methods – or new combinations of methods – will be required. Although subject to significant limitations, approaches that are currently being developed towards this goal include analysis of single molecule (e.g. PH domain) diffusion on membrane surfaces with different compositions (Knight et al., 2010), NMR-based micelle docking studies to assess lipid interactions (Dancea et al., 2008), and electron microscopy studies of domain-tubulated membranes (Frost et al., 2009). However, these studies are only beginning to scratch the surface of this important problem.
Finally, another consequence of coincidence detection at the level of the individual domain – and the ability of domains to recognize general properties of membranes – is that we need to think very carefully about approaches for screening for new domains. Traditional approaches utilizing immobilized lipid headgroups are unlikely to be effective, and so are most studies that focus on individual binding targets (rather than combinations). Clearly, significant challenges remain in this interesting field.
Highlights.
Some modules stereospecifically recognize lipid headgroups
PH, PX, FYVE, ENTH, Tubby, C1 and C2 domains are structurally well characterized
Screening for new phospholipid-interaction domains has not yielded major new classes
Few domain specifically recognize PtdIns4P, PtdIns5P or PtdIns(3,5)P2
Several domains recognize the PI3K products PtdIns(3,4,5)P3 and PtdIns(3,4)P2
PtdIns(4,5)P2-binding domains have less stringent specificity requirements
Several membrane-binding domains bind polyanions or curved surfaces
Target coincidence detection is a common theme in membrane-binding domains
Acknowledgements
We thank members of the Lemmon and Ferguson groups for helpful discussions and comments. Work in the Lemmon lab on phospholipid binding domains has been funded by R01 grants GM056846 and GM078345 to M.A.L. and by a predoctoral fellowship from the American Heart Association Great Rivers Affiliate (K.M.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Antonny B. Mechanisms of membrane curvature sensing. Annu. Rev. Biochem. 2011;80:101–123. doi: 10.1146/annurev-biochem-052809-155121. [DOI] [PubMed] [Google Scholar]
- Bravo J, Karathanassis D, Pacold CM, Pacold ME, Ellson CD, Anderson KE, Butler PJ, Lavenir I, Perisic O, Hawkins PT, Stephens L, Williams RL. The crystal structure of the PX domain from p40(phox) bound to phosphatidylinositol 3-phosphate. Mol. Cell. 2001;8:829–839. doi: 10.1016/s1097-2765(01)00372-0. [DOI] [PubMed] [Google Scholar]
- Catimel B, Schieber C, Condron M, Patsiouras H, Connolly L, Catimel J, Nice EC, Burgess AW, Holmes AB. The PI(3,5)P2 and PI(4,5)P2 interactomes. J. Proteome Res. 2008;7:5295–5313. doi: 10.1021/pr800540h. [DOI] [PubMed] [Google Scholar]
- Catimel B, Yin MX, Schieber C, Condron M, Patsiouras H, Catimel J, Robinson DE, Wong LS, Nice EC, Holmes AB, Burgess AW. PI(3,4,5)P3 Interactome. J. Proteome Res. 2009;8:3712–3726. doi: 10.1021/pr900320a. [DOI] [PubMed] [Google Scholar]
- Ceccarelli DF, Blasutig IM, Goudreault M, Li Z, Ruston J, Pawson T, Sicheri F. Non-canonical interaction of phosphoinositides with pleckstrin homology domains of Tiam1 and ArhGAP9. J. Biol. Chem. 2007;282:13864–13874. doi: 10.1074/jbc.M700505200. [DOI] [PubMed] [Google Scholar]
- Cheever ML, Sato TK, de Beer T, Kutateladze TG, Emr SD, Overduin M. Phox domain interaction with PtdIns(3)P targets the Vam7 t-SNARE to vacuole membranes. Nature Cell Biol. 2001;3:613–618. doi: 10.1038/35083000. [DOI] [PubMed] [Google Scholar]
- Cho W, Stahelin RV. Membrane binding and subcellular targeting of C2 domains. Biochim. Biophys. Acta. 2006;1761:838–849. doi: 10.1016/j.bbalip.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Collins BM, McCoy AJ, Kent HM, Evans PR, Owen DJ. Molecular architecture and functional model of the endocytic AP2 complex. Cell. 2002;109:523–535. doi: 10.1016/s0092-8674(02)00735-3. [DOI] [PubMed] [Google Scholar]
- Colon-Gonzalez F, Kazanietz MG. C1 domains exposed: from diacylglycerol binding to protein-protein interactions. Biochim. Biophys. Acta. 2006;1761:827–837. doi: 10.1016/j.bbalip.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Dancea F, Kami K, Overduin M. Lipid interaction networks of peripheral membrane proteins revealed by data-driven micelle docking. Biophys. J. 2008;94:515–524. doi: 10.1529/biophysj.107.115923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Camilli P, Chen H, Hyman J, Panepucci E, Bateman A, Brünger AT. The ENTH domain. FEBS Letts. 2002;513:11–18. doi: 10.1016/s0014-5793(01)03306-3. [DOI] [PubMed] [Google Scholar]
- Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- DiNitto JP, Lambright DG. Membrane and juxtamembrane targeting by PH and PTB domains. Biochim. Biophys. Acta. 2006;1761:850–867. doi: 10.1016/j.bbalip.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Dippold HC, Ng MM, Farber-Katz SE, Lee SK, Kerr ML, Peterman MC, Sim R, Wiharto PA, Galbraith KA, Madhavarapu S, Fuchs GJ, Meerloo T, Farquhar MG, Zhou H, Field SJ. GOLPH3 bridges phosphatidylinositol-4- phosphate and actomyosin to stretch and shape the Golgi to promote budding. Cell. 2009;139:337–351. doi: 10.1016/j.cell.2009.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon MJ, Gray A, Boisvert FM, Agacan M, Morrice NA, Gourlay R, Leslie NR, Downes CP, Batty IH. A screen for novel phosphoinositide 3-kinase effector proteins. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.M110.003178. M110.003178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dove SK, Cooke FT, Douglas MR, Sayers LG, Parker PJ, Michell RH. Osmotic stress activates phosphatidylinositol-3,5-bisphosphate synthesis. Nature. 1997;390:187–192. doi: 10.1038/36613. [DOI] [PubMed] [Google Scholar]
- Dove SK, Dong K, Kobayashi T, Williams FK, Michell RH. Phosphatidylinositol 3,5-bisphosphate and Fab1p/PIKfyve underPPIn endo-lysosome function. Biochem. J. 2009;419:1–13. doi: 10.1042/BJ20081950. [DOI] [PubMed] [Google Scholar]
- Dowler S, Currie RA, Campbell DG, Deak M, Kular G, Downes CP, Alessi DR. Identification of pleckstrin-homology-domain-containing proteins with novel phosphoinositide-binding specificities. Biochem. J. 2000;351:19–31. doi: 10.1042/0264-6021:3510019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas JJ, Merithew E, Sudharshan E, Rajamani D, Hayes S, Lawe D, Corvera S, Lambright DG. Multivalent endosome targeting by homodimeric EEA1. Mol. Cell. 2001;8:947–958. doi: 10.1016/s1097-2765(01)00385-9. [DOI] [PubMed] [Google Scholar]
- Fan W, Nassiri A, Zhong Q. Autophagosome targeting and membrane curvature sensing by Barkor/Atg14(L) Proc. Natl. Acad. Sci. USA. 2011;108:7769–7774. doi: 10.1073/pnas.1016472108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson KM, Kavran JM, Sankaran VG, Fournier E, Isakoff SJ, Skolnik EY, Lemmon MA. Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains. Mol. Cell. 2000;6:373–384. doi: 10.1016/s1097-2765(00)00037-x. [DOI] [PubMed] [Google Scholar]
- Ferguson KM, Lemmon MA, Schlessinger J, Sigler PB. Structure of the high affinity complex of inositol trisphosphate with a phospholipase C pleckstrin homology domain. Cell. 1995;83:1037–1046. doi: 10.1016/0092-8674(95)90219-8. [DOI] [PubMed] [Google Scholar]
- Ford MG, Mills IG, Peter BJ, Vallis Y, Praefcke GJ, Evans PR, McMahon HT. Curvature of clathrin-coated pits driven by epsin. Nature. 2002;419:361–366. doi: 10.1038/nature01020. [DOI] [PubMed] [Google Scholar]
- Ford MG, Pearse BM, Higgins MK, Vallis Y, Owen DJ, Gibson A, Hopkins CR, Evans PR, McMahon HT. Simultaneous binding of PtdIns(4,5)P2 and clathrin by AP180 in the nucleation of clathrin lattices on membranes. Science. 2001;291:1051–1055. doi: 10.1126/science.291.5506.1051. [DOI] [PubMed] [Google Scholar]
- Frame MC, Patel H, Serrels B, Lietha D, Eck MJ. The FERM domain: organizing the structure and function of FAK. Nat. Rev. Mol. Cell. Biol. 2010;11:802–814. doi: 10.1038/nrm2996. [DOI] [PubMed] [Google Scholar]
- Frost A, Unger VM, De Camilli P. The BAR domain superfamily: membrane-molding macromolecules. Cell. 2009;137:191–196. doi: 10.1016/j.cell.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo R, Ivarsson Y, Schymkowitz J, Rousseau F, Zimmermann P. Structural diversity of PDZ-lipid interactions. Chembiochem. 2010;11:456–467. doi: 10.1002/cbic.200900616. [DOI] [PubMed] [Google Scholar]
- Gallego O, Betts MJ, Gvozdenovic-Jeremic J, Maeda K, Matetzki C, Aguilar-Gurrieri C, Beltran-Alvarez P, Bonn S, Fernández-Tornero C, Jensen LJ, Kuhn M, Trott J, Rybin V, Müller CW, Bork P, Kaksonen M, Russell RB, Gavin AC. A systematic screen for protein-lipid interactions in Saccharomyces cerevisiae. Mol. Syst. Biol. 2010;6:430. doi: 10.1038/msb.2010.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godi A, Di Campli A, Konstantakopoulos A, Di Tullio G, Alessi DR, Kular GS, Daniele T, Marra P, Lucocq JM, De Matteis MA. FAPPs control Golgi-to-cell-surface membrane traffic by binding to ARF and PtdIns(4)P. Nature Cell Biol. 2004;6:393–404. doi: 10.1038/ncb1119. [DOI] [PubMed] [Google Scholar]
- Gokhale NA, Zaremba A, Shears SB. Receptor-dependent compartmentalization of PPIP5K1, a kinase with a cryptic polyphosphoinositide binding domain. Biochem. J. 2011;434:415–426. doi: 10.1042/BJ20101437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozani O, Karuman P, Jones DR, Ivanov D, Cha J, Lugovskoy AA, Baird CL, Zhu H, Field SJ, Lessnick SL, Villasenor J, Mehrotra B, Chen J, Rao VR, Brugge JS, Ferguson CG, Payrastre B, Myszka DG, Cantley LC, Wagner G, Divecha N, Prestwich GD, Yuan J. The PHD finger of the chromatin-associated protein ING2 functions as a nuclear phosphoinositide receptor. Cell. 2003;114:99–111. doi: 10.1016/s0092-8674(03)00480-x. [DOI] [PubMed] [Google Scholar]
- Guerrero-Valero M, Ferrer-Orta C, Querol-Audí J, Marin-Vicente C, Fita I, Gómez-Fernández JC, Verdaguer N, Corbalán-García S. Structural and mechanistic insights into the association of PKCalpha-C2 domain to PtdIns(4,5)P2. Proc. Natl. Acad. Sci. USA. 2009;106:6603–6607. doi: 10.1073/pnas.0813099106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han BK, Emr SD. Phosphoinositide [PI(3,5)P2] lipid-dependent regulation of the general transcriptional regulator Tup1. Genes Dev. 2011;25:984–995. doi: 10.1101/gad.1998611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlan JE, Hajduk PJ, Yoon HS, Fesik SW. Pleckstrin homology domains bind to phosphatidylinositol-4,5-bisphosphate. Nature. 1994;371:168–170. doi: 10.1038/371168a0. [DOI] [PubMed] [Google Scholar]
- Hasegawa J, Tokuda E, Tenno T, Tsujita K, Sawai H, Hiroaki H, Takenawa T, Itoh T. SH3YL1 regulates dorsal ruffle formation by a novel phosphoinositide-binding domain. J. Cell Biol. 2011;193:901–916. doi: 10.1083/jcb.201012161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Scott JL, Heroux A, Roy S, Lenoir M, Overduin M, Stahelin RV, Kutateladze TG. Molecular basis of phosphatidylinositol 4-phosphate and ARF1 GTPase recognition by the FAPP1 pleckstrin homology (PH) domain. J. Biol. Chem. 2011;286:18650–18657. doi: 10.1074/jbc.M111.233015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Dev. Cell. 2011;21:77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- Huang W, Zhang H, Davrazou F, Kutateladze TG, Shi X, Gozani O, Prestwich GD. Stabilized phosphatidylinositol-5-phosphate analogues as ligands for the nuclear protein ING2: chemistry, biology, and molecular modeling. J. Am. Chem. Soc. 2007;129:6498–6506. doi: 10.1021/ja070195b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH, Hanson PI. Membrane budding and scission by the ESCRT machinery: it's all in the neck. Nat. Rev. Mol. Cell Biol. 2010;11:556–566. doi: 10.1038/nrm2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman J, Chen H, Di Fiore PP, De Camilli P, Brunger AT. Epsin 1 undergoes nucleocytosolic shuttling and its eps15 interactor NH(2)-terminal homology (ENTH) domain, structurally similar to Armadillo and HEAT repeats, interacts with the transcription factor promyelocytic leukemia Zn(2)+ finger protein (PLZF) J. Cell Biol. 2000;149:537–546. doi: 10.1083/jcb.149.3.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyvönen M, Macias MJ, Nilges M, Oschkinat H, Saraste M, Wilmanns M. Structure of the binding site for inositol phosphates in a PH domain. EMBO J. 1995;14:4676–4685. doi: 10.1002/j.1460-2075.1995.tb00149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isakoff SJ, Cardozo T, Andreev J, Li Z, Ferguson KM, Abagyan R, Lemmon MA, Aronheim A, Skolnik EY. Identification and analysis of PH domain-containing targets of phosphatidylinositol 3-kinase using a novel in vivo assay in yeast. EMBO J. 1998;17:5374–5387. doi: 10.1093/emboj/17.18.5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janmey PA. Protein regulation by phosphatidylinositol lipids. Chem. Biol. 1995;2:61–65. doi: 10.1016/1074-5521(95)90276-7. [DOI] [PubMed] [Google Scholar]
- Janmey PA, Stossel TP. Modulation of gelsolin function by phosphatidylinositol 4,5-bisphosphate. Nature. 1987;325:362–364. doi: 10.1038/325362a0. [DOI] [PubMed] [Google Scholar]
- Karathanassis D, Stahelin RV, Bravo J, Perisic O, Pacold CM, Cho W, Williams RL. RL. Binding of the PX domain of p47(phox) to phosphatidylinositol 3,4-bisphosphate and phosphatidic acid is masked by an intramolecular interaction. EMBO J. 2002;21:5057–5068. doi: 10.1093/emboj/cdf519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight JD, Lerner MG, Marcano-Velázquez JG, Pastor RW, Falke JJ. Single molecule diffusion of membrane-bound proteins: window into lipid contacts and bilayer dynamics. Biophys. J. 2010;99:2879–2887. doi: 10.1016/j.bpj.2010.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutateladze TG. Phosphatidylinositol 3-phosphate recognition and membrane docking by the FYVE domain. Biochim. Biophys. Acta. 2006;1761:868–877. doi: 10.1016/j.bbalip.2006.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutateladze TG. Translation of the phosphoinositide code by PI effectors. Nature Chem. Biol. 2010;6:507–513. doi: 10.1038/nchembio.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassing I, Lindberg U. Specific interaction between phosphatidylinositol 4,5-bisphosphate and profilactin. Nature. 1985;314:472–474. doi: 10.1038/314472a0. [DOI] [PubMed] [Google Scholar]
- Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- Lemmon MA, Ferguson KM, O'Brien R, Sigler PB, Schlessinger J. Specific and high-affinity binding of inositol phosphates to an isolated pleckstrin homology domain. Proc. Natl. Acad. Sci. USA. 1995;92:10472–10476. doi: 10.1073/pnas.92.23.10472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenoir M, Coskun U, Grzybek M, Cao X, Buschhorn SB, James J, Simons K, Overduin M. Structural basis of wedging the Golgi membrane by FAPP pleckstrin homology domains. EMBO Rep. 2010;11:279–284. doi: 10.1038/embor.2010.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. SMART 6: recent updates and new developments. Nucl. Acids Res. 2008;37:D229–D232. doi: 10.1093/nar/gkn808. (Database issue), [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine TP, Munro S. Targeting of Golgi-specific pleckstrin homology domains involves both PtdIns 4-kinase-dependent and -independent components. Curr. Biol. 2002;12:695–704. doi: 10.1016/s0960-9822(02)00779-0. [DOI] [PubMed] [Google Scholar]
- Lewis AE, Sommer L, Arntzen MØ, Strahm Y, Morrice NA, Divecha N, D'Santos CS. Identification of nuclear phosphatidylinositol 4,5-bisphosphate-interacting proteins by neomycin extraction. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.M110.003376. M110.003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lietzke SE, Bose S, Cronin T, Klarlund J, Chawla A, Czech MP, Lambright DG. Structural basis of 3-phosphoinositide recognition by pleckstrin homology domains. Mol. Cell. 2000;6:385–394. doi: 10.1016/s1097-2765(00)00038-1. [DOI] [PubMed] [Google Scholar]
- McLaughlin S, Murray D. Plasma membrane phosphoinositide organization by protein electrostatics. Nature. 2005;438:605–611. doi: 10.1038/nature04398. [DOI] [PubMed] [Google Scholar]
- Medkova M, Cho W. Interplay of C1 and C2 domains of protein kinase C-alpha in its membrane binding and activation. J. Biol. Chem. 1999;274:19852–19861. doi: 10.1074/jbc.274.28.19852. [DOI] [PubMed] [Google Scholar]
- Michell RH, Heath VL, Lemmon MA, Dove SK. Phosphatidylinositol 3,5-bisphosphate: metabolism and cellular functions. Trends Biochem. Sci. 2006;31:52–63. doi: 10.1016/j.tibs.2005.11.013. [DOI] [PubMed] [Google Scholar]
- Milano SK, Kim YM, Stefano FP, Benovic JL, Brenner C. Nonvisual arrestin oligomerization and cellular localization are regulated by inositol hexakisphosphate binding. J. Biol. Chem. 2006;281:9812–9823. doi: 10.1074/jbc.M512703200. [DOI] [PubMed] [Google Scholar]
- Millard TH, Bompard G, Heung MY, Dafforn TR, Scott DJ, Machesky LM, Fütterer K. Structural basis of filopodia formation induced by the IRSp53/MIM homology domain of human IRSp53. EMBO J. 2005;24:240–250. doi: 10.1038/sj.emboj.7600535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra S, Hurley JH. Crystal structure of a phosphatidylinositol 3-phosphate-specific membrane-targeting motif, the FYVE domain of Vps27p. Cell. 1999;97:657–666. doi: 10.1016/s0092-8674(00)80776-x. [DOI] [PubMed] [Google Scholar]
- Moravcevic K, Mendrola JM, Schmitz KR, Wang YH, Slochower D, Janmey PA, Lemmon MA. Kinase associated-1 domains drive MARK/PAR1 kinases to membrane targets by binding acidic phospholipids. Cell. 2010;143:966–977. doi: 10.1016/j.cell.2010.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oancea E, Meyer T. Protein kinase C as a molecular machine for decoding calcium and diacylglycerol signals. Cell. 1998;95:307–318. doi: 10.1016/s0092-8674(00)81763-8. [DOI] [PubMed] [Google Scholar]
- Park WS, Heo WD, Whalen JH, O'Rourke NA, Bryan HM, Meyer T, Teruel MN. Comprehensive identification of PIP3-regulated PH domains from C. elegans to H. sapiens by model prediction and live imaging. Mol. Cell. 2008;30:381–392. doi: 10.1016/j.molcel.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, Evans PR, McMahon HT. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- Premkumar L, Bobkov AA, Patel M, Jaroszewski L, Bankston LA, Stec B, Vuori K, Côté JF, Liddington RC. Structural basis of membrane targeting by the Dock180 family of Rho family guanine exchange factors (Rho-GEFs) J. Biol. Chem. 2010;285:13211–13222. doi: 10.1074/jbc.M110.102517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qualmann B, Koch D, Kessels MM. Let's go bananas: revisiting the endocytic BAR code. EMBO J. 2011;30:3501–3515. doi: 10.1038/emboj.2011.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rameh LE, Tolias KF, Duckworth BC, Cantley LC. A new pathway for synthesis of phosphatidylinositol-4,5-bisphosphate. Nature. 1997;390:192–196. doi: 10.1038/36621. [DOI] [PubMed] [Google Scholar]
- Ramel D, Lagarrigue F, Pons V, Mounier J, Dupuis-Coronas S, Chicanne G, Sansonetti PJ, Gaits-Iacovoni F, Tronchère H, Payrastre B. Shigella flexneri infection generates the lipid PI5P to alter endocytosis and prevent termination of EGFR signaling. Science Signal. 2011;4:ra61. doi: 10.1126/scisignal.2001619. [DOI] [PubMed] [Google Scholar]
- Sanchez R, Zhou MM. The PHD finger: a versatile epigenome reader. Trends Biochem. Sci. 2011;36:364–372. doi: 10.1016/j.tibs.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Bautista S, Marín-Vicente C, Gómez-Fernández JC, Corbalán-García S. The C2 domain of PKCalpha is a Ca2+ -dependent PtdIns(4,5)P2 sensing domain: a new insight into an old pathway. J. Mol. Biol. 2006;362:901–914. doi: 10.1016/j.jmb.2006.07.093. [DOI] [PubMed] [Google Scholar]
- Santagata S, Boggon TJ, Baird CL, Gomez CA, Zhao J, Shan WS, Myszka DG, Shapiro L. G-protein signaling through tubby proteins. Science. 2001;292:2041–2050. doi: 10.1126/science.1061233. [DOI] [PubMed] [Google Scholar]
- Schmitz KR, Liu J, Li S, Setty TG, Wood CS, Burd CG, Ferguson KM. Golgi localization of glycosyltransferases requires a Vps74p oligomer. Dev. Cell. 2008;14:523–534. doi: 10.1016/j.devcel.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoebel S, Blankenfeldt W, Goody RS, Itzen A. High-affinity binding of phosphatidylinositol 4-phosphate by Legionella pneumophila DrrA. EMBO Rep. 2010;11:598–604. doi: 10.1038/embor.2010.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seet LF, Hong W. The Phox (PX) domain proteins and membrane traffic. Biochim. Biophys. Acta. 2006;1761:878–896. doi: 10.1016/j.bbalip.2006.04.011. [DOI] [PubMed] [Google Scholar]
- Shimada A, Niwa H, Tsujita K, Suetsugu S, Nitta K, Hanawa-Suetsugu K, Akasaka R, Nishino Y, Toyama M, Chen L, Liu ZJ, Wang BC, Yamamoto M, Terada T, Miyazawa A, Tanaka A, Sugano S, Shirouzu M, Nagayama K, Takenawa T, Yokoyama S. Curved EFC/F-BAR-domain dimers are joined end to end into a filament for membrane invagination in endocytosis. Cell. 2007;129:761–772. doi: 10.1016/j.cell.2007.03.040. [DOI] [PubMed] [Google Scholar]
- Stahelin RV, Karathanassis D, Murray D, Williams RL, Cho W. Structural and membrane binding analysis of the Phox homology domain of Bem1p: basis of phosphatidylinositol 4-phosphate specificity. J. Biol. Chem. 2007;282:25737–25747. doi: 10.1074/jbc.M702861200. [DOI] [PubMed] [Google Scholar]
- Stefan CJ, Audhya A, Emr SD. The yeast synaptojanin-like proteins control the cellular distribution of phosphatidylinositol (4,5)-bisphosphate. Mol. Biol. Cell. 2002;13:542–557. doi: 10.1091/mbc.01-10-0476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolt PC, Jeon H, Song HK, Herz J, Eck MJ, Blacklow SC. Origins of peptide selectivity and phosphoinositide binding revealed by structures of disabled-1 PTB domain complexes. Structure. 2003;11:569–579. doi: 10.1016/s0969-2126(03)00068-6. [DOI] [PubMed] [Google Scholar]
- Szentpetery Z, Balla A, Kim YJ, Lemmon MA, Balla T. Live cell imaging with protein domains capable of recognizing phosphatidylinositol 4,5-bisphosphate; a comparative study. BMC Cell Biol. 2009;10:67. doi: 10.1186/1471-2121-10-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi M, Audhya A, Parsons AB, Boone C, Emr SD. The phosphatidylinositol 4,5-biphosphate and TORC2 binding proteins Slm1 and Slm2 function in sphingolipid regulation. Mol. Cell. Biol. 2006;26:5861–5875. doi: 10.1128/MCB.02403-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo H, Gill DJ, Sun J, Perisic O, Veprintsev DB, Vallis Y, Emr SD, Williams RL. ESCRT-I core and ESCRT-II GLUE domain structures reveal role for GLUE in linking to ESCRT-I and membranes. Cell. 2006;125:99–111. doi: 10.1016/j.cell.2006.01.047. [DOI] [PubMed] [Google Scholar]
- Uchida Y, Hasegawa J, Chinnapen D, Inoue T, Okazaki S, Kato R, Wakatsuki S, Misaki R, Koike M, Uchiyama Y, Iemura S, Natsume T, Kuwahara R, Nakagawa T, Nishikawa K, Mukai K, Miyoshi E, Taniguchi N, Sheff D, Lencer WI, Taguchi T, Arai H. Intracellular phosphatidylserine is essential for retrograde membrane traffic through endosomes. Proc. Natl. Acad. Sci. USA. 2011;108:15846–15851. doi: 10.1073/pnas.1109101108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdaguer N, Corbalan-Garcia S, Ochoa WF, Fita I, Gomez-Fernandez JC. Ca(2+) bridges the C2 membrane-binding domain of protein kinase Calpha directly to phosphatidylserine. EMBO J. 1999;18:6329–6338. doi: 10.1093/emboj/18.22.6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood CS, Schmitz KR, Bessman NJ, Setty TG, Ferguson KM, Burd CG. PtdIns4P recognition by Vps74/GOLPH3 links PtdIns 4-kinase signaling to retrograde Golgi trafficking. J. Cell Biol. 2009;187:967–975. doi: 10.1083/jcb.200909063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung T, Gilbert GE, Shi J, Silvius J, Kapus A, Grinstein S. Membrane phosphatidylserine regulates surface charge and protein localization. Science. 2008;319:210–213. doi: 10.1126/science.1152066. [DOI] [PubMed] [Google Scholar]
- Yu JW, Mendrola JM, Audhya A, Singh S, Keleti D, DeWald DB, Murray D, Emr SD, Lemmon MA. Genome-wide analysis of membrane targeting by S. cerevisiae pleckstrin homology domains. Mol. Cell. 2004;13:677–688. doi: 10.1016/s1097-2765(04)00083-8. [DOI] [PubMed] [Google Scholar]
- Zhang G, Kazanietz MG, Blumberg PM, Hurley JH. Crystal structure of the cys2 activator-binding domain of protein kinase C delta in complex with phorbol ester. Cell. 1995;81:917–924. doi: 10.1016/0092-8674(95)90011-x. [DOI] [PubMed] [Google Scholar]
- Zhao C, Du G, Skowronek K, Frohman MA, Bar-Sagi D. Phospholipase D2-generated phosphatidic acid couples EGFR stimulation to Ras activation by Sos. Nature Cell Biol. 2006;9:706–712. doi: 10.1038/ncb1594. [DOI] [PubMed] [Google Scholar]
- Zheng J, Chen RH, Corblan-Garcia S, Cahill SM, Bar-Sagi D, Cowburn D. The solution structure of the pleckstrin homology domain of human SOS1. A possible structural role for the sequential association of diffuse B cell lymphoma and pleckstrin homology domains. J. Biol. Chem. 1997;272:30340–30344. doi: 10.1074/jbc.272.48.30340. [DOI] [PubMed] [Google Scholar]
- Zhu H, Bilgin M, Bangham R, Hall D, Casamayor A, Bertone P, Lan N, Jansen R, Bidlingmaier S, Houfek T, Mitchell T, Miller P, Dean RA, Gerstein M, Snyder M. Global analysis of protein activities using proteome chips. Science. 2001;293:2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]