Abstract
Transcription of genomic loci containing protein-coding genes often yields not only cognate mRNAs but also assorted non-coding RNAs (ncRNAs), which typically map in the vicinity of transcription start sites. A new study demonstrates that far from being random byproducts of gene expression, many long ncRNAs (lncRNAs) are synthesized in a coordinate fashion and control important cellular processes, such as survival in the face of DNA damage.
In 1990 Shirley Tilghman's lab made a puzzling discovery: the very abundant RNA encoded by the H19 gene was transcribed by RNA polymerase II, then spliced and polyadenylated, but unlike canonical messenger RNAs (mRNA) never associated with the translational machinery1. It was later confirmed that the H19 RNA is a fully functional molecule and plays the key role in the imprinting of its own locus2. These studies not only ushered in the long non-coding RNA (lncRNA) era but also set off a prolonged debate whether lncRNAs act locally (in cis) or globally (in trans). There is plenty of evidence in support of various cis modes of action. Up to 70% of protein coding transcripts are thought to be transcribed in both sense and antisense directions3 and the X-chromosome-encoded Xist RNA “coats” and silences its own chromosome 4. However, a new study from Howard Chang's laboratory appearing in this issue of Nature Genetics presents evidence for trans functions for lncRNAs5.
Transcribe locally, act globally
Hung and coauthors used ultrahigh-resolution microarray technology to identify more than 200 lncRNAs that are encoded in close proximity to 56 cell cycle-controlling genes (cyclins, cyclin-dependent kinases (cdks), cdk inhibitors, etc)5. Predictably, during cell cycle progression, self-renewal, and neoplastic transformation, levels of the cell-cycle-related mRNAs fluctuated - but so did levels of lncRNAs encoded in their vicinity.
When these fluctuating lncRNAs were grouped based on expression patterns, colocalized lncRNAs usually ended up in the same clusters, suggesting that adjacent ncRNAs are regulated in concert. In principle, they could act locally, for instance by regulating the nearby mRNA levels. However, the authors found that the expression of lncRNA clusters did not correlate either positively or negatively with expression of the nearest mRNAs. This finding led the authors to reject the idea that most of lncRNAs function in cis and challenged them to identify an alternative mode of action. They focused in particular on a lncRNA that is induced by p53, a master regulator of diverse cellular processes ranging from senescence to apoptosis.
Trans-fixed by p53
An emerging concept in the RNA field is that lncRNAs can function through binding to and altering the activity of transcription factors 6 and the broader the function of the transcription factor, the longer the reach of the interacting lncRNA. For example, the maternally expressed gene-3 (MEG3) was found to increase p53 levels and activity via direct interaction with p53 or indirectly by inhibiting the dedicated ubiquitin ligase MDM2 7,8). Also, a locus encoding the long intergenic non-coding (linc) RNA lincRNA-p21 was discovered 15 kilobases upstream of the CDKN1A gene. Notably, it clearly impinges on the p53 pathway9. Its transcription (along with that of CDKN1A ) is induced after exposure to DNA damaging agents such as doxorubicin9, the effects of which are mostly mediated by p53. Once activated, lincRNA-p21 binds to the heterogenous nuclear ribonucleoprotein K (hnRPN-K) known to interact with repressive transcriptional complexes, and in doing so assists p53 in inhibiting gene expression9. Thus, p53 both regulates and is regulated by the CDKN1A locus. Now it turns out that this feedback loop has another kink.
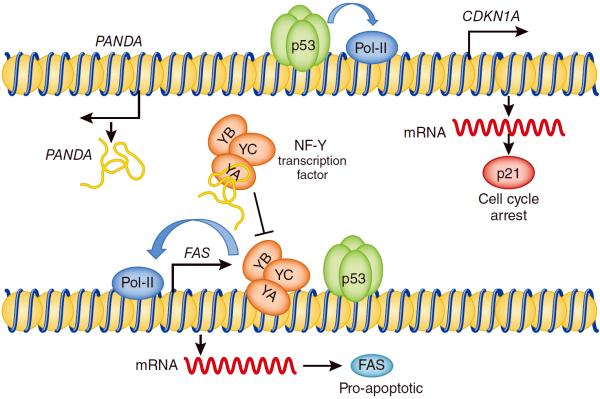
Hung et al show that between the protein-coding CDKN1A gene and lincRNA-p21 there is a gene for yet another lncRNA, which they named PANDA5 (P21 Associated ncRNA DNA damage Activated). PANDA is one of 12 lncRNAs that displayed expression changes in response to p53 activation via DNA damage. Its close proximity to CDKN1A (only 5 kilobases upstream on the antisense strand) suggested that perhaps it could regulate CDKN1A expression. However, upon DNA damage PANDA was induced appreciably earlier than CDKN1A mRNA . Also, knockdown of PANDA had no effect on p21 expression. Rather, knockdown of PANDA selectively enhanced induction of p53-regulated pro-apoptotic genes such as FAS and APAF1. These p53 targets are distinguished from cell cycle-related p53 targets by the presence of binding sites for the transcription factor NF-Y10.
NF-Y connection
NF-Y is a heterotrimeric complex composed of NF-YA, NF-YB and NF-YC, of which NF-YA is an important regulatory subunit11. There is a close and complex relationship between NF-Y and p53. On the one hand, p53 seems to rely on NFY to repress transcription of many of its target genes12. On the other hand, NF-Y has been shown to function as a trans-activator of a subset of p53 targets, such as FAS, that lack a TATA box in their promoter region and require binding of both NF-Y and p5310.
Hung and collaborators hypothesized that PANDA might function through sequestration of NF-YA away from NF-Y/p53 co-regulated promoters. Using RNA chromatography and chromatin immunoprecipitation, they showed that PANDA indeed binds to NF-YA and its knockdown increases the presence of NF-YA at promoter regions of p53-dependent pro-apoptotic target genes 5 (Figure 1). This could lead to increased cell death in response to DNA damage. Consistent with this idea, reducing PANDA levels with siRNA resulted in increased rates of apoptosis5. Whether pro-survival effects of PANDA during DNA damage response are mediated through “eviction” of NF-YA and ensuing repression of FAS remains to be determined. At the very least, it is consistent with the prevailing view that NF-Y transcription factors help orchestrate p53-dependent responses to DNA damage13. Thus, while PANDA might be little more that “junk” RNA for the purpose of p53-dependent cell cycle arrest, it plays a key role in protecting the stressed cell from apoptotic death.
Figure 1.
Model of pro-survival effects of PANDA lncRNA during the DNA damage response. Upon DNA damage, p53 activates expression of target genes such as CDKN1A which encodes the cell cycle regulator p21. p53 also activates expression of PANDA lncRNA encoded upstream of the CDKN1A..PANDA physically interacts with the NF-YA subunit of the NF-Y transcription factor and prevents it from cooperating with p53 on the promoters of p53-dependent pro-apoptotic targets, such as FAS.
Acknowledgments
The authors are supported by the National Institutes of Health grant CA122334 (ATT) and Alex's Lemonade Stand Foundation Innovation (ATT) and Young Investigator (ES) awards.
References
- 1.Brannan CI, Dees EC, Ingram RS, Tilghman SM. The product of the H19 gene may function as an RNA. Mol. Cell Biol. 1990;10:28–36. doi: 10.1128/mcb.10.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pfeifer K, Leighton PA, Tilghman SM. The structural H19 gene is required for transgene imprinting. Proc. Natl. Acad. Sci. U. S. A. 1996;93:13876–13883. doi: 10.1073/pnas.93.24.13876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Katayama S, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 4.Herzing LB, Romer JT, Horn JM, Ashworth A. Xist has properties of the X-chromosome inactivation centre. Nature. 1997;386:272–275. doi: 10.1038/386272a0. [DOI] [PubMed] [Google Scholar]
- 5.Hung T, et al. Extensive and coordinated transcription of noncoding RNAs within cell cycle promotes. Nature Genetics. 2011 doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orom UA, Shiekhattar R. Long non-coding RNAs and enhancers. Curr. Opin. Genet. Dev. 2011;21:194–198. doi: 10.1016/j.gde.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou Y, et al. Activation of p53 by MEG3 non-coding RNA. J Biol. Chem. 2007;282:24731–24742. doi: 10.1074/jbc.M702029200. [DOI] [PubMed] [Google Scholar]
- 8.Benetatos L, Vartholomatos G, Hatzimichael E. MEG3 imprinted gene contribution in tumorigenesis. Int. J Cancer. 2011 doi: 10.1002/ijc.26052. [DOI] [PubMed] [Google Scholar]
- 9.Huarte M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morachis JM, Murawsky CM, Emerson BM. Regulation of the p53 transcriptional response by structurally diverse core promoters. Genes Dev. 2010;24:135–147. doi: 10.1101/gad.1856710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manni I, et al. Posttranslational regulation of NF-YA modulates NF-Y transcriptional activity. Mol. Biol. Cell. 2008;19:5203–5213. doi: 10.1091/mbc.E08-03-0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ceribelli M, Alcalay M, Vigano MA, Mantovani R. Repression of new p53 targets revealed by ChIP on chip experiments. Cell Cycle. 2006;5:1102–1110. doi: 10.4161/cc.5.10.2777. [DOI] [PubMed] [Google Scholar]
- 13.Benatti P, et al. A balance between NF-Y and p53 governs the pro- and anti-apoptotic transcriptional response. Nucleic Acids Res. 2008;36:1415–1428. doi: 10.1093/nar/gkm1046. [DOI] [PMC free article] [PubMed] [Google Scholar]