Abstract
Most cancers are characterized by multiple molecular alterations, but identification of the key proteins involved in these signaling pathways is currently beyond reach. We show that the inhibitor PU-H71 preferentially targets tumor-enriched Hsp90 complexes and affinity captures Hsp90-dependent oncogenic client proteins. We have used PU-H71 affinity capture to design a proteomic approach that, when combined with bioinformatic pathway analysis, identifies dysregulated signaling networks and key oncoproteins in chronic myeloid leukemia. The identified interactome overlaps with the well-characterized altered proteome in this cancer, indicating that this method can provide global insights into the biology of individual tumors, including primary patient specimens. In addition, we show that this approach can be used to identify previously uncharacterized oncoproteins and mechanisms, potentially leading to new targeted therapies. We further show that the abundance of the PU-H71-enriched Hsp90 species, which is not dictated by Hsp90 expression alone, is predictive of the cell’s sensitivity to Hsp90 inhibition.
Most cancers arise from multiple molecular lesions, and functional redundancy of affected pathways limits the utility of specific molecularly targeted drugs. A better understanding of the molecular aberrations that maintain the malignant phenotype of cancer cells would enable more efficient targeting of tumor-promoting molecules and aid the development of more effective and less toxic anticancer treatments.
Application of genomics technologies, including large-scale genome sequencing, has led to the identification of many gene mutations in various cancers, emphasizing the complexity of this disease1,2. However, such genetic analyses intrinsically lack the ability to elucidate the functional complexity of signaling networks that are aberrantly activated as a consequence of the identified genetic defect(s). Thus, the development of complementary proteomic methodologies to identify molecular lesions intrinsic to tumors in a patient- and disease stage–specific manner must follow.
Most proteomic strategies are limited to measuring protein expression in a particular tumor but are unable to provide information on the functional importance of such findings3. Some functional information can be obtained using antibodies directed at specific proteins or post-translational modifications, and by activity-based protein profiling using small molecules targeting the active site of certain enzymes4–7. Although such methods allow one to query a specific pathway or post-translational modification, they are not well suited to capture more global information regarding the malignant state3.
To maintain homeostasis, cells use intricate molecular machineries comprising thousands of proteins that are programmed to execute well-defined functions. Dysregulation of these pathways, through protein misexpression or mutation, provides biological advantages that confer the malignant phenotype. At the molecular level, this requires cells to invest energy in maintaining the stability and function of these proteins, and for this reason cancer cells co-opt molecular chaperones, including Hsp90 (refs. 8,9).
Hsp90 has important roles in maintaining the transformed phenotype8,9. Hsp90 and its associated cochaperones assist in the correct folding of cellular proteins, collectively referred to as ‘client proteins’, many of which are effectors of signal transduction pathways controlling cell growth, differentiation, the DNA-damage response and cell survival. Tumor cell addiction to these proteins (that is, through mutations, aberrant expression, improper cellular translocation and so on) thus makes them critically reliant on Hsp90 (ref. 9).
Although Hsp90 is expressed in all cells and tissues, tumors preferentially contain Hsp90 in a higher-order multi-chaperone complex with high affinity for certain Hsp90 inhibitors, whereas normal tissues harbor a latent, uncomplexed Hsp90 that has low affinity for these inhibitors10.
Based on these data, we hypothesize that small molecules able to target tumor-enriched Hsp90 complexes can be used to affinity capture Hsp90-dependent oncogenic client proteins. When combined with bioinformatic analysis, this should enable the creation of a detailed molecular map of transformation-specific lesions that can guide the development of combination therapies that are optimally effective for a specific patient.
Here we describe an Hsp90 inhibitor-based chemical biology–proteomics–bioinformatics approach to discover oncogenic proteins and pathways in chronic myeloid leukemia (CML). We show that the method provides a global overview of the Hsp90 interactome in malignant cells, and that this interactome represents a substantial fraction of the functional malignant proteome8,9.
Results
Heterogeneous Hsp90 presentation in cancer cells
To investigate the interaction of small-molecule Hsp90 inhibitors with tumor Hsp90 complexes, we used agarose beads that were covalently attached to either geldanamycin or PU-H71 (referred to as GM and PU beads, respectively). Both geldanamycin and PU-H71, which are chemically distinct agents, interact with and inhibit Hsp90 by binding to a regulatory pocket in its N-terminal domain11. For comparison, we generated agarose beads coupled to an anti-Hsp90 antibody (H9010).
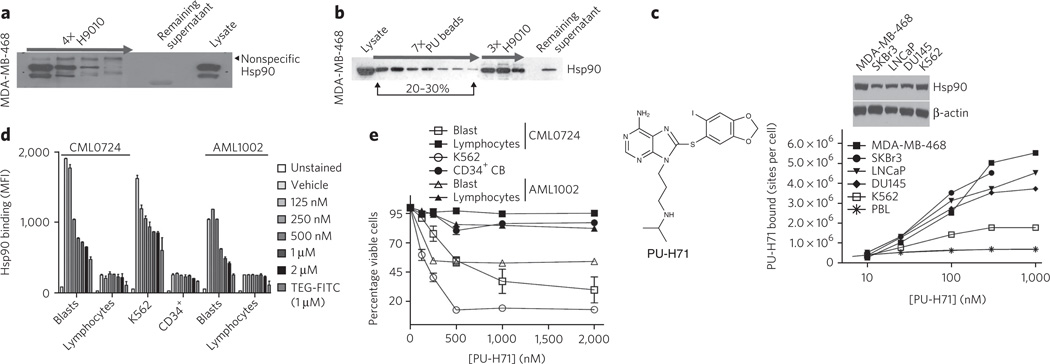
First, we evaluated the binding of these agents to Hsp90 in lysates from breast cancer cells and CML cells. Four consecutive immunoprecipitation steps with H9010, but not with a nonspecific IgG, quantitatively depleted Hsp90 from these extracts (Fig. 1a). In contrast, sequential pull-downs with PU or GM beads removed only a limited fraction of total cellular Hsp90 (Fig. 1b and Supplementary Results, Supplementary Fig. 1a,b). Specifically, in MDA-MB-468 breast cancer cells, the combined PU bead fractions represented ~20–30% of the total Hsp90 pool, and further addition of fresh PU bead aliquots failed to precipitate the Hsp90 remaining in the lysate (Fig. 1b, PU beads). This PU-H71-depleted, remaining Hsp90 fraction, although inaccessible to the small molecule, maintained affinity for H9010 (Fig. 1b, H9010). From this we conclude that a substantial fraction of Hsp90 in the MDA-MB-468 cell extracts was still in a native conformation but not reactive with PU-H71.
Figure 1. PU-H71 interacts with a restricted fraction of Hsp90 that is more abundant in cancer cells.
(a) Sequential immunopurification steps, as indicated by the arrow, with H9010 (a Hsp90-specific antibody) deplete Hsp90 in the MDA-MB-468 cell extract. Lysate, control cell extract. (b) Hsp90 from MDA-MB-468 extracts was isolated through sequential chemical-purification and immunopurification steps. (c) Saturation studies were performed with 131I-PU-H71 in the indicated cells (below). Expression of Hsp90 in the indicated cells was analyzed by western blotting (above). Representative data of four separate repeats are presented. (d) Binding of PU-FITC, presented as mean fluorescence intensity (MFI), to primary AML and CML, CD34+ cord blood cells (CB), or K562 cells pretreated with the indicated doses of PU-H71 for 24 h. TEG-FITC is a nonspecific binding control. (e) Percent viability relative to untreated control for the indicated cells after treatment for 96 h with the indicated doses of PU-H71. Data are presented as means ± s.e.m. (n = 3).
To exclude the possibility that changes in Hsp90’s configuration in cell lysates make it unavailable for binding to immobilized PU-H71 but not to the antibody, we analyzed binding of radiolabeled 131I-PU-H71 to Hsp90 in intact cancer cells (Fig. 1c). Binding of 131I-PU-H71 to Hsp90 in several cancer cell lines saturated at a well-defined, although distinct, number of sites per cell (Fig. 1c).
We quantified the fraction of cellular Hsp90 bound by PU-H71 in MDA-MB-468 cells. First, we determined that Hsp90 represented 2.66–3.33% of the total protein in these cells, a value in close agreement with the reported abundance of Hsp90 in other tumor cells9. Approximately 41.65 × 106 MDA-MB-468 cells were lysed to yield 3,875 µg of protein, of which between 103.07 µg and 129.04 µg was Hsp90. One cell, therefore, contained (2.47–3.09) × 10−6 µg, (2.74–3.43) × 10−11 µmols or (1.64–2.06) × 107 molecules of Hsp90. In MDA-MB-468 cells, 131I-PU-H71 bound to, at most, 5.5 × 106 of the available cellular binding sites (Fig. 1c), which amounts to 26.6–33.5% of the total Hsp90 (calculated as 5.5 × 106/(1.64–2.06) × 107*100). This value is markedly similar to that obtained with PU bead pull-downs in cell extracts (Fig. 1b), confirming that PU-H71 binds to a fraction of Hsp90 in MDA-MB-468 cells that represents approximately 30% of the total Hsp90 pool, and validating the use of PU beads to efficiently isolate this pool. In K562 and other established t(9;22)+ CML cell lines, PU-H71 bound 10.3–23% of the total cellular Hsp90 (Fig. 1c and Supplementary Fig. 1b,c).
Next, we extended our studies to several primary leukemia cells and to normal blood cells. Among these were primary chronic- and blast-phase CML and acute myeloid leukemia (AML) samples that contained both blasts (malignant cell population) and lymphocytes (normal cell population), CD34+ cells isolated from the cord blood of healthy donors, total mononuclear cells from peripheral blood and also peripheral blood leukocytes (PBLs) (Fig. 1c–e and Supplementary Figs. 1d and 2a–h). We used a fluorescein-labeled PU-H71 (PU-FITC) and flow cytometric analysis to determine PU-H71 binding to distinct cell populations.
PU-H71 efficiently bound to Hsp90 in K562 cells and in CML and AML blasts with a half-maximal inhibitory concentration (IC50) of 116 nM, 201 nM and 425 nM, respectively (Fig. 1d). In contrast, its affinity for Hsp90 in normal blood cells was weaker, with IC50 values higher than 2,000 nM (Fig. 1d and Supplementary Fig. 1d). Hsp90 is highly expressed in these normal blood cells, as indicated by substantial binding of the Hsp90 antibody (Supplementary Fig. 1d).
Cells with the highest avidity for PU-H71 were also the most sensitive to killing by this agent (Fig. 1e and Supplementary Figs. 1e and 2). When evaluated in a panel of CML and AML cell lines and primary samples, we noted a good correlation between PU-H71 binding to Hsp90 and its cell-killing potential (Supplementary Figs. 1e and 2).
Collectively, these data confirm that certain Hsp90 inhibitors bind preferentially to a subset of Hsp90 species that is more abundant in cancer cells than in normal cells (Supplementary Fig. 3a). Abundance of this species is not dictated solely by the amount of Hsp90 expression and is predictive of cellular sensitivity to Hsp90 inhibition.
PU-H71 selects for oncoprotein–Hsp90 complex species
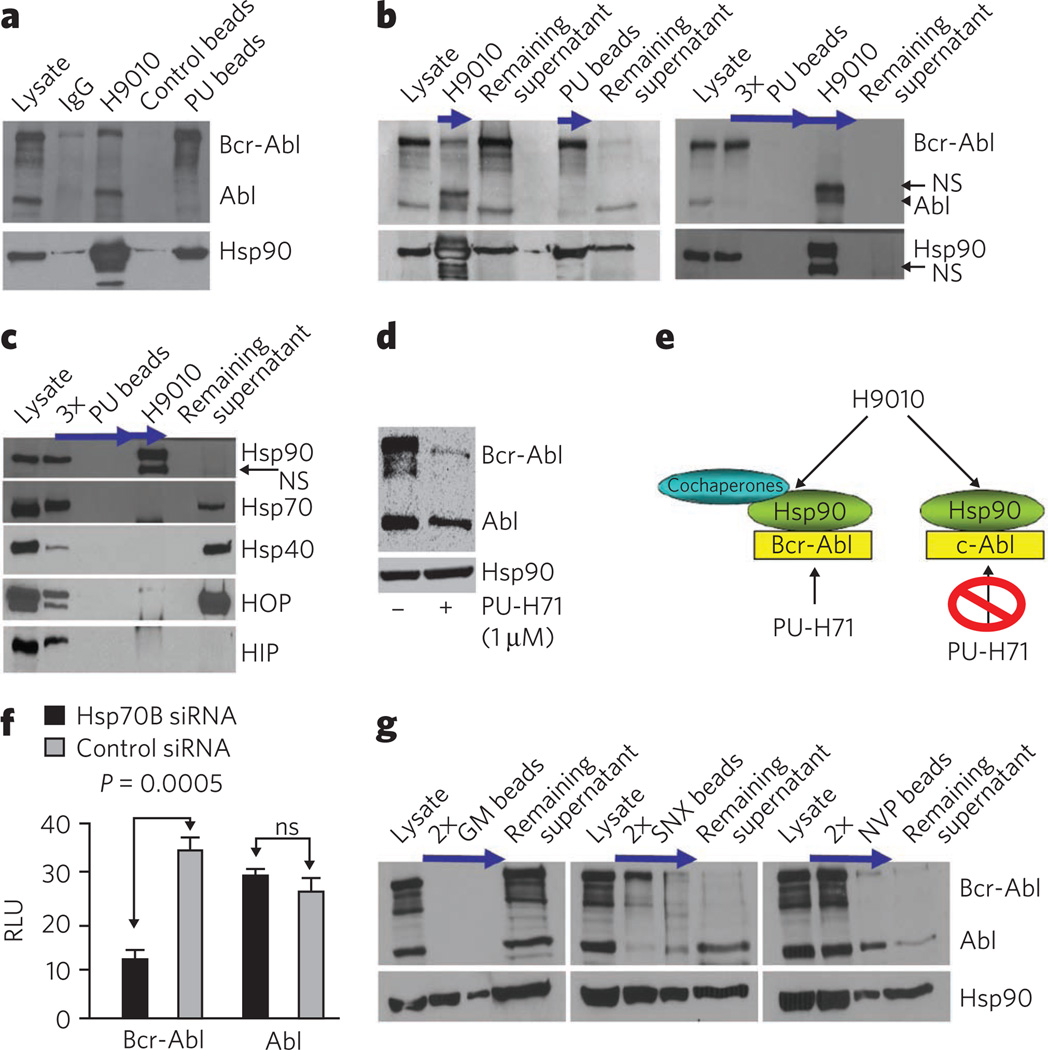
Next we performed immunoprecipitations and chemical precipitations with beads coated with H9010-specific antibody and those carrying PU, and analyzed the associated client cargo in each case. We first investigated K562 CML cells because this cell line coexpresses the aberrant Bcr-Abl protein, a constitutively active kinase, and its normal counterpart c-Abl. These two Abl species are clearly separable by molecular weight and are therefore easily distinguishable by western blot (Fig. 2a, lysate). We observed that H9010, but not a nonspecific IgG, isolated Hsp90 complexed with both Bcr-Abl and Abl (Fig. 2a and Supplementary Fig. 3b,c, H9010). Comparison of immunoprecipitated Bcr-Abl and Abl (Fig. 2a,b, H9010) with the fraction of each protein remaining in the supernatant (Fig. 2b, remaining supernatant), indicated that H9010 did not preferentially enrich for Hsp90 bound to either mutant or wild-type Abl in K562 cells.
Figure 2. PU-H71 is selective for and isolates Hsp90 in complex with oncoproteins and cochaperones.
(a) Representative western blot of Hsp90 complexes in K562 extracts isolated by precipitation with H9010, a nonspecific IgG, PU beads or control beads containing 2-methoxyethylamine, an Hsp90-inert molecule. (b,c) Single or sequential immunoprecipitations and chemical precipitations, as indicated by arrows, conducted in K562 extracts with H9010 and PU beads. NS, nonspecific. (d) Representative western blot of K562 cells treated for 24 h with vehicle (−) or PU-H71 (+). (e) Proposed Hsp90 species in K562 cells, in complex with both aberrant, Bcr-Abl, and normal, c-Abl, proteins. PU-H71, but not H9010, selects for the Hsp90 population that is Bcr-Abl oncoprotein bound. (f) Expression of proteins in Hsp70–knocked down K562 cells. Changes in protein concentrations are presented in relative luminescence units (RLU). Control, scrambled small interfering RNA (siRNA). Data are presented as means ± s.e.m. (n = 3). ns, nonsignificant. (g) Representative western blot of sequential chemical precipitations, as indicated by arrows, conducted in K562 extracts with GM, SNX and NVP beads.
In contrast, PU-H71-bound Hsp90 preferentially isolated Bcr-Abl protein (Fig. 2a,b, PU beads). Following depletion of the Hsp90–Bcr-Abl species by PU beads (Fig. 2b,c, PU beads), H9010 precipitated the remaining Hsp90–Abl species (Fig. 2b,c, H9010). PU beads retained selectivity for Hsp90–Bcr-Abl at substantially saturating conditions (that is, excess of lysate and beads; Supplementary Fig. 4a). As further confirmation of the biochemical selectivity of PU-H71 for the Bcr-Abl–Hsp90 complex, Bcr-Abl was more susceptible to degradation by PU-H71 than was Abl (Fig. 2d). We confirmed the selectivity of PU-H71 for the aberrant Abl in other established t(9;22)+ CML cell lines (Supplementary Fig. 4b) and in primary CML samples (Supplementary Fig. 4c).
PU-H71 selects for Hsp90 bound to cochaperones
To further differentiate between the PU-H71-isolated and antibody-isolated Hsp90 fractions, we evaluated the cochaperone constituency of both species8. The fraction of Hsp90 bound to Bcr-Abl also associated with several cochaperones, including Hsp70, Hsp40, HOP and HIP (Fig. 2c, PU beads), as well as additional cochaperones (Supplementary Data Set 1a–d). These findings strongly suggest that PU-H71 recognizes cochaperone-associated Hsp90. After depletion by PU beads, the Hsp90 pool detected by H9010, shown to include Hsp90–Abl complexes, did not contain cochaperones (Fig. 2c, H9010), although their abundant expression was detected in the lysate (Fig. 2c, remaining supernatant). Cochaperones were isolated by H9010 in total cell extract (Supplementary Fig. 3b,c).
These findings suggest the existence of distinct pools of Hsp90 that are preferentially bound to either Bcr-Abl or Abl in CML cells (Fig. 2e). H9010 recognizes both pools, whereas PU-H71 selects for the Bcr-Abl–Hsp90 species. Our data also suggest that Hsp90 may use and require more acutely the classical cochaperones Hsp70, Hsp40 and HOP when it modulates the activity of aberrant (that is, Bcr-Abl) but not normal (that is, Abl) proteins (Supplementary Fig. 3a). In accord with this hypothesis, we find that Bcr-Abl is more sensitive than Abl to knockdown of Hsp70 in K562 cells (Fig. 2f and Supplementary Fig. 3d).
Not all Hsp90 inhibitors trap Hsp90–oncoprotein complexes
We next evaluated whether other inhibitors that interact with the N-terminal ATP pocket of Hsp90, including the synthetic inhibitors SNX-2112 and NVP-AUY922 and the natural product geldanamycin11, could selectively isolate similar Hsp90 species (Fig. 2g). SNX beads showed selectivity for Bcr-Abl–Hsp90, whereas NVP beads behaved similarly to H9010 and did not discriminate between Bcr-Abl–Hsp90 and Abl–Hsp90 species (see SNX versus NVP beads, respectively; Fig. 2g). Although GM beads also recognized a subpopulation of Hsp90 in cell lysates (Supplementary Fig. 1a), they were much less efficient than were PU beads in co-precipitating Bcr-Abl (Fig. 2g, GM beads). The ineffectiveness of GM in trapping Hsp90–client protein complexes was previously reported12.
PU-H71 identifies additional Hsp90 oncoclient proteins
To determine whether selectivity towards oncoprotein–Hsp90 complexes was a general property of PU-H71, we tested several additional well-defined Hsp90 client proteins in other tumor cell lines13,14 (Supplementary Fig. 4d–f). In agreement with our results in K562 cells, H9010 precipitated Hsp90 bound to both mutant B-Raf expressed in SKMel28 melanoma cells and to wild-type B-Raf expressed in CCD18Co normal colon fibroblasts (Supplementary Fig. 4e). PU and GM beads, however, selectively recognized Hsp90 bound to mutant B-Raf, showing little recognition of Hsp90 bound to wild-type B-Raf (Supplementary Fig. 4e). Still, GM beads were measurably less efficient than PU beads in co-precipitating the mutant client protein. We obtained similar results for other Hsp90 clients12 (Supplementary Fig. 4d–f).
PU beads identify the aberrant signalosome in CML
The data presented above suggest that PU-H71, which specifically interacts with Hsp90 (ref. 15; Supplementary Fig. 5), preferentially selects for oncoprotein–Hsp90 species and traps Hsp90 in a client-binding conformation (Fig. 2). Therefore, we examined whether PU beads could be used as a tool to investigate the cellular complement of oncogenic Hsp90 client proteins. Because the aberrant Hsp90 clientele is hypothesized to comprise proteins most crucial for the maintenance of the tumor phenotype8,9,16, this approach could potentially identify critical signaling pathways in a tumor-specific manner. To test this hypothesis, we performed an unbiased analysis of the protein cargo isolated by PU beads in K562 cells, in which at least some of the key functional lesions are known17,18.
We subjected the protein cargo isolated from cell lysate using PU beads or control beads to proteomic analysis by nano–liquid chromatography coupled to tandem mass spectrometry (nano-LC-MS/MS). We carried out initial protein identification using the Mascot search engine, and further evaluated our results using Scaffold Proteome Software (Supplementary Data Set 1a–f). Among the PU bead–interacting proteins was Bcr-Abl (see Bcr and Abl1, Supplementary Data Set 1a), confirming our previous data (Fig. 2).
We then used Ingenuity Pathway Analysis (IPA) to build biological networks from the identified proteins (Fig. 3a, Supplementary Fig. 6a–g and Supplementary Data Set 1e,f). IPA assigned PU-H71-isolated proteins to 13 networks associated with cell death, cell cycle, cell growth and proliferation. These networks overlap well with known canonical CML signaling pathways (Supplementary Fig. 6a).
Figure 3. PU-H71 identifies the aberrant signalosome in CML cells.
(a) Pathway diagram highlighting the PU bead–identified CML signalosome with focus on networks 1 (Raf-MAPK and PI3K-AKT-mTO R pathway), 2 (NF-κB pathway) and 8 (STAT5 pathway). Key nodal proteins in the identified networks are shown in yellow. A detailed list of identified protein networks and component proteins is shown in Supplementary Figure 6 and Supplementary Data Set 1f. (b) Left, representative western blot of a subset of MS-identified protein complexes. No proteins were detected in the control bead pull-downs, and those data are omitted for simplicity of presentation. Right, representative western blot of K562 cells treated for 24 h with vehicle (−) or PU-H71 (+). (c) Representative western blot of single chemical precipitations conducted in primary CML cell extracts with PU and control beads.
In addition to signaling proteins, we identified proteins that regulate carbohydrate and lipid metabolism, protein synthesis, gene expression, and cellular assembly and organization. These findings are in accord with the postulated broad roles of Hsp90 in maintaining cellular homeostasis and in being an important mediator of cell transformation8,9,16,19.
Following identification by MS, we further validated a number of key proteins by chemical precipitation and western blotting, in both K562 cells and in primary CML blasts (Fig. 3b,c and Supplementary Fig. 4b,c). We also queried the effect of PU-H71 on the steady-state concentrations of these proteins to further support their Hsp90-regulated expression and stability8 (Fig. 3b).
The top-scoring networks enriched on the PU beads were those used by Bcr-Abl to propagate aberrant signaling in CML: the PI3K-AKT-mTOR-, Raf-MAPK- and NFκB-mediated signaling pathways (network 1, 22 focus molecules, score = 38, and network 2, 22 focus molecules, score = 36; Supplementary Data Set 1f). We created connectivity maps for these networks to investigate the relationship between component proteins (Supplementary Fig. 6b,c). These maps were simplified for clarity, retaining only major pathway components and relationships (Fig. 3a).
The PI3K-AKT-mTOR pathway
Activation of the PI3K-AKT-mTOR pathway has emerged as an essential signaling mechanism in Bcr-Abl leukemogenesis17. Of particular interest in this pathway is the mammalian target of rapamycin (mTOR), which is constitutively activated in Bcr-Abl-transformed cells. A recent study reported that both mTORC1 and mTORC2 are activated in Bcr-Abl cells20. We identified both mTOR and key activators of mTOR, such as RICTOR, RAPTOR, Sin1 (MAPKAP1) and the class 3 PI3Ks PIK3C3 (hVps34) and PIK3R4 (VPS15)21, in the PU-H71–Hsp90 pull-downs (Fig. 3b,c, Supplementary Figs. 4c and 6b, and Supplementary Data Set 1a,d).
The NF-κB pathway
Activation of nuclear factor-κB (NF-κB) is required for Bcr-Abl transformation of primary bone marrow cells and for Bcr-Abl-transformed hematopoietic cells to form tumors in nude mice22. PU-H71-isolated proteins enriched in this pathway include NF-κB as well as NF-κB activators, including IKBKAP, TANK-binding kinase 1 (TBK-1) and TAK1-binding protein 1 (TAB1)23 (Supplementary Data Set 1a,d). Recently, Bcr-Abl-induced activation of the NF-κB cascade in myeloid leukemia cells was shown to be largely mediated by tyrosine-phosphorylated PKD2 (or PRKD2)24 which we identify here to be a PU-H71–Hsp90 interactor (Fig. 3b,c, Supplementary Figs. 4c and 6c, and Supplementary Data Set 1a,d).
The Raf–MAPK pathway
Key effectors of the MAPK pathway, also activated in CML17,22, including Raf-1, A-Raf, ERK, p90RSK, Vav and several MAPKs, are included in the PU-H71–Hsp90-bound pool (Fig. 3b,c, Supplementary Figs. 4c and 6b, and Supplementary Data Set 1a,d). IPA connects the MAPK pathway to key elements of many signal transduction pathways including the PI3K-AKT-mTOR, STAT and focal adhesion pathways (Fig. 3a and Supplementary Fig. 6b–e).
The STAT pathway
The STAT pathway is also activated in CML and confers cytokine independence and protection against apoptosis22, and was enriched by PU-H71 chemical precipitation (network 8, 20 focus molecules, score = 14; Supplementary Fig. 6d and Supplementary Data Set 1f). Both STAT5 and STAT3 were associated with PU-H71–Hsp90 complexes (Fig. 3b,c and Supplementary Data Set 1a,d). In CML, STAT5 activation by phosphorylation is driven by Bcr-Abl17. Bruton agammaglobulinemia tyrosine kinase (BTK), constitutively phosphorylated and activated by Bcr-Abl in pre–B lymphoblastic leukemia cells25, can also signal through STAT5 (ref. 26). BTK is another Hsp90-regulated protein that we identified in CML (Fig. 3b,c, Supplementary Fig. 4c and Supplementary Data Set 1a,d). In addition to phosphorylation, STATs can be activated in myeloid cells by calpain (CAPN1)-mediated proteolytic cleavage, leading to truncated STAT species27. CAPN1 is also found in PU-H71-bound Hsp90 pull-downs, as is activated Ca2+–calmodulin-dependent protein kinase IIγ (CaMKIIγ) activated in CML by Bcr-Abl28 (Supplementary Data Set 1a,d).
The focal adhesion pathway
The focal adhesion pathway was well represented in PU-H71 pull-downs (network 12, 16 focus molecules, score = 13; Supplementary Fig. 6e and Supplementary Data Set 1f). The focal adhesion–associated proteins paxillin, FAK, vinculin, talin and tensin are constitutively phosphorylated in Bcr-Abl-transfected cell lines29,30, and these too were isolated in PU-H71–Hsp90 complexes (Fig. 3a).
Other pathways
Other important transforming pathways in CML, driven by MYC31 (network 7, 15 focus molecules, score = 22) and TGF-β32 (network 10, 13 focus molecules, score = 18) were identified here as well (Supplementary Fig. 6a,f,g and Supplementary Data Set 1f). Among the identified networks were also those important for disease progression and aberrant cell cycle and proliferation of CML (networks 3–6, 9, 11 and 13; Supplementary Fig. 6a and Supplementary Data Set 1f).
In summary, PU-H71 pull-down enriched for a broad cross-section of proteins that participate in signaling pathways vital to the malignant phenotype of CML. Interaction of PU-H71-bound Hsp90 with the aberrant CML signalosome was retained in primary CML samples (Fig. 3c and Supplementary Fig. 4c).
PU-H71 cargo contributes to the malignant phenotype
To demonstrate that the networks identified by PU beads are important for transformation in K562, we next showed that inhibitors of key nodal proteins from individual networks (Fig. 3a, yellow boxes—Bcr-Abl, NFκB, mTOR, MEK and CAMKII) diminish the growth and proliferation potential of K562 cells (Table 1 and Supplementary Fig. 7a).
Table 1.
IC50 values for the indicated compounds in cell proliferation assays
| Drug or cell line | PU-H71 | Gleevec | PP242 | AS703026 | BMS-345541 | KN-93 |
|---|---|---|---|---|---|---|
| K562 | 0.13 ± 0.02 | 0.33 ± 0.07 | 2.00 ± 0.10 | 1.07 ± 0.10 | 8.60 ± 0.30 | 5.75 ± 0.60 |
| Mia-PaCa-2 | 0.15 ± 0.01 | >10.00 | 0.70 ± 0.14 | NA | NA | NA |
Cells were treated for 72 h with the indicated inhibitors, and the effect on cell growth was analyzed. The half-maximal inhibitory concentration (IC50) is indicated in µM. Data are presented as means ± s.d. (n = 3). NA, not available.
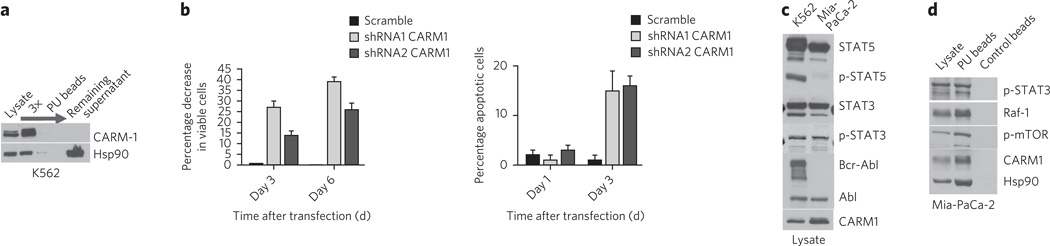
Next we showed that PU beads identified a set of Hsp90 interactors with no known role in CML, but which also contribute to the transformed phenotype. The histone-arginine methyltransferase CARM1, a transcriptional coactivator of many genes33, was validated in the PU bead pull-downs from CML cell lines and primary CML cells (Fig. 3b,c and Supplementary Fig. 4b,c). This is the first reported link between Hsp90 and CARM1, although other arginine methyltransferases, such as PRMT5, have been shown to be Hsp90 clients in ovarian cancer cells34. Although elevated amounts of CARM1 are implicated in the development of prostate and breast cancers, little is known about the importance of CARM1 in CML leukomogenesis33. CARM1 was essentially entirely captured by the Hsp90 species recognized by PU beads (Fig. 4a), and it was also sensitive to degradation by PU-H71 (Fig. 3b). Therefore, CARM1 may be a previously unreported Hsp90 oncoprotein in CML. Indeed, knockdown experiments with CARM1 but not control short hairpin RNAs (shRNAs), showed reduced viability and enhanced apoptosis in K562 (Fig. 4b).
Figure 4. PU-H71-identified proteins and networks are those important for the malignant phenotype.
(a) Representative western blot of sequential chemical precipitations, as indicated by the arrow, conducted in K562 extracts with the PU beads. (b) The effect of CARM1 knockdown on cell viability using tryptan blue (left) or acridine orange-ethidium bromide (right) staining was evaluated in K562 cells. Data are presented as means ± s.e.m. (n = 3). (c) The expression of select potential Hsp90-interacting proteins was analyzed by western blotting in K562 leukemia and Mia-PaCa-2 pancreatic cancer cells. (d) Representative western blot of single chemical precipitations conducted in Mia-PaCa-2 cell extracts with PU and control beads.
To demonstrate that the presence of proteins in PU-H71 pull-downs is due to their participation in aberrantly activated signaling and not merely their abundant expression, we compared PU bead pull-downs from K562 and Mia-PaCa-2, a pancreatic cancer cell line (Supplementary Data Set 1a). Although both cell lines express large amounts of STAT5 protein (Fig. 4c), we noted activation of the STAT5 pathway, as demonstrated by STAT5 phosphorylation (Fig. 4c) and DNA binding35, in only the K562 cells. In accordance, this protein was identified only in the K562 PU bead pull-downs (Supplementary Data Set 1a and Supplementary Fig. 7b). In contrast, we identified activated STAT3 in PU-H71–Hsp90 complexes from both K562 (Fig. 3b) and Mia-PaCa-2 cell extracts (Fig. 4d).
We identified the mTOR pathway using PU beads in both K562 and Mia-PaCa-2 cells (Supplementary Fig. 7b) and, indeed, its pharmacological inhibition by PP242, a selective inhibitor that targets the ATP domain of mTOR36, was toxic to both cells. In contrast, the Abl inhibitor Gleevec37 was toxic to only K562 cells (Table 1 and Supplementary Fig. 7a,c). Both cell lines express Abl but only K562 has the oncogenic Bcr-Abl (Fig. 4c), and PU beads identified Abl, as Bcr-Abl, in K562 but not in Mia-PaCa-2 cells (Supplementary Fig. 7b).
PU-H71 identifies a new STAT activation mechanism
PU bead pull-downs contain several proteins, including Bcr-Abl17, CAMKIIγ28, FAK29, vav-1 (ref. 38) and PRKD2 (ref. 24), that are constitutively activated in CML. These are classical Hsp90-regulated clients that depend on Hsp90 for their stability because their steady-state concentrations decrease upon Hsp90 inhibition8,9 (Fig. 3b). Constitutive activation of STAT3 and STAT5 is also reported in CML17,22. These proteins, however, do not fit the criteria of classical Hsp90 client proteins, because STAT5 and STAT3 concentrations remain essentially unchanged upon Hsp90 inhibition (Fig. 3b). The PU-H71 pull-downs also contained proteins that may constitute active signaling megacomplexes, such as mTOR, VPS32, VPS15 and RAPTOR20. Furthermore, PU-H71–Hsp90 complexes contained adapter proteins such as GRB2, DOCK, CRKL and EPS15, which link Bcr-Abl to key effectors of multiple aberrantly activated signaling pathways in K562 (refs. 6,17; Fig. 3a). Their expression also remained unchanged upon Hsp90 inhibition (Fig. 3b). We therefore wondered whether the contribution of Hsp90 to certain oncogenic pathways extends beyond its classical folding and stabilizing activity. Specifically, we hypothesized that Hsp90 might also act as a scaffolding molecule to maintain signaling complexes in an active configuration, as has been previously postulated16,39.
Hsp90 binds to and influences the conformation of STAT5
To investigate this hypothesis further we focused on STAT5, which is constitutively phosphorylated in CML40. The overall concentration of p-STAT5 is determined by the balance of phosphorylation and dephosphorylation events. The high concentrations of p-STAT5 in K562 cells may reflect either an increase in upstream kinase activity or a decrease in protein tyrosine phosphatase (PTPase) activity. A direct interaction between Hsp90 and p-STAT5 could also modulate the amount of p-STAT5 in cells.
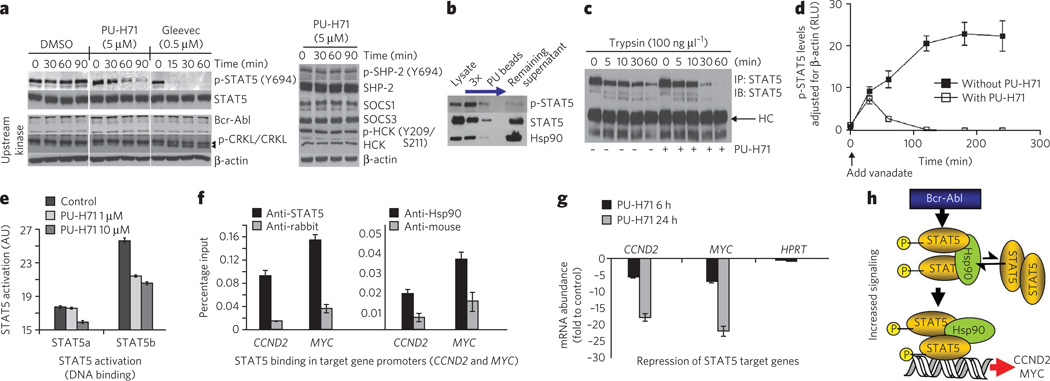
To dissect the relative contributions of these potential mechanisms, we first investigated the effect of PU-H71 on the main kinases and PTPases that regulate STAT5 phosphorylation in K562 cells. Bcr-Abl directly activates STAT5 without the need for JAK phosphorylation40. Concordantly, STAT5 phosphorylation rapidly decreased in the presence of the Bcr-Abl inhibitor Gleevec (Fig. 5a, Gleevec). While Hsp90 regulates Bcr-Abl stability, the reduction in steady-state Bcr-Abl concentrations following Hsp90 inhibition requires more than 3 h41. Indeed, we observed no change in Bcr-Abl expression (Fig. 5a, PU-H71, Bcr-Abl) or function, as evidenced by no decrease in CRKL phosphorylation (Fig. 5a, PU-H71, p-CRKL and CRKL), with PU-H71 in the time interval during which p-STAT5 concentrations were reduced (Fig. 5a, PU-H71, p-STAT5). Also, we saw no change in the activity and expression of HCK, a kinase activator of STAT5 in 32Dcl3 cells transfected with Bcr-Abl42 (Fig. 5a, HCK and p-HCK). We also excluded the possibility of a change in PTPase activity, because the expression and activity of SHP2, the major cytosolic STAT5 phosphatase43, was not altered in this time interval (Fig. 5a, SHP2 and p-SHP2), nor were the concentrations of SOCS1 and SOCS3, which form a negative-feedback loop that switches off STAT signaling37 (Fig. 5a, SOCS1 and SOCS3). Thus, no effect on STAT5 phosphorylation in the 0–90-min interval can be attributed to a change in kinase or phosphatase activity upon Hsp90 inhibition.
Figure 5. Hsp90 facilitates an enhanced STAT5 activity in CML.
(a) Representative western blot of K562 cells treated for the indicated times with PU-H71 (5 µM), Gleevec (0.5 µM) or DMSO (vehicle). (b) Representative western blot of sequential chemical precipitations conducted in K562 cells with PU and control beads, as indicated by the blue arrow. (c) Representative western blot of STAT5 immunocomplexes from cells pretreated with vehicle or PU-H71, and then treated for the indicated times with trypsin. (d) p-STAT5 concentrations in K562 cells treated for the indicated times with vanadate (1 mM) in the presence and absence of PU-H71 (5 µM). Data are presented as mean ± s.d. (n = 3). (e) The DNA-binding capacity of STAT5 in K562 cells treated for 24 h with the indicated concentrations of PU-H71. (f) Quantitative ChIP performed with STAT5 or Hsp90 antibodies versus an IgG control for two known STAT5 target genes. A primer that amplifies an intergenic region was used as negative control. Results are expressed as a percentage of the input for the specific antibody (STAT5 or Hsp90) over the respective IgG control. (g) The transcript abundance of CCND2 and MYC in K562 cells exposed to µM of PU-H71. Results are expressed as fold change compared to baseline (time 0 h) and were normalized to RPL13A. HPRT was used as negative control. Data are presented as means ± s.e.m. (h) Proposed mechanism for Hsp90-facilitated increased STAT5 signaling in CML. Hsp90 binds to and influences the conformation of STAT5 and maintains STAT5 in an active conformation directly within STAT5-containing transcriptional complexes.
Because most p-STAT5, but not STAT5, is bound to Hsp90 in CML cells (Fig. 5b), we hypothesized that the cellular concentrations of activated STAT5 are fine-tuned by direct binding to Hsp90. The activation and inactivation cycle of STATs entails their transition between different dimer conformations44. We found that STAT5 is more susceptible to trypsin cleavage when bound to Hsp90 (Fig. 5c), indicating that binding of Hsp90 directly modulates the conformational state of STAT5, potentially keeping it in a conformation unfavorable for dephosphorylation and/or favorable for phosphorylation. To investigate this possibility, we used a pulse-chase strategy in which orthovanadate (Na3VO4), a nonspecific PTPase inhibitor, was added to cells to block dephosphorylation of STAT5. We then determined the residual concentration of p-STAT5 at several later time points (Fig. 5d and Supplementary Fig. 7d). In the absence of PU-H71, p-STAT5 accumulated rapidly, whereas in its presence, cellular p-STAT5 concentrations were diminished. The kinetics of this process (Fig. 5d) were similar to the rate of p-STAT5 steady-state reduction (Fig. 5a, PU-H71).
Hsp90 supports STAT5-mediated transcription
The biological activity of STAT5 also requires its nuclear translocation and direct binding to its various target genes40,44. We wondered whether Hsp90 might also facilitate the transcriptional activation of STAT5-regulated genes, and thus participate in promoter-associated STAT5 transcription complexes. We found that STAT5 is constitutively active in K562 cells and binds to a STAT5-binding consensus sequence. STAT5 activation and DNA binding were partially abrogated, in a dose-dependent manner, upon Hsp90 inhibition with PU-H71 (Fig. 5e). Furthermore, quantitative chromatin immunoprecipitation (ChIP) assays in K562 cells revealed the presence of both Hsp90 and STAT5 at the critical STAT5 targets MYC and CCND2 (Fig. 5f). Neither protein was present at intergenic control regions (data not shown). Accordingly, PU-H71 (1 µM) decreased the mRNA abundance of the STAT5 target genes CCND2, MYC, CCND1, BCL-XL and MCL1 (ref. 45), but not of the control genes HPRT and GAPDH (Fig. 5g).
Collectively, these data show that STAT5 activity is positively regulated by Hsp90 in CML cells (Fig. 5h). Our findings are consistent with a scenario whereby Hsp90 binding to STAT5 modulates the conformation of the protein to alter STAT5 phosphorylation and dephosphorylation kinetics. In addition, Hsp90 maintains STAT5 in an active conformation in STAT5-containing transcriptional complexes. Considering the complexity of the STAT pathway, other potential mechanisms cannot be excluded. Therefore, in addition to its role in promoting protein stability, Hsp90 promotes oncogenesis by maintaining client proteins in an active configuration.
Discussion
We present herein a rapid and simple chemical-proteomics method for surveying tumor oncoproteins (Supplementary Fig. 8). The method takes advantage of PU-H71’s ability to (i) preferentially bind to a pool of Hsp90 associated with oncogenic client proteins, and (ii) trap Hsp90 in an oncoclient-bound configuration. We propose that this approach provides a powerful tool to dissect, tumor-by-tumor, molecular lesions that are characteristic of distinct cancers. Because of the initial chemical-precipitation step, which purifies and enriches the aberrant protein population as part of PU bead–bound Hsp90 complexes, the method does not require expensive SILAC labeling or two-dimensional gel separation of samples. Instead, protein cargo from PU bead pull-downs is simply eluted in SDS buffer and submitted to standard SDS-PAGE, and then the separated proteins are extracted and trypsinized for LC-MS/MS analysis.
Although this method presents a unique approach to identifying the oncoproteins that maintain the malignant phenotype of tumor cells, one needs to be aware that, similar to other chemical or antibody-based proteomics techniques, it has potential limitations46. For example, ‘sticky’ or abundant proteins may bind in a nondiscriminatory way to proteins isolated by PU beads. Second, although we have presented several lines of evidence that PU-H71 is specific for Hsp90, one must consider that, at the high concentration of PU-H71 present on the beads, some nonspecific binding to a few non-Hps90 proteins is unavoidable.
Despite these potential limitations, we have used this method to carry out the first global evaluation of Hsp90-dependent aberrant signaling pathways in CML. The Hsp90 interactome identified by PU-H71 affinity purification overlaps substantially with the well-characterized CML signalosome, indicating that this method can identify a large part of the complex web of pathways and proteins that define the molecular basis of this leukemia. When applied to less well-characterized tumor types, this method may provide unpredicted targets for combinatorial therapy.
We believe the functional proteomics method described here will assist identification of the critical proteome subset that is dysregulated in individual tumors, including primary patient specimens. Thus, tumor-specific Hsp90 client profiling could ultimately yield an approach for personalized therapeutic targeting of tumors (Supplementary Fig. 8).
Our work also proposes that Hsp90 forms biochemically distinct complexes in cancer cells (Supplementary Fig. 3a). In this view, a major fraction of cancer cell Hsp90 retains ‘housekeeping’ chaperone functions similar to normal cells, whereas a functionally distinct Hsp90 pool that is enriched or expanded in cancer cells specifically interacts with the oncogenic proteins required to maintain tumor cell survival. Perhaps this Hsp90 fraction represents a cell stress–specific form of chaperone complex that is expanded and constitutively maintained in the tumor cell context. Our data suggest that it may execute functions necessary to maintain the malignant phenotype. One such role is to regulate the folding of mutated (that is, mB-Raf) or chimeric (that is, Bcr-Abl) proteins8,9. We now present experimental evidence for an additional role; that is, to facilitate scaffolding and complex formation of molecules involved in aberrantly activated signaling complexes.
What distinguishes the PU-H71-binding fraction of Hsp90 from the non–PU-H71-binding fraction? This is a complex question that remains under active investigation. Although both Hsp90α and Hsp90β isoforms are recognized by PU-H71, our data provide evidence for at least one difference between Bcr-Abl–Hsp90 (PU-H71 preferring) and Abl–Hsp90 (PU-H71 nonpreferring) chaperone complexes. That is, Bcr-Abl–Hsp90 chaperone complexes contain a number of cochaperones (suggesting that an active chaperoning process is underway, further supported by the sensitivity of Bcr-Abl to the silencing of Hsp70), whereas Abl–Hsp90 complexes lack associated cochaperones (probably representing sequestered but not actively chaperoned Abl, supported by the insensitivity of Abl to Hsp70 knockdown). Finally, we have observed a differential impact of Hsp90 phosphorylation on PU-H71 and geldanamycin binding. These findings, which are being pursued further, suggest that various Hsp90 inhibitors may be uniquely affected by specific post-translational modifications to the chaperone. Taken together, these preliminary observations suggest that PU-H71 recognizes an Hsp90 fraction that is participating in an active chaperone cycle, and that this characteristic is not necessarily shared by other Hsp90 inhibitors.
Our work uses chemical tools to provide new insights into the heterogeneity of tumor-associated Hsp90 and harnesses the biochemical features of a particular Hsp90 inhibitor to identify tumor-specific biological pathways and proteins. We believe the functional proteomics method described here will allow identification of the critical proteome subset that becomes dysregulated in distinct tumors. This will allow for the identification of new cancer mechanisms, as exemplified by the STAT5 mechanism, the identification of new oncoproteins, as exemplified by CARM1, and the identification of therapeutic targets for the development of rationally combined targeted therapies complementary to Hsp90.
METHODS
Cell lines and primary cells
The CML cell lines K562, Kasumi-4, MEG-01 and KU182, triple-negative breast cancer cell line MDA-MB-468, HER2+ breast cancer cell line SKBr3, melanoma cell line SK-Mel-28, prostate cancer cell lines LNCaP and DU145, pancreatic cancer cell line Mia-PaCa-2 and colon fibroblast cell line CCCD18Co were obtained from the American Type Culture Collection. The CML cell line KCL-22 was obtained from the Japanese Collection of Research Bioresources. The NIH-3T3 fibroblast cells were transfected as described41. Cells were cultured in DMEM in F12 (MDA-MB-468, SKBr3 and Mia-PaCa-2), RPMI (K562, SK-Mel-28, LNCaP, DU145 and NIH-3T3) or MEM (CCD18Co) supplemented with 10% (v/v) fetal bovine serum (FBS), 1× l-glutamine and 1× penicillin and streptomycin (Pen/Strep). Kasumi-4 cells were maintained in IMDM supplemented with 20% (v/v) FBS, 10 ng ml−1 granulocyte macrophage colony-stimulating factor (GM-CSF) and 1× Pen/Strep. PBLs (n = 3) and cord blood (n = 5) were obtained from patient blood purchased from the New York Blood Center. We layered 35 ml of the cell suspension over 15 ml of Ficoll-Paque plus (GE Healthcare). Samples were centrifuged at 2,000 r.p.m. for 40 min at 4 °C, and the leukocyte interface was collected. Cells were plated in RPMI medium with 10% (v/v) FBS and used as indicated. Primary human chronic and blast crisis CML and AML cells were obtained with informed consent. The manipulation and analysis of specimens was approved by the University of Rochester, Weill Cornell Medical College and University of Pennsylvania Institutional Review Boards. Mononuclear cells were isolated using Ficoll-Paque (Pharmacia Biotech) density gradient separation. Cells were cryopreserved in freezing medium consisting of IMDM, 40% (v/v) FBS, and 10% (v/v) DMSO or in CryoStor CS-10 (Biolife). When cultured, cells were kept in a humidified atmosphere of 5% CO2 at 37 °C.
Cell lysis for chemical precipitation and immunoprecipitation
Cells were lysed by collecting them in Felts buffer (20 mM HEPES, 50 mM KCl, 5 mM MgCl2, 0.01% (w/v) NP-40, freshly prepared 20 mM Na2MoO4 (pH 7.2–7.3)) with added 1 µg µl−1 protease inhibitors (leupeptin and aprotinin), followed by three successive freeze (in dry ice) and thaw steps. Total protein concentration was determined using the BCA kit (Pierce) according to the manufacturer’s instructions.
Immunoprecipitation
The Hsp90 antibody (H9010) or normal IgG (Santa Cruz Biotechnology) was added at a volume of 10 µl to the indicated amount of cell lysate, together with 40 µl of protein G agarose beads (Upstate), and the mixture incubated at 4 °C overnight. The beads were washed five times with Felts lysis buffer and separated by SDS-PAGE, followed by a standard western blotting procedure.
Chemical precipitation
Hsp90 inhibitors beads or control beads, containing an Hsp90 inactive chemical (2-methoxyethylamine) conjugated to agarose beads, were washed three times in lysis buffer. Unless otherwise indicated, the bead conjugates (80 µl) were then incubated at 4 °C with the indicated amounts of cell lysate (120–500 µg), and the volume was adjusted to 200 µl with lysis buffer. Following incubation, bead conjugates were washed five times with the lysis buffer and proteins in the pull-down were analyzed by western blotting. For depletion studies, two to four successive chemical precipitations were performed, followed by immunoprecipitation steps, where indicated.
Statistical analysis
Unless otherwise indicated, data were analyzed by unpaired two-tailed t-tests as implemented in GraphPad Prism (version 4; GraphPad Software). A P-value of less than 0.05 was considered significant. Unless otherwise noted, data are presented as the mean ± s.d or mean ± s.e.m. of duplicate or triplicate replicates. Error bars represent the s.d or s.e.m. of the mean. If a single panel is presented, data are representative of two or three individual experiments.
Additional methods
Detailed methodology is described in the Supplementary Methods section on line.
Supplementary Material
Acknowledgments
This work was supported in part by the Geoffrey Beene Cancer Research Center of the Memorial Sloan-Kettering Cancer Center (G.C.), Leukemia and Lymphoma Society (G.C., M.L.G., S.D.N., X.Z. and R.L.), Breast Cancer Research Fund (G.C.), the SPORE Pilot Award and Research & Therapeutics Program in Prostate Cancer (G.C.), the Hirshberg Foundation for Pancreatic Cancer (G.C.), the Byrne Fund (G.C.), 1U01 AG032969-01A1 (G.C.), 1R01 CA155226-01 (G.C. and A.M.) and US National Cancer Institute (NCI) Cancer Center Support Grant P30 CA08748 (H.E.B.). K.B. and L.N. were supported by funds from the Intramural Program of the NCI. S.M.L. and P.M.S.-J. are supported by the Ludwig Center for Cancer Immunotherapy at MSKCC and by NCI Grant P50-CA86483. M.L.G. is funded by the US National Institutes of Health (NIH) through the NIH Director’s New Innovator Award Program, 1 DP2 OD007399-01 and the V foundation. F.P. is funded by the American Italian Cancer Foundation. We thank D. Toft (Mayo Clinic) and M. Cox (University of Texas) for the gifts of H9010 Hsp90-specific antibodies, L.A. Fabrizio, A.M. Morrishow, H. Deng and J. Fernandez for help with MS analysis, A. Perl, C.T. Jordan, M. Becker and J. Nicoll for providing the primary CML samples or suggestions on their use, and B. Clarkson, J. Bromberg and P. Gregor for suggestions with the manuscript.
Footnotes
Author contributions
K.M., J.H.A., H.Z., L.C., A.R., K.B., P.S.-J., F.P., K.H., L.P.V., X.Z., H.E.-B., N.P. and T.K. performed experiments and D.Z., T.T., A.R., R.L., S.M.L., M.L.G. and S.S.G. provided reagents. All authors participated in the design and analysis of various experiments and G.C., A.M., S.D.N., M.L.G. and L.N. wrote the paper.
Competing financial interests
The authors declare competing financial interests: details accompany the full-text HTML version of the paper at http://www.nature.com/naturechemicalbiology/.
Additional information
Supplementary information is available online at http://www.nature.com/naturechemicalbiology/. Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.Ley TJ, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanash S, Taguchi A. The grand challenge to decipher the cancer proteome. Nat. Rev. Cancer. 2010;10:652–660. doi: 10.1038/nrc2918. [DOI] [PubMed] [Google Scholar]
- 4.Kolch W, Pitt A. Functional proteomics to dissect tyrosine kinase signalling pathways in cancer. Nat. Rev. Cancer. 2010;10:618–629. doi: 10.1038/nrc2900. [DOI] [PubMed] [Google Scholar]
- 5.Nomura DK, Dix MM, Cravatt BF. Activity-based protein profiling for biochemical pathway discovery in cancer. Nat. Rev. Cancer. 2010;10:630–638. doi: 10.1038/nrc2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brehme M, et al. Charting the molecular network of the drug target Bcr-Abl. Proc. Natl. Acad. Sci. USA. 2009;106:7414–7419. doi: 10.1073/pnas.0900653106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashman K, Villar EL. Phosphoproteomics and cancer research. Clin. Transl. Oncol. 2009;11:356–362. doi: 10.1007/s12094-009-0369-z. [DOI] [PubMed] [Google Scholar]
- 8.Zuehlke A, Johnson JL. Hsp90 and co-chaperones twist the functions of diverse client proteins. Biopolymers. 2010;93:211–217. doi: 10.1002/bip.21292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Workman P, Burrows F, Neckers L, Rosen N. Drugging the cancer chaperone HSP90: combinatorial therapeutic exploitation of oncogene addiction and tumor stress. Ann. NY Acad. Sci. 2007;1113:202–216. doi: 10.1196/annals.1391.012. [DOI] [PubMed] [Google Scholar]
- 10.Kamal A, et al. A high-affinity conformation of Hsp90 confers tumour selectivity on Hsp90 inhibitors. Nature. 2003;425:407–410. doi: 10.1038/nature01913. [DOI] [PubMed] [Google Scholar]
- 11.Janin YL. ATPase inhibitors of heat-shock protein 90, second season. Drug Discov. Today. 2010;15:342–353. doi: 10.1016/j.drudis.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 12.Tsaytler PA, Krijgsveld J, Goerdayal SS, Rudiger S, Egmond MR. Novel Hsp90 partners discovered using complementary proteomic approaches. Cell Stress Chaperones. 2009;14:629–638. doi: 10.1007/s12192-009-0115-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.da Rocha Dias S, et al. Activated B-RAF is an Hsp90 client protein that is targeted by the anticancer drug 17-allylamino-17-demethoxygeldanamycin. Cancer Res. 2005;65:10686–10691. doi: 10.1158/0008-5472.CAN-05-2632. [DOI] [PubMed] [Google Scholar]
- 14.Grbovic OM, et al. V600E B-Raf requires the Hsp90 chaperone for stability and is degraded in response to Hsp90 inhibitors. Proc. Natl. Acad. Sci. USA. 2006;103:57–62. doi: 10.1073/pnas.0609973103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taldone T, Chiosis G. Purine-scaffold hsp90 inhibitors. Curr. Top. Med. Chem. 2009;9:1436–1446. doi: 10.2174/156802609789895737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dezwaan DC, Freeman BC. HSP90: the Rosetta stone for cellular protein dynamics? Cell Cycle. 2008;7:1006–1012. doi: 10.4161/cc.7.8.5723. [DOI] [PubMed] [Google Scholar]
- 17.Ren R. Mechanisms of BCR-ABL in the pathogenesis of chronic myelogenous leukaemia. Nat. Rev. Cancer. 2005;5:172–183. doi: 10.1038/nrc1567. [DOI] [PubMed] [Google Scholar]
- 18.Burke BA, Carroll M. BCR-ABL: a multi-faceted promoter of DNA mutation in chronic myelogeneous leukemia. Leukemia. 2010;24:1105–1112. doi: 10.1038/leu.2010.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McClellan AJ, et al. Diverse cellular functions of the Hsp90 molecular chaperone uncovered using systems approaches. Cell. 2007;131:121–135. doi: 10.1016/j.cell.2007.07.036. [DOI] [PubMed] [Google Scholar]
- 20.Carayol N, et al. Critical roles for mTORC2- and rapamycin-insensitive mTORC1-complexes in growth and survival of BCR-ABL-expressing leukemic cells. Proc. Natl. Acad. Sci. USA. 2010;107:12469–12474. doi: 10.1073/pnas.1005114107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nobukuni T, Kozma SC, Thomas G. hvps34, an ancient player, enters a growing game: mTOR Complex1/S6K1 signaling. Curr. Opin. Cell Biol. 2007;19:135–141. doi: 10.1016/j.ceb.2007.02.019. [DOI] [PubMed] [Google Scholar]
- 22.McCubrey JA, et al. Targeting survival cascades induced by activation of Ras/Raf/MEK/ERK, PI3K/PTEN/Akt/mTOR and Jak/STAT pathways for effective leukemia therapy. Leukemia. 2008;22:708–722. doi: 10.1038/leu.2008.27. [DOI] [PubMed] [Google Scholar]
- 23.Häcker H, Karin M. Regulation and function of IKK and IKK-related kinases. Sci. STKE. 2006;2006:re13. doi: 10.1126/stke.3572006re13. [DOI] [PubMed] [Google Scholar]
- 24.Mihailovic T, et al. Protein kinase D2 mediates activation of nuclear factor kappaB by Bcr-Abl in Bcr-Abl+ human myeloid leukemia cells. Cancer Res. 2004;64:8939–8944. doi: 10.1158/0008-5472.CAN-04-0981. [DOI] [PubMed] [Google Scholar]
- 25.Hendriks RW, Kersseboom R. Involvement of SLP-65 and Btk in tumor suppression and malignant transformation of pre-B cells. Semin. Immunol. 2006;18:67–76. doi: 10.1016/j.smim.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 26.Mahajan S, et al. Transcription factor STAT5A is a substrate of Bruton’s tyrosine kinase in B cells. J. Biol. Chem. 2001;276:31216–31228. doi: 10.1074/jbc.M104874200. [DOI] [PubMed] [Google Scholar]
- 27.Oda A, Wakao H, Fujita H. Calpain is a signal transducer and activator of transcription (STAT) 3 and STAT5 protease. Blood. 2002;99:1850–1852. doi: 10.1182/blood.v99.5.1850. [DOI] [PubMed] [Google Scholar]
- 28.Si J, Collins SJ. Activated Ca2+/calmodulin-dependent protein kinase IIgamma is a critical regulator of myeloid leukemia cell proliferation. Cancer Res. 2008;68:3733–3742. doi: 10.1158/0008-5472.CAN-07-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salgia R, et al. Increased tyrosine phosphorylation of focal adhesion proteins in myeloid cell lines expressing p210BCR/ABL. Oncogene. 1995;11:1149–1155. [PubMed] [Google Scholar]
- 30.Le Y, et al. FAK silencing inhibits leukemogenesis in BCR/ABL-transformed hematopoietic cells. Am. J. Hematol. 2009;84:273–278. doi: 10.1002/ajh.21381. [DOI] [PubMed] [Google Scholar]
- 31.Sawyers CL. The role of myc in transformation by Bcr-Abl. Leuk. Lymphoma. 1993;11:45–46. doi: 10.3109/10428199309047862. [DOI] [PubMed] [Google Scholar]
- 32.Naka K, et al. TGF-beta-FOXO signaling maintains leukemia-initiating cells in chronic myeloid leukemia. Nature. 2010;463:676–680. doi: 10.1038/nature08734. [DOI] [PubMed] [Google Scholar]
- 33.Bedford MT, Clarke SG. Protein arginine methylation in mammals: who, what, and why. Mol. Cell. 2009;33:1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maloney A, et al. Gene and protein expression profiling of human ovarian cancer cells treated with the heat shock protein 90 inhibitor 17-allylamino-17-demethoxygeldanamycin. Cancer Res. 2007;67:3239–3253. doi: 10.1158/0008-5472.CAN-06-2968. [DOI] [PubMed] [Google Scholar]
- 35.Jaganathan S, Yue P, Turkson J. Enhanced sensitivity of pancreatic cancer cells to concurrent inhibition of aberrant signal transducer and activator of transcription 3 and epidermal growth factor receptor or Src. J. Pharmacol. Exp. Ther. 2010;333:373–381. doi: 10.1124/jpet.109.162669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Apsel B, et al. Targeted polypharmacology: discovery of dual inhibitors of tyrosine and phosphoinositide kinases. Nat. Chem. Biol. 2008;4:691–699. doi: 10.1038/nchembio.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deininger MW, Druker BJ. Specific targeted therapy of chronic myelogenous leukemia with imatinib. Pharmacol. Rev. 2003;55:401–423. doi: 10.1124/pr.55.3.4. [DOI] [PubMed] [Google Scholar]
- 38.Katzav S. Flesh and blood: the story of Vav1, a gene that signals in hematopoietic cells but can be transforming in human malignancies. Cancer Lett. 2007;255:241–254. doi: 10.1016/j.canlet.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 39.Pratt WB, Morishima Y, Osawa Y. The Hsp90 chaperone machinery regulates signaling by modulating ligand binding clefts. J. Biol. Chem. 2008;283:22885–22889. doi: 10.1074/jbc.R800023200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Groot RP, Raaijmakers JA, Lammers JW, Jove R, Koenderman L. STAT5 activation by BCR-Abl contributes to transformation of K562 leukemia cells. Blood. 1999;94:1108–1112. [PubMed] [Google Scholar]
- 41.An WG, Schulte TW, Neckers LM. The heat shock protein 90 antagonist geldanamycin alters chaperone association with p210bcr-abl and v-src proteins before their degradation by the proteasome. Cell Growth Differ. 2000;11:355–360. [PubMed] [Google Scholar]
- 42.Klejman A, et al. The Src family kinase Hck couples BCR/ABL to STAT5 activation in myeloid leukemia cells. EMBO J. 2002;21:5766–5774. doi: 10.1093/emboj/cdf562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu D, Qu CK. Protein tyrosine phosphatases in the JAK/STAT pathway. Front. Biosci. 2008;13:4925–4932. doi: 10.2741/3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lim CP, Cao X. Structure, function and regulation of STAT proteins. Mol. Biosyst. 2006;2:536–550. doi: 10.1039/b606246f. [DOI] [PubMed] [Google Scholar]
- 45.Paukku K, Silvennoinen O. STATs as critical mediators of signal transduction and transcription: lessons learned from STAT5. Cytokine Growth Factor Rev. 2004;15:435–455. doi: 10.1016/j.cytogfr.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 46.Rix U, Superti-Furga G. Target profiling of small molecules by chemical proteomics. Nat. Chem. Biol. 2009;5:616–624. doi: 10.1038/nchembio.216. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.