Abstract
The Transforming growth factor β (Tgf-β) pathway, by signaling via the activation of Smad transcription factors, induces the expression of many diverse downstream target genes thereby regulating a vast array of cellular events essential for proper development and homeostasis. In order for a specific cell type to properly interpret the Tgf-β signal and elicit a specific cellular response, cell-specific transcriptional co-factors often cooperate with the Smads to activate a discrete set of genes in the appropriate temporal and spatial manner. Here, via a conditional knockout approach, we show that mice mutant for Forkhead Box O transcription factor FoxO1 exhibit an enamel hypomaturation defect which phenocopies that of the Smad3 mutant mice. Furthermore, we determined that both the FoxO1 and Smad3 mutant teeth exhibit changes in the expression of similar cohort of genes encoding enamel matrix proteins required for proper enamel development. These data raise the possibility that FoxO1 and Smad3 act in concert to regulate a common repertoire of genes necessary for complete enamel maturation. This study is the first to define an essential role for the FoxO family of transcription factors in tooth development and provides a new molecular entry point which will allow researchers to delineate novel genetic pathways regulating the process of biomineralization which may also have significance for studies of human tooth diseases such as amelogenesis imperfecta.
Introduction
The process of biomineralization is observed throughout metazoans and results in the generation of biologically important tissues such as shells, carapaces, spicules, bones and teeth [1]. The biomineralization of the mammalian tooth is a particularly striking case as dental enamel contains less than 1% organic matter by weight and is predominantly composed of a highly ordered lattice of calcium hydroxyapatite (Ca10(PO4)6(OH)2) crystals making it the hardest mineralized tissue known [2]. Prior to the development of the mature protein-free enamel structure, enamel formation consists of specific cellular events termed the secretory, transition and maturation stages [3], [4]. During the secretory stage, specialized, ectodermally-derived cells called the ameloblasts deposit a complex extracellular matrix composed of amelogenin, ameloblastin, enamelin and other proteins [5], [6], [7]. These enamel matrix proteins are thought to promote the formation and elongation of thin ribbons of hydroxyapatite crystallites which lengthen parallel to one another while not growing in width [8]. At the end of the secretory stage, the enamel ribbons have reached their full-length thus defining the ultimate thickness of the mature enamel layer. Next, during the transition stage, the ameloblasts reduce their deposition of extracellular matrix proteins and convert from the columnar-shaped secretory ameloblasts to the more shortened maturation ameloblasts. It is during the maturation stage that minerals are deposited on the sides of the fully elongated crystallites resulting in an increase in thickness and width. This structure is further elaborated until the thickening parallel crystals come into contact with adjacent crystals [8], [9].
Genetic studies of amelogenesis imperfecta patients and mice with mutations in the genes encoding the enamel matrix proteins show that these proteins are critical for the proper growth and maturation of enamel crystals [10], [11], [12], [13], [14], [15], [16], [17]. However, these proteins are not part of the mature enamel structure. To complete mineralization of the enamel matrix, in which enamel rods grow firmly against one another and become mechanically interlocked, the extracellular matrix components must be digested and reabsorbed by the ameloblasts [8]. In the mouse, both Matrix metalloproteinase 20 (Mmp-20) and Kallikrein 4 (Klk4) have been shown to have enamel matrix protein protease activity in vivo and both Mmp-20 and Klk4 knockout mice exhibit malformed enamel [18], [19]. Also, mutations in MMP-20 and KLK-4 were reported in amelogenesis imperfecta patients [20], [21], [22], [23], [24]. Interestingly, Mmp-20 is expressed and deposited within the extracellular matrix by the ameloblasts during the secretory stage coincident with Amelogenin, Ameloblastin and Enamelin. It is believed that Mmp-20 cleavage of the extracellular matrix permits hydroxyapatite crystal elongation rather than thickening [8]. Klk4 is expressed beginning at the transition stage and into the maturation stage; however, it has not been detected in secretory ameloblasts [19]. Klk4 is thought to catalyze the proteolytic degradation of the residual extracellular matrix thereby providing additional free space for the elongated enamel crystals to expand in width, contact adjacent crystals and interlock [19]. Overall, the process of enamel biomineralization requires tight spatial and temporal control of numerous genes which also interact post-translationally. Thus, we hypothesized that an ameloblast-specific mechanism of transcriptional regulation likely coordinates this highly specialized developmental process.
Previously, the Smad3 transcriptional co-factor was implicated in the regulation of biomineralization in vivo, as mice with a targeted mutation in Smad3 exhibit a hypomineralized tooth phenotype [25], [26]. Given the well-documented role of Smad3 (together with the co-Smad, Smad4) as an intracellular molecule that mediates signaling from the Transforming growth factor-β (Tgf-β) receptor, we reasoned that Tgf-β signaling might activate a repertoire of genes necessary for the coordination of biomineralization [27]. Consistent with this idea, many studies have already implicated Tgf-β signaling as being important for proper craniofacial development, including tooth formation [28]. However, due to various functional redundancies between Tgf-β signaling molecules, receptors, Smads and Smad transcriptional co-factors, a complete picture of Tgf-β signaling as it relates to specific cellular events during tooth development and maturation remains elusive. Further complicating the issue is the multifunctional role Tgf-β signaling can serve in any given tissue. Thus, targeted deletion of Tgf-β components in mice often results in early embryonic lethality or other pleiotropic effects which make it difficult to assign specific functions to specific components of the pathway.
When considering the broad transcriptional changes which generally occur in response to Tgf-β, how is the Tgf-β signal interpreted by differentiated ameloblasts as a cue to coordinate the expression of specific genes essential for the completion of such specialized developmental processes as enamel biomineralization? Due to studies suggesting that the Smad complex alone is usually insufficient for target gene binding and activation [29], transcription target specificity likely depends on the association with specific Smad transcriptional co-factors expressed coincidentally within the ameloblasts. Members of the forkhead box O family of transcription factors, which serve diverse roles such as cellular growth and proliferation, development, metabolism and longevity [30], have been shown to function as Smad co-factors in vitro [31], [32], [33]. Here, via a conditional knockout approach, we show that ameloblast-specific loss of Forkhead box O transcription factor 1 (FoxO1) results in an enamel hypomaturation phenotype that is reminiscent of the Smad3 mutant [25]. Furthermore, we also show that both FoxO1 and Smad3 mutants exhibit a reduction in the expression of a similar cohort of genes encoding enamel matrix proteins. These data raise the possibility that FoxO1 acts as a Smad co-factor, within an ameloblast-expressed transcriptional complex, to regulate a specific set of genes required for proper enamel biomineralization.
Results
Ameloblast-specific loss of FoxO1 results in a white tooth phenotype
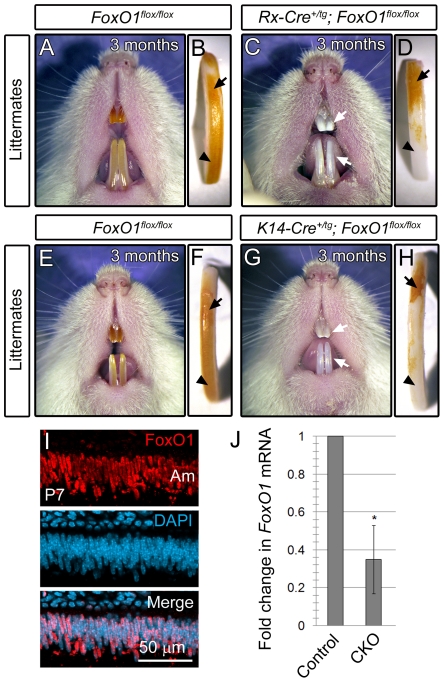
In an independent study, we aimed to address the role of FoxO1 in retinal development by crossing the FoxO1 floxed allele [34] to the Rx-Cre recombinase line [35]. The Rx-Cre+/tg; FoxO flox/flox mice were born at a normal Mendelian frequency and were viable and fertile. However, the retinae in these mice had normal thickness and the cellular composition was indistinguishable from controls (data not shown). Upon further examination of the mutant mice, we found that 100% of the FoxO1 conditional knockouts exhibited a chalky, white tooth phenotype that was first apparent by 1 month postnatally and more pronounced by 3 months (Figure 1A, 1C). Extracted maxillary incisors showed that the proximal, immature enamel of the un-erupted mutant incisor appeared yellowish-brown in color similar to controls (Compare Figure 1B to 1D, arrows). However, in the more distal incisor where the tooth has erupted, we noticed that the FoxO1 mutants were progressively whiter toward the incisal edge (Compare Figure 1B to 1D, arrowhead). This finding suggested that the enamel on FoxO1 mutant teeth experienced extensive attrition due to chewing. Importantly, we also analyzed two control groups, Rx-Cre+/tg; FoxO+/flox and Rx-Cre+/tg mice and they were found to be phenotypically normal (data not shown).
Figure 1. Gross phenotype of the FoxO1 mutant teeth.
Rx-Cre and K14-Cre-mediated knockout of FoxO1 results in viable, fertile mice exhibiting abnormally white, chalky incisors (compare A to C and E to G). Extracted incisors had intact enamel on the un-erupted surface of the incisor that appeared similar to controls (arrows in B, D, F and H). However, the most distal, erupted region of the mutant incisors was white in appearance suggesting enamel attrition due to occlusal forces (arrowheads in B, D, F and H). FoxO1 is expressed in differentiated postnatal ameloblasts (Am) (I). qrtPCR analysis of postnatal conditional knockout whole incisors confirmed the expected reduction of FoxO1 transcript (J).
Given the documented forebrain and retinal-specific activity of Rx-Cre [35], a tooth phenotype was surprising. Since the Rx-Cre is expressed within the forebrain early in development, it is possible that Cre expression is broader than previously appreciated, encompassing precursors of the enamel producing ameleoblasts. To show that FoxO1 is required in ameloblasts, we crossed the FoxO1 floxed allele to mice carrying the Keratin14 (K14)-Cre transgene, which has been shown to drive Cre activity within ectodermally-derived tissues including the ameloblasts [36], [37]. As expected, K14-Cre+/tg; FoxO1flox/flox mice also exhibited a white tooth phenotype similar to the Rx-Cre+/tg; FoxO1flox/flox mutants (Compare Figure 1E to 1G, arrows). These mutants also showed the same progressive proximal to distal whitening of the incisor surface (Compare Figure 1F to 1H).
To further confirm a role for FoxO1 in the mouse ameloblasts, we labeled cryosections of postnatal day 7 (P7) incisors with anti-FoxO1 antibodies and 4′,6-diamidino-2-phenylindole (DAPI) nuclear dye. Upon confocal microscopy, we found that FoxO1 was expressed ubiquitously throughout the mouse incisors and molars and co-localized with the DAPI signal (Figure 1I and data not shown). The nuclear expression of FoxO1 was consistent with the known role of FoxO1 as a transcription factor. Furthermore, we performed quantitative reverse transcriptase polymerase chain reaction (qrtPCR) on mRNA isolated from adult FoxO1 conditional mutant and control incisor enamel organs and found that the FoxO1 conditional mutants exhibited a 65% reduction (P<0.001) in FoxO1 mRNA levels relative to the controls (Figure 1J). Given the broad expression of FoxO1, the residual FoxO1 mRNA observed in the conditional mutants is likely due to the presence of cells outside the ameloblast layer which did not undergo Cre-mediated recombination.
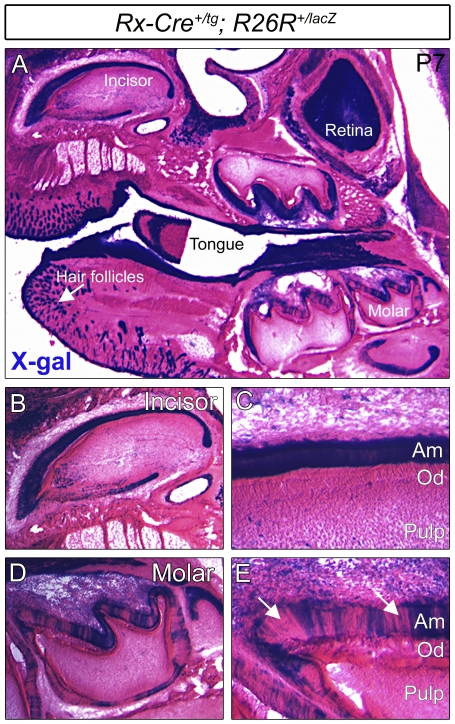
For final confirmation that the Rx-Cre+/tg; FoxO1flox/flox mutant phenotype was due to specific loss of FoxO1 within the ameloblasts, we crossed the Rx-Cre line to mice expressing the ROSA26+/lacZ Cre reporter [38], thus generating Rx-Cre+/tg; ROSA26R+/lacZ progeny. Postnatal day 7 heads were cryosectioned and ROSA26+/lacZ Cre reporter activity was assessed via 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-gal) staining. As expected, we observed lacZ activity in a wide array of tissues within the head and facial regions including hair follicles, retina, skin, oral epithelium, tongue, bone and teeth (Figure 2A and data not shown). Within the teeth, lacZ expression was confined predominantly to the ameloblast layer and we did not observe expression within the odontoblasts (Figure 2B–C). Furthermore, the ameloblasts of the incisors exhibited uniform expression of lacZ whereas the molars had a variable mosaic pattern of expression in which regions of the ameloblast layer were negative for lacZ (Compare Figure 2C to 2E, arrows). The basis for the mosaic Cre activity in the molars is not known.
Figure 2. Within the tooth, Rx-Cre activity is confined to the ameloblast layer.
Rx-Cre+/tg mice were crossed to the ROSA26+/lacZ Cre reporter line and Cre activity was assessed via X-gal staining. Rx-Cre exhibited broad activity throughout the anterior head in such tissues as the retina, tongue, skin, hair follicles, and teeth (A). Closer inspection of the incisors (B and C) and the molars (D and E) revealed that Cre activity was confined to the ameloblast layer (Am) and completely absent from the odontoblast layer (Od). The Cre activity in the molars was more mosaic than the incisors (arrows in E).
FoxO1 mutant tooth enamel is softer than controls
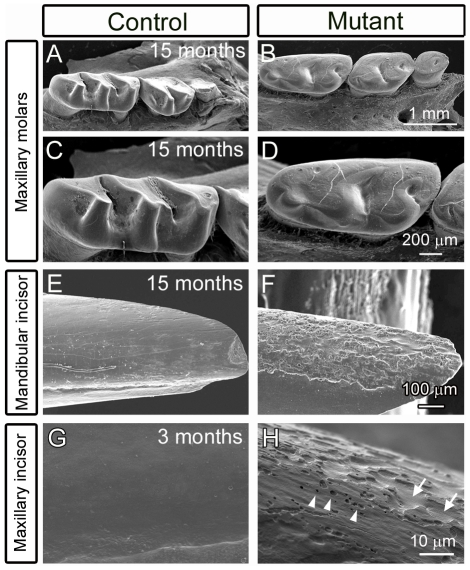
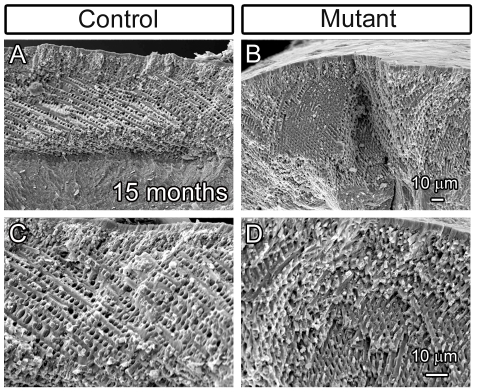
Given the white tooth phenotype of the FoxO1 mutants and activity of Rx-Cre and K14-Cre within the ameloblasts, we reasoned that FoxO1 deletion within the ameloblasts led to hypomaturation of the enamel layer. In order to examine the enamel structure in greater detail, control and FoxO1 mutant maxillae and mandibles were removed and the periradicular bone was dissected away. The exposed molars and incisors were analyzed via scanning electron microscopy (SEM). At 15 months, the FoxO1 mutant molars exhibited dramatic attrition of the enamel surface such that the cusps of the molars were smooth in appearance as compared to controls (Figure 3A–D). We also examined the surface of the mandibular incisors and found that the FoxO1 mutants had a rough appearance in stark contrast to the smooth surface of the control incisors (Compare Figure 3E to 3F). Analysis of maxillary incisors of 3 month old mutants also revealed a similar defect (Figure 3G and 3H). Upon close inspection, regions of the enamel surface appeared to erode away thereby creating the appearance of valleys (Figure 3H, arrows). We also observed small, discrete holes within the enamel surface ranging from approximately 0.5 to 2.0 microns in diameter (Figure 3H, arrowheads). Taken together, these data suggest that the FoxO1 mutant tooth enamel is weaker than the control mice and erodes away over time. However, subsequent imaging of mutant enamel layer cross-sections in fractured incisor preparations revealed that the thickness of the enamel layer as well as the decussating pattern of the enamel rods appeared similar to the controls (Figure 4A–D). This finding suggested that the secretory stage ameloblasts of the FoxO1 mutants are at least partially functional. However, despite substantial enamel matrix deposition, our data indicated that the FoxO1 mutant enamel does not undergo normal maturation resulting in compromised enamel strength.
Figure 3. FoxO1 mutant teeth suffer from severe enamel attrition.
Scanning electron microscopy revealed that 15 month old FoxO1 mutant molars displayed a pronounced wearing of the enamel layer to the extent that the molar cusps had almost completely eroded away (compare A and B and high magnification views in C and D). The 15 month old mutant incisors had a similar phenotype in which the labial surfaces of both the maxillary (shown) and mandibular (not shown) incisors had a dramatic chipping of the enamel from the dentin layer giving the incisors a rough appearance (compare E to F). This phenotype was also seen as early as 3 months (compare G to H) and higher magnification revealed discrete valleys (arrows in H) and holes (arrowheads in H) in the enamel surface.
Figure 4. FoxO1 mutant teeth deposit an enamel matrix and form enamel rods.
Scanning electron microscopy of fractured 15 month old FoxO1 mutant incisors revealed that, in regions where enamel had not chipped away, the mutant teeth initially had an enamel thickness that was comparable to their littermate controls (compare A and B). Higher magnification revealed that the mutant enamel also exhibited the typical decussating pattern of enamel rods (compare C and D).
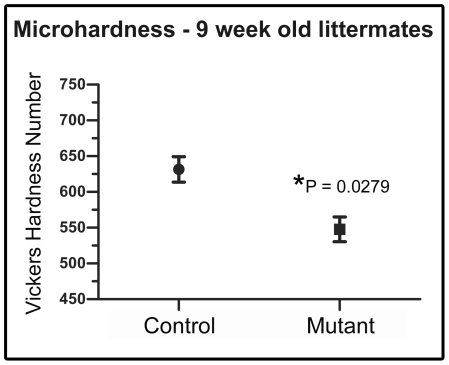
In order to confirm our interpretation of the FoxO1 mutant SEM data as indicating an enamel hypomaturation phenotype, we next attempted to determine whether the FoxO1 mutant enamel is softer than controls. To do this, we subjected littermate control and mutant teeth to an enamel microhardness test. Micro-indentations were created on maxillary incisors from 9 week old FoxO1 mutant and control mice (n = 3 per group). The average hardness for control enamel was found to be 631.4 VHN (SEM ± 17.78) whereas the FoxO1 mutant enamel was 547.7 VHN (SEM ± 17.28). The difference was statistically significant (p = 0.0279) and the results demonstrated that the mature mutant enamel was approximately 13.3% softer than control enamel (Figure 5). We attempted the same analysis on 15 month old teeth, but we were unable to identify large enough, intact enamel surfaces on which to perform the test.
Figure 5. FoxO1 mutant enamel is softer than controls.
The Vickers microhardness test revealed that the adult FoxO1 mutant teeth are significantly softer than their littermate controls.
FoxO1 loss-of-function does not affect gross ameloblast development or differentiation
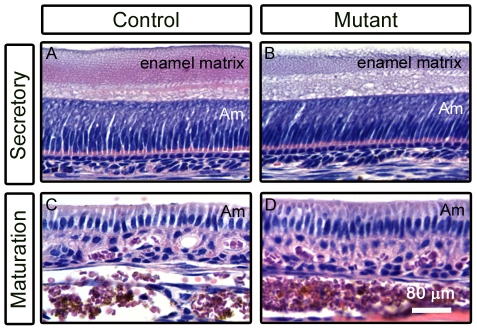
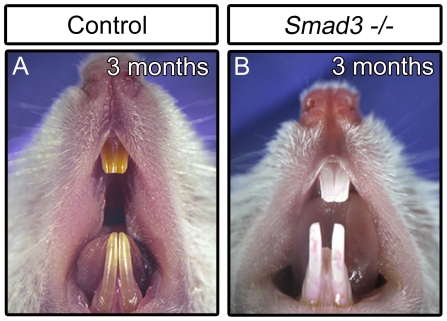
Since our data support a requirement for FoxO1 in ameloblasts, we next sought to determine whether the FoxO1 mutants exhibited a decrease in ameloblast density, failure of polarization or a failure to transition between different stages of amelogenesis that could explain the resulting weakened enamel structure. To assess ameloblast cytoarchitecture, we dissected and decalcified maxillary and mandibular incisors from 15 month old control and FoxO1 mutant littermates and performed hematoxylin and eosin-Y (H&E) staining on paraffin sections. Interestingly, in control and mutant mice, both the secretory and maturation stage ameloblast layers appeared very similar in terms of overall cell density, organization and polarization (Figure 6 A–D). Thus, it is unlikely that the observed FoxO1 mutant soft enamel phenotype is due to defects in ameloblast genesis and/or differentiation. Interestingly, Smad3 mutants show a very similar white tooth phenotype yet also fail to exhibit obvious defects in the morphology of the ameloblast layer (Figure 7) [25].
Figure 6. The morphology of FoxO1 mutant ameloblasts appears unaffected.
Hematoxylin and EosinY staining of paraffin sections of decalcified control and FoxO1 adult mutant teeth showed that mutant secretory stage (A and B) and maturation stage (C and D) ameloblasts (Am) appeared structurally similar to controls.
Figure 7. Smad3 mutants suffer from a white, chalky tooth phenotype reminiscent of the FoxO1 mutants.
FoxO1 and Smad3 mutants exhibit dysregulation of a similar cohort of genes necessary for biomineralization
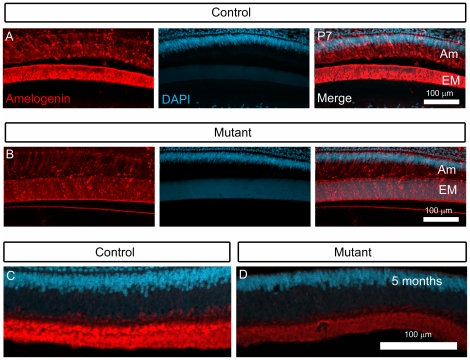
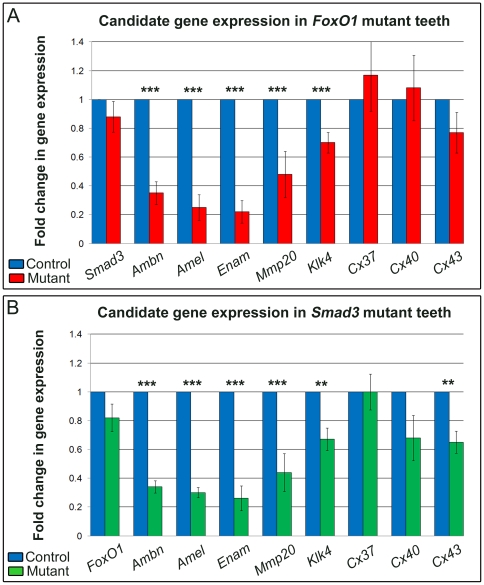
When considering the known role of FoxO1 as a transcription factor, we next sought to address whether FoxO1 regulates a set of genes that might provide insight into the enamel hypomaturation phenotype. Since the FoxO1 mutants failed to exhibit an obvious change in ameloblast specification and differentiation, we decided to focus our analysis predominantly on genes that have been directly implicated in the process of enamel biomineralization. Since Amelogenin proteins constitute 90% of the ameloblast extracellular matrix and are essential for normal enamel organization and maturation [11], [39], [40], we began our analysis by assessing Amelogenin protein expression in FoxO1 mutant and control teeth. Using antibodies against Amelogenin, we performed immunofluorescent analysis of cryosections from 7 day and 5 month old FoxO1 mutant and control incisors. Confocal microscopy suggested that the FoxO1 mutant Amelogenin protein expression levels were reduced as compared to littermate controls (compare Figure 8A to 8B and Figure 8C to 8D). While immunofluorescence provides spatial information regarding gene product expression, it cannot be reliably used as quantitative assessment. Thus, we next performed qrtPCR analysis of adult FoxO1 mutant and control incisors. Given the previously documented role of FoxO transcription factors as Smad transcriptional co-activators [31], [32], [33], the expression of FoxO1 and Smads within endogenous ameloblasts [41], [42], [43] and the white tooth phenotype of the FoxO1 and Smad3 mutant mice (Figure 1 and Figure 7) [25], we decided to include Smad3 mutant teeth in our qrtPCR analysis. We extracted adult maxillary and mandibular incisors from FoxO1 mutant, Smad3 mutant and control mice and purified mRNA for subsequent qrtPCR analysis. It should be noted that littermate controls were used in every case so that genetic background was consistent with that of the respective mutant. Upon analysis of transcript levels, we found that Ameloblastin (Ambn), Amelogenin (Amel), Enamelin (Enam), Mmp20 and Klk4 mRNA levels were all significantly reduced in both the FoxO1 and Smad3 mutant mice as compared to controls with the enamel matrix proteins (including amelogenin) showing the greatest reduction (Figure 9A–B). Furthermore, the relative reduction of Ameloblastin, Amelogenin, Enamelin, Mmp20 and Klk4 expression had a very similar trend in both FoxO1 and Smad3 mutants with Enamelin being the most reduced and Klk4 being the least reduced. Since FoxO1 loss-of-function in other tissues during development has been shown to cause a dramatic reduction in the expression of Connexin-37 (Cx37) and Connexin-40 (Cx40) mRNA levels (47), we included these genes in our analysis [44]. We also included Connexin-43 (Cx43) as Cx43 mutants exhibit an enamel maturation defect [45], [46], [47]. Loss of FoxO1 or Smad3 has no effect on the levels of Cx37 or Cx40 mRNA (Figure 9A–B). The level of Cx43 was modestly down-regulated in the Smad3 mutants but not significantly reduced in the FoxO1 mutants. Finally, we determined that Smad3 is normally expressed in the FoxO1 mutants and FoxO1 is normally expressed in the Smad3 mutants. Taken together, these data suggest that FoxO1 and Smad3 potentially regulate a common set of genes necessary for enamel formation and maturation.
Figure 8. FoxO1 mutant incisors exhibit an apparent reduction in amelogenin protein expression.
Immunofluorescent analysis of cryosections derived from P7 and 5 month old FoxO1 and littermate control mice revealed that FoxO1 mutants exhibit a qualitative reduction in Amelogenin protein expression. This reduction in fluorescent signal was observed in both the ameloblasts (Am) and the enamel matrix (EM).
Figure 9. Loss of FoxO1 and Smad3 results in the down-regulation of a common set of genes.
Quantitative rtPCR was performed on mRNA isolated from adult FoxO1 mutant (A) and Smad3 mutant (B) incisors and compared to control littermates. Both mutants exhibited a similar trend in the down-regulation of genes known to be necessary for proper enamel development and maturation (*p<0.05, **p<0.01, ***p<0.001).
In silico analysis of the enamel matrix proteins revealed conserved FoxO and Smad binding elements
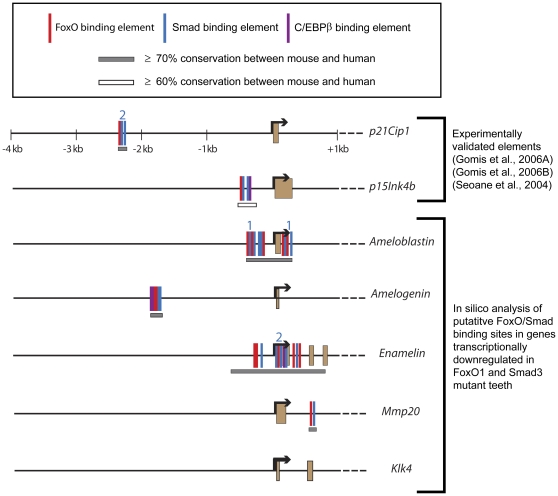
Next, using a method similar to previous studies [31], [32], [33], we performed in silico analysis to map putative FoxO and Smad binding elements within the genes that showed a transcriptional decrease in the FoxO1 and Smad3 mutants (Figure 10). As a positive control, to ensure our search method was comparable to previous methods, we included searches of regions upstream of p21Cip1 and p15Ink4a as these have been previously mapped and were validated experimentally [31], [32], [33]. Also, since its binding sites are occasionally located within FoxO/Smad binding site clusters [32], [33], we searched for elements recognized by the transcription factor C/EBPβ (CBEs). For this analysis, we used mouse genomic sequence from 4 kb upstream of the transcription start site to the stop codon of each gene. FoxO binding elements (also know as forkhead-binding elements or FHBEs) [(G/A)(T/C)AAA(T/A)A] and Smad binding elements (SBEs) [AGAC] (within 100 nucleotides upstream and downstream of the FoxO elements) were then identified using the dual site matching program. Putative FoxO and Smad elements were localize upstream of the transcription start site of Ameloblastin, Amelogenin and Enamelin. Furthermore, Amelogenin contained a single C/EBPβ element within the same FoxO/Smad cluster. Ameloblastin and Enamelin contained additional sites within the first exon and intron. Mmp20 contained only a single FoxO/Smad cluster and it was localized to the first intron. Klk4 did not contain any putative FoxO, Smad or C/EBPβ elements. These data suggest that at least a subset of the genes down-regulated in the FoxO1 and Smad mutants may be direct targets of the FoxO/Smad transcriptional complex that has been previously identified in epithelial cells [31], [32], [33] and that, in some cases, C/EBPβ may provide additional regulation.
Figure 10. Mapping of putative FoxO/Smad genomic binding sites.
In silico analysis of putative FoxO, Smad, and CEBPβ binding elements, conserved between mice and humans and residing within several genes down-regulated in the FoxO1 and Smad mutants (see text for further details). The colored numbers indicate additional SBEs or CBEs that, due to scaling, could not be represented on the gene tracks.
Discussion
Here, we have uncovered a novel role for FoxO1 in enamel maturation as well as a molecular entry point from which to further elucidate the mechanism of transcriptional control over mammalian biomineralization. While the FoxO1 mutants exhibit soft tooth enamel which undergoes severe attrition over time, we did not observe any obvious dysplasia of the ameloblasts. This phenotype is strikingly similar to that of the Smad3 mutant mice [25]. Based on such a close phenocopy between these two mutants, we hypothesized that these genes play similar roles in enamel development. Indeed, we demonstrated that the FoxO1 and Smad3 mutants exhibit down-regulation of a similar cohort of genes known to be important for enamel development and maturation. Previously, in in vitro studies of epithelial cells, it was shown that FoxO and Smad proteins exist in a TGF-β-dependent transcriptional complex which regulates the expression of a common set of genes termed a synexpresison group [31], [32], [33]. These genes comprise a functionally diverse group including mediators of cellular stress and cystostatic responses. This finding raises the possibility that a similar transcription factor complex functions in ameloblasts (the epithelial component of the tooth) to regulate enamel matrix protein expression. Consistent with this idea, in silico mapping of FoxO and Smad binding sites uncovered clusters of conserved regulatory elements located upstream of the transcriptional start sites of Ameloblastin, Amelogenin and Enamelin. Interestingly, in the FoxO1 and Smad mutants, these three genes also showed a greater reduction in expression levels as compared to Mmp20 and Klk4 proteases which did not contain upstream FoxO or Smad binding elements. These data raise the possibility that a FoxO1/Smad3 complex may directly regulate a subset of genes that must be expressed in an overlapping temporal and spatial manner and serve a similar role during enamel development. However, unequivocal demonstration of such a complex directly regulating the expression of enamel matrix proteins in ameloblasts in vivo awaits FoxO1/Smad3 co-immunoprecipitation and chromatin immnoprecipitation analyses of adult mouse incisors.
Surprisingly, loss-of-function studies of genes encoding the enamel matrix proteins (Ameloblastin, Amelogenin, and Enamelin) exhibit a phenotype which is more severe than the FoxO1 or Smad3 mutants. These mutants have dramatic defects in the development of the enamel in which the normal prismatic enamel structure is severely disrupted or the enamel completely fails to form [10], [11], [14], [48], [49]. In the FoxO1 mutant teeth, the enamel prisms form but fail to become fully mineralized resulting in a weakened structure. Furthermore, while the Ameloblastin, Amelogenin, and Enamelin mutant ameloblasts show morphological abnormalities such as loss of cell polarity, cyst-like structures and detachment from the extracellular matrix [10], [11], [14], [48], [49], the FoxO1 and Smad3 ameloblasts appear morphologically normal [25]. The relatively mild defects observed in the FoxO1 and Smad3 mutants are likely explained by the finding that these mutants still express appreciable levels of the major enamel matrix proteins. However, the levels are significantly reduced from wild type. It is worth noting that the Enamelin knockout mice exhibit an autosomal dominant enamel phenotype that appears less severe than the FoxO1 and Smad3 mutants [13]. The mandibular incisors of these mice were white and chalky in appearance and exhibited attrition of the erupted portions. However, as in the FoxO1 mutant mice, the enamel prisms looked normal. These data suggests that the Enamelin heterozygous mutant enamel forms but is softer than normal. Interestingly, among all the putative FoxO/Smad target genes analyzed in this study, Enamelin mRNA levels showed the greatest reduction. Thus, it is formally possible that the reduction of Enamelin expression is the main contributing factor to the FoxO1 and Smad3 mutant enamel phenotypes.
In a recent study, FoxO1 was shown to directly induce the expression of transcription factor Runx2 (Cbfa1) as well as physically associate with Runx2 protein to regulate genes essential for osteoblast differentiation and skeletogenesis [50]. Of further significance is the finding that Runx2 has the ability to bind to regions of the Ameloblastin promoter and possibly participate in the control of Ameloblastin transcription [51]. Also, Runx2 was shown to be endogenously expressed in secretory and maturation ameloblasts, and loss of Runx2 expression results in severely hypoplastic teeth lacking definitive odontoblast and ameloblast differentiation [52], [53], [54], [55]. Thus, it is possible that FoxO1 and Runx2 may be functionally related during enamel maturation. However, we performed qrtPCR to assess the levels of Runx2 expression in FoxO1 mutant incisors and found that Runx2 (and Runx1) levels were the same as controls (not shown). It is also important to recognize that the documented interaction of FoxO1 and Runx2 during osteoblast differentiation occurs in mesenchymal cells [50] while enamel formation is driven by the ameloblasts, which are epithelial cells. Thus, one can envision dramatic differences in terms of Runx2 function depending on the specific cellular context. Indeed, it has been shown that TGF-β signaling to osteoblasts, via Smad3, has the ability to repress the transcription of Runx2 as well as Runx2 protein activity [56], [57]. However, whether this particular function of Smad3 is FoxO-dependent was not addressed. Other transcription factors potentially functioning in concert with FoxOs or Smads in enamel maturation are C/EBPβ and related family members. One member, C/EBPα, was shown to be a transcriptional activator of the mouse Amelogenin gene in vitro. Interestingly, ameloblast specific ablation of C/EBPα did not result in an enamel phenotype or loss of Amelogenin expression and this was shown to be due to redundancy with C/EBPδ [58], [59]. By employing future bioinformatic experiments to map FoxO/Smad binding elements, we should be able to uncover more putative target genes. By combining this approach with subsequent conditional knockout of target genes and functional validation of direct binding sites, we will gain increasingly better insight into the process of enamel formation and maturation in vivo.
Our data show that FoxO1 mutant ameloblasts are morphologically normal yet the enamel is weaker than controls. This suggests to us that FoxO1 likely does not function in ameloblast development, but rather in the production of mature enamel. While it is formally possible that low or mosaic Cre activity at early tooth developmental stages might preclude the discovery of a subtle, earlier function of FoxO1 in ameloblast specification and differentiation, we do not believe this scenario to be likely. The K14-Cre transgene has been shown by several groups to be active at the E12 tooth bud stage which is several days before ameloblast differentiation occurs [60], [61] and K14-Cre-mediated conditional knockouts of other genes have been shown to disrupt ameloblast differentiation and organization [61], [62]. Furthermore, it is well-known that rodent incisors contain a stem cell niche called the cervical loop from which ameloblasts continuously renew throughout the lifetime of the animal [63], [64]. It is also known that K14-Cre and Rx-Cre (shown in our study, Fig. 2B) are active in the cervical loop [36]. Thus, the ameloblast stem cell population would be expected to lose FoxO1 expression thereby giving rise to FoxO1-deficient ameloblasts in the adult. Since, we only observed an enamel maturation defect in the mutant mice, this would argue against a role for FoxO1 in ameloblast differentiation.
Our study of the role of FoxO1 in mouse ameloblasts has established a critical molecular entry point which will allow researchers to delineate novel genetic pathways regulating the process of biomineralization. FoxO1 and Smads, either directly or indirectly, likely control the expression of numerous genes, possibly in addition to enamel matrix proteins, which are essential for the completion of tooth mineralization. Some of these genes may also be dysregulated in human diseases such as amelogenesis imperfecta.
Materials and Methods
Mouse Strains
FoxO1flox/flox [34], Rx-Cre flox/flox [35], K14-Cre flox/flox [65], and ROSA26R+/lacZ [38] mice were maintained on a mixed C57BL/6J, FVB/NJ background. Smad3−/− mice were maintained on a BALB/c background [26]. PCR genotyping was performed as described in the references indicated above. All animal research was conducted according to protocols approved by the Institutional Animal Care and Use Committee (IACUC) of Baylor College of Medicine (assurance number 3823-01).
Scanning Electron Microscopy
Mandibular and maxillary incisors from 3 animals at each stage and genotype were mounted whole or were fractured and then mounted on aluminum stubs to expose the fractured plane. Samples were sputter-coated for analyzing enamel structure on a scanning electron microscope (JEOL).
Microhardness Test
Erupted portions of maxillary incisors from 3 FoxO1 mutant and 3 control littermates were fixed overnight in 100 mg/ml formaldehyde, 10 g/L zinc sulfate, washed and dehydrated with grade alcohol and acetone. Incisors were embedded sagittally in hard-formulation epoxy embedding medium (EpoFix, EMS). Samples were ground and polished to 0.25 µm with a diamond suspension (EMS). The polished samples were tested for enamel microhardness on a Leco M 400 HI testing machine (Leco). Testing was performed with a load of 25 g for 5 sec with a Vickers tip. Twenty-five indents per sample were measured for hardness. Statistical significance was determined by one-way student's t-test (GraphPad Prism 5).
RNA extraction and quantitative rtPCR
Adult control and mutant incisors (n = 3 per genotype), removed from the surrounding bone and with the enamel organ intact, were homogenized in liquid nitrogen using a mortar and pestle. The ground samples were further homogenized in Trizol (Invitrogen) and total RNA was extracted according to the manufacturer's protocol. RNA was subsequently DNAse (Invitrogen) digested to eliminate genomic DNA, and purified using the RNeasy RNA clean up kit (Qiagen). The purified RNA was reverse transcribed using the Superscript III first strand synthesis kit with Oligo(dT)20 and random hexamer priming (Invitrogen). The Taqman® gene expression assay from Applied Biosystems was used for qrtPCR, and primer pairs from Applied Biosystems were used to detect gene expression: Gja1/Cx43 (Mm00439105_m1), Gja5/Cx40 (Mm01265686_m1), Gja4/Cx37 (Mm01179783_m1), Mmp20 (Mm00600244_m1), FoxO1 (Mm00490671_m1), Klk4 (Mm00517338_m1), Amelogenin (Mm00711644_g1), Ameloblastin (Mm00477485_m1), Enamelin (Mm00516922_m1), and Smad3 (Mm00489638_m1). qrtPCR was performed on an Applied Biosystems' ABI PRISM 7000 Real time PCR System under the following PCR conditions: 50°C for 2 min., 95°C for 2 min., 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. To determine relative quantification of gene expression, qrtPCR was performed at different dilutions (0.1 ng–10 ng) in quintuplicate between control and mutant cDNA samples. The data was normalized to a housekeeping gene, Gapdh (Applied Biosystems Cat #4352932E). For data analysis, the Pfaffl method was used to determine relative gene expression ratios (Pfaffl MW, 2001).
H&E staining
Maxillary and mandibular incisors from control and FoxO1 mutant littermate mice were fixed overnight in Zinc-Formalin, decalcified in 5% formic acid, dehydrated and embedded in paraffin. For staining, sections were re-hydrated and stained with Hematoxylin and EosinY.
X-gal staining and immunofluorescence
Whole postnatal day 7 mouse heads were fixed in 4% paraformaldehyde for 3 hours at 4°C. After fixation, the heads were washed in 1× phosphate-buffered saline (PBS, pH 7.3) 3 times for 10 minutes at 4°C. Next, the samples were cryoprotected by immersing in 15% and then 30% sucrose until the tissue sank to the bottom of the tubes. Subsequently, the tissue was immersed in a 1∶1 solution of 30% sucrose and OCT medium and left at 4°C for a couple of hours. Then, the tissue was embedded in OCT on dry ice and stored at −80°C prior to sectioning. Cryosections were cut at 20 µm on a cryostat and mounted on Superfrost Plus slides (VWR Brand, Westchester, PA). X-gal staining was performed at 37°C for 6 hours as previously described [66]. For immunofluorescence of cryosections, slides were post-fixed in 4% PFA for 10 minutes and then washed in 1× PBS-T (PBS+0.1% TritonX-100) 3 times for 10 minutes at room temperature. Next, the slides were blocked in 2% normal donkey serum diluted in 1× PBS for 1 hour at room temperature. anti-FoxO1 (sc49437, Santa Cruz) or anti-Amelogenin (sc32892, Santa Cruz) primary antibodies were diluted (1∶200) in the same blocking solution and incubated on the slides overnight at 4°C in a humid chamber. Next, the slides were washed 4 times at room temperature in 1× PBS. Labeling with donkey, anti-rabbit Alexa Fluor 488 secondary antibodies (1∶400) (Molecular Probes) was performed using the same blocking solution (2% donkey serum in 1× PBS) and slides were incubated for 1 hour at room temperature. Slides were then stained with DAPI (1∶500) and mounted with FluoroMount-G (Southern Biotech).
In silico analysis of FoxO/Smad binding sites
Mouse and human genomic sequences were obtained from the University of California Santa Cruz genome browser (http://genome.ucsc.edu). Genomic sequence from 4 kb upstream of the transcription start site to the stop codon of each gene were extracted from either mouse genome release mm9 (NCBI Build 37, July 2007) or human genome release hg19 (NCBI Build 37, Feb. 2009). Forkhead binding elements (FHBEs) [(G/A)(T/C)AAA(T/A)A] and Smad binding elements (SBEs) (AGAC) within 100 nucleotides upstream and downstream of the FHBE(s) were then identified in the mouse sequences using the dual site matching program (http://cbio.mskcc.org/cgi-bin/lash/dualsite). Binding elements were identified in the dual site program after the following steps were taken: i) The “Invert Sites?” option marked as “yes;” ii) the known 5′-to-3′ sequence of the binding element and its reverse were added into the Primary of Secondary Sites fields (therefore, noting the SBE sequence to be AGAC, one would enter this sequence plus the sequence CAGA into the Secondary Sites field). The identified FHBE-SBE sites detected in the mouse sequences were BLAST-compared against the homologous human gene sequences using NCBI bl2seq (http://blast.ncbi.nlm.nih.gov/Blast.cgi) to confirm conservation between the two species. Conservation of a FHBE-SBE(s) was only noted on Figure 9 if i) the FHBE was completely conserved or the conservation was incomplete by a single nucleotide, ii) if the FHBE conserved between the two species was not identical by nucleotide character but exactly matched published FHBE sequences, iii) if the SBE was completely conserved or the conservation was incomplete by a single nucleotide. The criteria were formulated after determining the nature of published and functionally validated FHBE-SBEs [31], [32], [33]. CBEs [T(G/T)NNG(A/C)AA(G/T)] and SBEs within 100 nucleotides upstream and downstream of the CBE were then identified in a similar manner. CBEs that were located within previously identified FHBE-SBE clusters were singled out and the sequence identity between mouse and human was again detected using the NCBI bl2seq program. As for the FHBE and SBE sequences, CBE sequences were only noted if i) the conservation between mouse and human sequences was complete, ii) incomplete by a single nucleotide, or iii) if the CBE conserved between the two species was not identical by nucleotide character but exactly matched published CBE sequences [32], [33]. Regions of ≥60% or 70% sequence identity larger than the regions encompassing the FHBE-SBE(s) were determined by comparing the entire promoter region of each mouse gene to the human gene using the NCBI bl2seq program. The sequence identity recorded for the Mmp20 and Cx40 genes come from the dual site program rather than the NCBI bl2seq program. Sequences for p21CIP1 and p15INK4b were extracted from releases mm6 and hg17, and mm8 and hg17, respectively, to mimic previously published results. It should be noted that the binding sites represented for p15INK4b were acquired from published results and were not obtained using the methods presented above because of the imperfect conservation of the binding elements between mouse and human.
Acknowledgments
We thank Christina Stuelten for providing Smad3 mutant tissue and Eric Swindell for providing the Rx-Cre mice. We also thank James Culver for critical reading of the manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: These studies were supported by funding from the National Institutes of Health R01EB005173 (to MED) and P20EB0070769 (to MED) as well as Institutional Core Grant #CA16672 High Resolution Electron Microscopy Facility, University of Texas, M.D. Anderson Cancer Center. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Wilt FH. Developmental biology meets materials science: Morphogenesis of biomineralized structures. Dev Biol. 2005;280:15–25. doi: 10.1016/j.ydbio.2005.01.019. [DOI] [PubMed] [Google Scholar]
- 2.Deakins M, Volker JF. Amount of organic matter in enamel from several types of human teeth. Journal of Dental Research. 1941;20:117–121. [Google Scholar]
- 3.Smith CE, Nanci A. Overview of morphological changes in enamel organ cells associated with major events in amelogenesis. Int J Dev Biol. 1995;39:153–161. [PubMed] [Google Scholar]
- 4.Warshawsky H, Smith CE. Morphological classification of rat incisor ameloblasts. Anat Rec. 1974;179:423–446. doi: 10.1002/ar.1091790403. [DOI] [PubMed] [Google Scholar]
- 5.Eastoe JE. Enamel protein chemistry–past, present and future. J Dent Res. 1979;58:753–764. doi: 10.1177/00220345790580022701. [DOI] [PubMed] [Google Scholar]
- 6.Krebsbach PH, Lee SK, Matsuki Y, Kozak CA, Yamada KM, et al. Full-length sequence, localization, and chromosomal mapping of ameloblastin. A novel tooth-specific gene. J Biol Chem. 1996;271:4431–4435. doi: 10.1074/jbc.271.8.4431. [DOI] [PubMed] [Google Scholar]
- 7.Hu CC, Fukae M, Uchida T, Qian Q, Zhang CH, et al. Cloning and characterization of porcine enamelin mRNAs. J Dent Res. 1997;76:1720–1729. doi: 10.1177/00220345970760110201. [DOI] [PubMed] [Google Scholar]
- 8.Bartlett JD, Simmer JP. Proteinases in developing dental enamel. Crit Rev Oral Biol Med. 1999;10:425–441. doi: 10.1177/10454411990100040101. [DOI] [PubMed] [Google Scholar]
- 9.Robinson C, Brookes SJ, Bonass WA, Shore RC, Kirkham J. Enamel maturation. Ciba Found Symp. 1997;205:156–170; discussion 170–154. doi: 10.1002/9780470515303.ch11. [DOI] [PubMed] [Google Scholar]
- 10.Barron MJ, Brookes SJ, Kirkham J, Shore RC, Hunt C, et al. A mutation in the mouse Amelx tri-tyrosyl domain results in impaired secretion of amelogenin and phenocopies human X-linked amelogenesis imperfecta. Hum Mol Genet. 2010;19:1230–1247. doi: 10.1093/hmg/ddq001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gibson CW, Yuan ZA, Hall B, Longenecker G, Chen E, et al. Amelogenin-deficient mice display an amelogenesis imperfecta phenotype. J Biol Chem. 2001;276:31871–31875. doi: 10.1074/jbc.M104624200. [DOI] [PubMed] [Google Scholar]
- 12.Smith CE, Wazen R, Hu Y, Zalzal SF, Nanci A, et al. Consequences for enamel development and mineralization resulting from loss of function of ameloblastin or enamelin. Eur J Oral Sci. 2009;117:485–497. doi: 10.1111/j.1600-0722.2009.00666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu JC, Hu Y, Smith CE, McKee MD, Wright JT, et al. Enamel defects and ameloblast-specific expression in Enam knock-out/lacz knock-in mice. J Biol Chem. 2008;283:10858–10871. doi: 10.1074/jbc.M710565200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fukumoto S, Kiba T, Hall B, Iehara N, Nakamura T, et al. Ameloblastin is a cell adhesion molecule required for maintaining the differentiation state of ameloblasts. J Cell Biol. 2004;167:973–983. doi: 10.1083/jcb.200409077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lagerstrom M, Dahl N, Nakahori Y, Nakagome Y, Backman B, et al. A deletion in the amelogenin gene (AMG) causes X-linked amelogenesis imperfecta (AIH1). Genomics. 1991;10:971–975. doi: 10.1016/0888-7543(91)90187-j. [DOI] [PubMed] [Google Scholar]
- 16.Aldred MJ, Crawford PJ, Roberts E, Thomas NS. Identification of a nonsense mutation in the amelogenin gene (AMELX) in a family with X-linked amelogenesis imperfecta (AIH1). Hum Genet. 1992;90:413–416. doi: 10.1007/BF00220469. [DOI] [PubMed] [Google Scholar]
- 17.Rajpar MH, Harley K, Laing C, Davies RM, Dixon MJ. Mutation of the gene encoding the enamel-specific protein, enamelin, causes autosomal-dominant amelogenesis imperfecta. Hum Mol Genet. 2001;10:1673–1677. doi: 10.1093/hmg/10.16.1673. [DOI] [PubMed] [Google Scholar]
- 18.Caterina JJ, Skobe Z, Shi J, Ding Y, Simmer JP, et al. Enamelysin (matrix metalloproteinase 20)-deficient mice display an amelogenesis imperfecta phenotype. J Biol Chem. 2002;277:49598–49604. doi: 10.1074/jbc.M209100200. [DOI] [PubMed] [Google Scholar]
- 19.Simmer JP, Hu Y, Lertlam R, Yamakoshi Y, Hu JC. Hypomaturation enamel defects in Klk4 knockout/LacZ knockin mice. J Biol Chem. 2009;284:19110–19121. doi: 10.1074/jbc.M109.013623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim JW, Simmer JP, Hart TC, Hart PS, Ramaswami MD, et al. MMP-20 mutation in autosomal recessive pigmented hypomaturation amelogenesis imperfecta. J Med Genet. 2005;42:271–275. doi: 10.1136/jmg.2004.024505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ozdemir D, Hart PS, Ryu OH, Choi SJ, Ozdemir-Karatas M, et al. MMP20 active-site mutation in hypomaturation amelogenesis imperfecta. J Dent Res. 2005;84:1031–1035. doi: 10.1177/154405910508401112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Papagerakis P, Lin HK, Lee KY, Hu Y, Simmer JP, et al. Premature stop codon in MMP20 causing amelogenesis imperfecta. J Dent Res. 2008;87:56–59. doi: 10.1177/154405910808700109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee SK, Seymen F, Kang HY, Lee KE, Gencay K, et al. MMP20 hemopexin domain mutation in amelogenesis imperfecta. J Dent Res. 2010;89:46–50. doi: 10.1177/0022034509352844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wright JT, Daly B, Simmons D, Hong S, Hart SP, et al. Human enamel phenotype associated with amelogenesis imperfecta and a kallikrein-4 (g.2142G>A) proteinase mutation. Eur J Oral Sci. 2006;114(Suppl 1):13–17; discussion 39–41, 379. doi: 10.1111/j.1600-0722.2006.00291.x. [DOI] [PubMed] [Google Scholar]
- 25.Yokozeki M, Afanador E, Nishi M, Kaneko K, Shimokawa H, et al. Smad3 is required for enamel biomineralization. Biochem Biophys Res Commun. 2003;305:684–690. doi: 10.1016/s0006-291x(03)00806-4. [DOI] [PubMed] [Google Scholar]
- 26.Yang X, Letterio JJ, Lechleider RJ, Chen L, Hayman R, et al. Targeted disruption of SMAD3 results in impaired mucosal immunity and diminished T cell responsiveness to TGF-beta. Embo J. 1999;18:1280–1291. doi: 10.1093/emboj/18.5.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moustakas A, Heldin CH. The regulation of TGFbeta signal transduction. Development. 2009;136:3699–3714. doi: 10.1242/dev.030338. [DOI] [PubMed] [Google Scholar]
- 28.Huang XF, Chai Y. TGF-beta signalling and tooth development. Chin J Dent Res. 2010;13:7–15. [PubMed] [Google Scholar]
- 29.Massague J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- 30.Arden KC. FOXO animal models reveal a variety of diverse roles for FOXO transcription factors. Oncogene. 2008;27:2345–2350. doi: 10.1038/onc.2008.27. [DOI] [PubMed] [Google Scholar]
- 31.Seoane J, Le HV, Shen L, Anderson SA, Massague J. Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell. 2004;117:211–223. doi: 10.1016/s0092-8674(04)00298-3. [DOI] [PubMed] [Google Scholar]
- 32.Gomis RR, Alarcon C, He W, Wang Q, Seoane J, et al. A FoxO-Smad synexpression group in human keratinocytes. Proc Natl Acad Sci U S A. 2006;103:12747–12752. doi: 10.1073/pnas.0605333103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gomis RR, Alarcon C, Nadal C, Van Poznak C, Massague J. C/EBPbeta at the core of the TGFbeta cytostatic response and its evasion in metastatic breast cancer cells. Cancer Cell. 2006;10:203–214. doi: 10.1016/j.ccr.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 34.Paik JH, Kollipara R, Chu G, Ji H, Xiao Y, et al. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128:309–323. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swindell EC, Bailey TJ, Loosli F, Liu C, Amaya-Manzanares F, et al. Rx-Cre, a tool for inactivation of gene expression in the developing retina. Genesis. 2006;44:361–363. doi: 10.1002/dvg.20225. [DOI] [PubMed] [Google Scholar]
- 36.Takamori K, Hosokawa R, Xu X, Deng X, Bringas P, Jr, et al. Epithelial fibroblast growth factor receptor 1 regulates enamel formation. J Dent Res. 2008;87:238–243. doi: 10.1177/154405910808700307. [DOI] [PubMed] [Google Scholar]
- 37.Xu X, Han J, Ito Y, Bringas P, Jr, Deng C, et al. Ectodermal Smad4 and p38 MAPK are functionally redundant in mediating TGF-beta/BMP signaling during tooth and palate development. Dev Cell. 2008;15:322–329. doi: 10.1016/j.devcel.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 39.Termine JD, Belcourt AB, Christner PJ, Conn KM, Nylen MU. Properties of dissociatively extracted fetal tooth matrix proteins. I. Principal molecular species in developing bovine enamel. J Biol Chem. 1980;255:9760–9768. [PubMed] [Google Scholar]
- 40.Smith CE. Cellular and chemical events during enamel maturation. Crit Rev Oral Biol Med. 1998;9:128–161. doi: 10.1177/10454411980090020101. [DOI] [PubMed] [Google Scholar]
- 41.Tsuchiya M, Sharma R, Tye CE, Sugiyama T, Bartlett JD. Transforming growth factor-beta1 expression is up-regulated in maturation-stage enamel organ and may induce ameloblast apoptosis. Eur J Oral Sci. 2009;117:105–112. doi: 10.1111/j.1600-0722.2009.00612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu X, Jeong L, Han J, Ito Y, Bringas P, Jr, et al. Developmental expression of Smad1-7 suggests critical function of TGF-beta/BMP signaling in regulating epithelial-mesenchymal interaction during tooth morphogenesis. Int J Dev Biol. 2003;47:31–39. doi: 10.1387/14. [DOI] [PubMed] [Google Scholar]
- 43.Ito Y, Zhao J, Mogharei A, Shuler CF, Weinstein M, et al. Antagonistic effects of Smad2 versus Smad7 are sensitive to their expression level during tooth development. J Biol Chem. 2001;276:44163–44172. doi: 10.1074/jbc.M011424200. [DOI] [PubMed] [Google Scholar]
- 44.Furuyama T, Kitayama K, Shimoda Y, Ogawa M, Sone K, et al. Abnormal angiogenesis in Foxo1 (Fkhr)-deficient mice. J Biol Chem. 2004;279:34741–34749. doi: 10.1074/jbc.M314214200. [DOI] [PubMed] [Google Scholar]
- 45.Flenniken AM, Osborne LR, Anderson N, Ciliberti N, Fleming C, et al. A Gja1 missense mutation in a mouse model of oculodentodigital dysplasia. Development. 2005;132:4375–4386. doi: 10.1242/dev.02011. [DOI] [PubMed] [Google Scholar]
- 46.Dobrowolski R, Sasse P, Schrickel JW, Watkins M, Kim JS, et al. The conditional connexin43G138R mouse mutant represents a new model of hereditary oculodentodigital dysplasia in humans. Hum Mol Genet. 2008;17:539–554. doi: 10.1093/hmg/ddm329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Toth K, Shao Q, Lorentz R, Laird DW. Decreased levels of Cx43 gap junctions result in ameloblast dysregulation and enamel hypoplasia in Gja1Jrt/+ mice. J Cell Physiol. 2010;223:601–609. doi: 10.1002/jcp.22046. [DOI] [PubMed] [Google Scholar]
- 48.Sawada T, Sekiguchi H, Uchida T, Yamashita H, Shintani S, et al. Histological and immunohistochemical analyses of molar tooth germ in enamelin-deficient mouse. Acta Histochem. 2012 doi: 10.1016/j.acthis.2010.05.007. In Press. [DOI] [PubMed] [Google Scholar]
- 49.Masuya H, Shimizu K, Sezutsu H, Sakuraba Y, Nagano J, et al. Enamelin (Enam) is essential for amelogenesis: ENU-induced mouse mutants as models for different clinical subtypes of human amelogenesis imperfecta (AI). Hum Mol Genet. 2005;14:575–583. doi: 10.1093/hmg/ddi054. [DOI] [PubMed] [Google Scholar]
- 50.Teixeira CC, Liu Y, Thant LM, Pang J, Palmer G, et al. Foxo1, a novel regulator of osteoblast differentiation and skeletogenesis. J Biol Chem. 2010;285:31055–31065. doi: 10.1074/jbc.M109.079962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dhamija S, Krebsbach PH. Role of Cbfa1 in ameloblastin gene transcription. J Biol Chem. 2001;276:35159–35164. doi: 10.1074/jbc.M010719200. [DOI] [PubMed] [Google Scholar]
- 52.Aberg T, Cavender A, Gaikwad JS, Bronckers AL, Wang X, et al. Phenotypic changes in dentition of Runx2 homozygote-null mutant mice. J Histochem Cytochem. 2004;52:131–139. doi: 10.1177/002215540405200113. [DOI] [PubMed] [Google Scholar]
- 53.Bronckers AL, Engelse MA, Cavender A, Gaikwad J, D'Souza RN. Cell-specific patterns of Cbfa1 mRNA and protein expression in postnatal murine dental tissues. Mech Dev. 2001;101:255–258. doi: 10.1016/s0925-4773(00)00562-1. [DOI] [PubMed] [Google Scholar]
- 54.D'Souza RN, Aberg T, Gaikwad J, Cavender A, Owen M, et al. Cbfa1 is required for epithelial-mesenchymal interactions regulating tooth development in mice. Development. 1999;126:2911–2920. doi: 10.1242/dev.126.13.2911. [DOI] [PubMed] [Google Scholar]
- 55.Kobayashi I, Kiyoshima T, Wada H, Matsuo K, Nonaka K, et al. Type II/III Runx2/Cbfa1 is required for tooth germ development. Bone. 2006;38:836–844. doi: 10.1016/j.bone.2005.10.026. [DOI] [PubMed] [Google Scholar]
- 56.Alliston T, Choy L, Ducy P, Karsenty G, Derynck R. TGF-beta-induced repression of CBFA1 by Smad3 decreases cbfa1 and osteocalcin expression and inhibits osteoblast differentiation. Embo J. 2001;20:2254–2272. doi: 10.1093/emboj/20.9.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kang JS, Alliston T, Delston R, Derynck R. Repression of Runx2 function by TGF-beta through recruitment of class II histone deacetylases by Smad3. Embo J. 2005;24:2543–2555. doi: 10.1038/sj.emboj.7600729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu Y, Zhou YL, Gonzalez FJ, Snead ML. CCAAT/enhancer-binding protein delta (C/EBPdelta) maintains amelogenin expression in the absence of C/EBPalpha in vivo. J Biol Chem. 2007;282:29882–29889. doi: 10.1074/jbc.M702097200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou YL, Snead ML. Identification of CCAAT/enhancer-binding protein alpha as a transactivator of the mouse amelogenin gene. J Biol Chem. 2000;275:12273–12280. doi: 10.1074/jbc.275.16.12273. [DOI] [PubMed] [Google Scholar]
- 60.Jarvinen E, Salazar-Ciudad I, Birchmeier W, Taketo MM, Jernvall J, et al. Continuous tooth generation in mouse is induced by activated epithelial Wnt/beta-catenin signaling. Proc Natl Acad Sci U S A. 2006;103:18627–18632. doi: 10.1073/pnas.0607289103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Michon F, Tummers M, Kyyronen M, Frilander MJ, Thesleff I. Tooth morphogenesis and ameloblast differentiation are regulated by micro-RNAs. Dev Biol. 2010;340:355–368. doi: 10.1016/j.ydbio.2010.01.019. [DOI] [PubMed] [Google Scholar]
- 62.Kurosaka H, Islam MN, Kuremoto K, Hayano S, Nakamura M, et al. Core binding factor Beta functions in the maintenance of stem cells and orchestrates continuous proliferation and differentiation in mouse incisors. Stem Cells. 2010;29:1792–1803. doi: 10.1002/stem.722. [DOI] [PubMed] [Google Scholar]
- 63.Harada H, Toyono T, Toyoshima K, Yamasaki M, Itoh N, et al. FGF10 maintains stem cell compartment in developing mouse incisors. Development. 2002;129:1533–1541. doi: 10.1242/dev.129.6.1533. [DOI] [PubMed] [Google Scholar]
- 64.Harada H, Kettunen P, Jung HS, Mustonen T, Wang YA, et al. Localization of putative stem cells in dental epithelium and their association with Notch and FGF signaling. J Cell Biol. 1999;147:105–120. doi: 10.1083/jcb.147.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jonkers J, Meuwissen R, van der Gulden H, Peterse H, van der Valk M, et al. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet. 2001;29:418–425. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- 66.Nagy A, Gertsenstein M, Vintersten K, Behringer RR. Manipulating the Mouse Embryo. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2003. [Google Scholar]