Abstract
Mycobacteria are able to enter into a state of non-replication or dormancy, which may result in their chronic persistence in soil, aquatic environments, and permissive hosts. Stresses such as nutrient deprivation and hypoxia provide environmental cues to enter a persistent state; however, a clear definition of the mechanism that mycobacteria employ to achieve this remains elusive. While the concept of sporulation in mycobacteria is not novel, it continues to spark controversy and challenges our perceptions of a non-replication. We investigated the potential role of sporulation in one-year old broth cultures of Mycobacterium subsp. paratuberculosis (MAP).
We show that dormant cultures of MAP contain a mix of vegetative cells and a previously unknown morphotype resembling a spore. These spore-like structures can be enriched for using sporulating media. Furthermore, purified MAP spore forms survive exposure to heat, lysozyme and proteinase K. Heat-treated spores are positive for MAP 16SrRNA and IS900. MAP spores display enhanced infectivity as well as maintain acid-fast characteristics upon germination in a well-established bovine macrophage model. This is the first study to demonstrate a new MAP morphotype possessing spore-like qualities. Data suggest that sporulation may be a viable mechanism by which MAP accomplishes persistence in the host and/or environment. Thus, our current understanding of mycobacterial persistence, pathogenesis, epidemiology and rational drug and vaccine design may need to be reevaluated.
Introduction
Mycobacteria represent a group of highly successful organisms that range from free-living saprophytes to those that have adapted full dependence on a living host [1], [2]. During their life cycle, mycobacterial species may encounter a number of stresses including nutrient deprivation, hypoxia, acidic pH, and even competition with other organisms for limited resources and occupation of a specific niche, such as soil and water [3], [4], [5], [6]. In order to survive in such unfavorable conditions, mycobacteria have developed mechanisms to achieve dormancy, latency and persistence [7], [8], [9]. While several studies have investigated persistence in mycobacteria, the definition remains loosely explained and the mechanisms that lead to and sustain this state of non-replication are poorly understood. A recent study by Ghosh et al. stated the formation of endospores in two month old cultures of M. marinum and M. bovis, which may serve as an unprecedented method employed by mycobacteria to withstand harsh conditions [10]. The concept of sporulation in mycobacteria continues to spark controversy and challenges our current perceptions of the facets involved in mycobacterial persistence. Follow-up studies conducted by Traag et al. could not reproduce endospore formation in 4 week to ∼8.5 month liquid cultures of M. marinum, which questioned the purity of cultures used in ultrastructural characterization by Ghosh et al. [11]. The current research trend focusing on sporulation in mycobacteria remains to reproduce findings by Ghosh et al. using identical isolation methods; however, we investigated whether the potential for sporulation was limited to M. marinum or may encompass another saprophyte and animal pathogen, Mycobacterium avium subsp. paratuberculosis (MAP).
MAP, the causative agent of Johne's disease (JD) in ruminants, is one of the most prevalent and well-documented infections of dairy cattle worldwide [12]. To date, JD eradication remains implausible and control efforts are hampered due to MAP's persistence within soil and water as well as shedding by subclinical and clinical cattle [12], [13], [14], [15]. Therefore, it is critical to augment our knowledge of the events that precede non-replication as well as the various mechanisms used to attain it. Our data showed that one year old broth cultures of MAP strains K-10, 7565, Linda and Ben have the potential to produce a previously undocumented morphotype possessing a spore-like structure given optimal sporulation conditions. All isolated MAP spore-like morphotypes shared appropriate spore ultrastructures, presence of dipicolinic acid and the MAP specific insertion sequence, IS900, and heat resistance. More importantly, heat treated MAP spore structures retained macrophage infectivity as well as acid-fast characteristics upon germination. These data suggest that sporulation may be a viable route by which MAP accomplishes persistence in the environment.
Results and Discussion
MAP produces a new morphology when grown on Arret-Kirshbaum sporulation agar under physiologically relevant temperature
One year old Middlebrook 7H9 (MB7H9) broth cultures (herein referred to as dormant cultures) of MAP strains K-10 (cattle isolate) and 7565 (sheep isolate) were used to examine sporulation activity and isolate spores. It is important to note that the culture medium was never changed and agitation was not supplied for ∼6 months (mo.); therefore, MAP strains were assumed to be subject to nutrient starvation and hypoxia. These methods differ from those utilized by Ghosh et al. in which M. marinum and M. bovis were grown on 7H10 agar plates for 2 and 12 weeks, respectively [10]. Methods also varied from those described by Traag et al. since MAP broth cultures were kept at a constant 37°C as opposed to intermittent incubations at 4°C and 33°C. Furthermore, we included a spore enrichment process in which MAP broth cultures were separately plated on Arret-Kirshbaum (A–K) sporulating agar and grown at 37°C and 39°C. Since A–K sporulating agar is used for induction of sporulation, particularly from Bacillus spp., we hypothesized that MAP may also produce spores upon nutrient exhaustion in this medium. We showed that MAP is capable of growth and sporulation on A–K agar after 72 h of incubation at 39°C, which was validated by differential staining for spores using malachite green and safranin (Figure 1A and B). These results are in stark contrast to E. coli K-12 A–K growth, which is negative for malachite green staining (Figure 1B). It is interesting to note that successful MAP growth on A–K medium occurred at 39°C and not at 37°C, the physiological body temperature of cattle in contrast to the B. subtilis control which grew at 37°C and 39°C (Figure 1A). Previous studies from our laboratory demonstrate that physiologically relevant temperatures greatly influence MAP gene expression profiles and speed of macrophage invasion [16]. It is well recognized that mycobacteria are sensitive to changes in temperature, which influence growth, cell morphology and pathogenesis [16], [17], [18], [19]. Temperature also impacts sporulation efficiency [20], [21], [22]. As previously mentioned, dormant cultures of M. marinum used by Traag et al. were stored at 4°C for an unspecified period of time, which may provide one explanation to the lack of spore detection [11]. Thus, additional pressures like host related temperature may be one of several contributing stressors capable of inducing differential rates of sporulation.
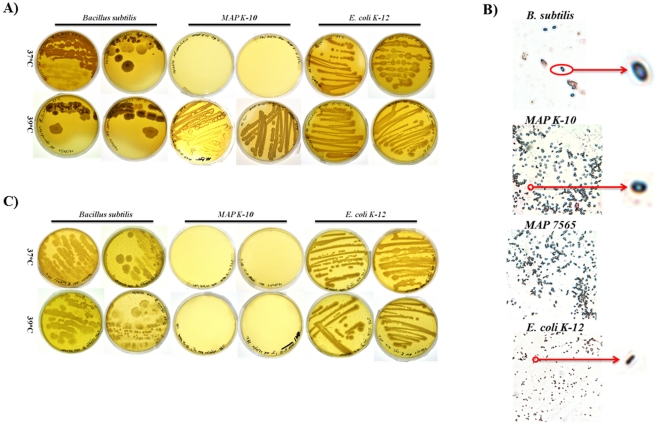
Figure 1. MAP morphotype induction is dependent upon temperature.
One year old MB7H9 MAP broth cultures were inoculated on A) A–K agar and C) BHI agar for 72 h at 37°C and 39°C. MAP showed growth only at 39°C compared with B. subtilis and E. coli K-12 controls, which had substantial growth at both temperatures (A). Spore enrichment was determined by malachite green staining (B). In order to confirm purity of MAP culture, the year old MAP culture was grown on BHI agar and determined to be free of any contaminating organisms (C).
Due to the age of MAP cultures, a valid concern arose that isolated morphotypes were not of a MAP origin but a known endospore contaminant. In order to confirm MAP purity, dormant MAP K-10 cultures used for spore enrichment were streaked on Brain-Heart Infusion (BHI) agar. While B. subtilis produced visible colonies, MAP K-10 showed no growth and was determined to be devoid of any contaminating organisms (Figure 1C). Also, MAP K-10 was revived on MB7H9 agar indicating that dormant cultures remained viable. Therefore, MAP K-10 has the potential to produce spore-like structures given sufficient and appropriate cues. Environmental cues that are involve nutrient and moisture limitation, temperature, hypoxia, and competing microbes are hypothesized to be sufficient to cause mycobacterial sporulation but may not all be necessary at the same time.
Transmission Electron Microscopy (TEM) shows presence of spore-like forms
Since malachite green primarily functions by permeating the mother cell through heat and is retained within the cell coat layers due to their thickness, the possibility remained that malachite green may be bound to the complex, waxy mycobacterial cell wall. In fact, malachite green has been used to stain flagella and leprosy bacilli in addition to endospores [23], [24]. To validate A–K agar and malachite green staining results and characterize the MAP spore-like structure, we examined the spore forms during different phases of growth: log, dormant, and induced (A–K isolated) spore phases (Figure 2A). Dormant MAP K-10 cultures displayed a mix of bacilli and spores, while the induced phase contained only spores (Figure 2A). Next, we increased the stringency of our testing for possible contamination and conducted duplex and normal polymerase chain reactions (PCRs) to demonstrate the presence and/or absence of IS900 integration sites in MAP, Bacillus spoIVA gene and Clostridium 16SrDNA gene in all MAP and control samples (Figure 2B). MAP dormant, log phase, germinated and A–K isolated spore cultures showed the presence of IS900 integration sites but did not display amplification of specific Bacillus and Clostridium gene elements, which suggested the unlikelihood of isolated MAP morphotypes containing Bacillus and Clostridium species (Figure 2B). As expected, control spore samples from B. subtilis and C. perfringens did not amplify the MAP specific IS900 integration sites (Figure 2B).
Figure 2. Ultrastructural characterization of MAP morphologies.
Fine MAP morphotype structure was determined by TEM (A). TEM images were taken of log-phase, dormant and A–K MAP cultures. While the dormant MAP culture showed a mix of vegetative cells and spores, A–K MAP cultures displayed typical spore characteristics, including a cortex, plasma membrane and coat layers. (B) All MAP cultures were assessed for contamination of duplex and normal PCR of IS900, spoIVA and Clostridium 16SrDNA. Only MAP samples contained the IS900 element and did not amplify Bacillus and Clostridium related genes. (C) Spore formation was confirmed by the detection of dipicolnic acid (DPA) using a colorimetric assay. DPA is a chemical found within the spore core of endospores. Intact and autoclaved mycobactin J (250.0 µg/mL) were used as controls. Each sample was conducted in triplicate.
We tested if MAP dormant and A–K spore cultures represented true spores and not a simple thickening of the cell wall as has been reported by Cunningham et al. by determining the presence of dipicolinic acid (DPA; pyridine-2,6 dicarboxylic acid), a chemical commonly found in spores [25]. Calcium bound DPA is found within the spore core and contributes to spore DNA resistance to wet and dry heat, desiccation and hydrogen peroxide [26], [27], [28]. In order to detect DPA, we used a colorimetric assay that releases DPA after autoclaving and acetic acid treatment and is subsequently available to bind to iron under an acidic pH [29]. Since mycobacteria secrete, contain, and are supplemented with siderophores, which form complexes with iron, we included a mycobactin J control to ensure abrogation of potential reactions with iron involved in DPA detection (Figure 2C) [30]. Elevated levels of DPA were found in MAP dormant and spore cultures and B. subtilis spores (matched for wet pellet weight) (Figure 2C). Not surprisingly, isolated MAP spores contained increased amounts of DPA (470.0 µg/mL) compared to any other MAP culture, which was not due to reactive mycobactin J (Figure 2C). It has been noted that DPA synthesis in addition to endospore formation occurs with low guanine (G) and cytosine (C) % content among Firmicutes, which conflicts with the high percentage of G+C found in mycobacteria [11]. Although rare, members of the Streptomyces (S. globisporus and S. avermitilis) are capable of endospore production given conditions conducive to endsporulation such as incubation in submerged cultures and phosphate limitation [31], [32]. Also, the presence of DPA was found by Stastná et al. despite the lack of the spoVF locus, which encodes a DPA synthetase [32]. Orsburn et al. has described that the spoVF locus is also absent in certain clostridia and that an electron transfer flavoprotein may compensate for DPA generation [33]. Therefore, spoVF may not be a strict requirement for DPA production and endosporulation may infrequently encompass genera from Actionbacteria and may not be restricted solely to low G+C% bacteria. Future studies should involve mechanistic based experiments to determine if DPA production in MAP depends upon an electron transfer flavoprotein. It is possible that novel morphotypes of MAP may not be strict endspores and are more reminiscent of Streptomyces spp. spore structures.
Sporulation in MAP is reproducible and consistent in a different culture, medium and multiple strains
Our enrichment technique using A–K agar was validated by another laboratory (National Animal Disease Center (NADC), USDA) using a separate MB7H9 liquid culture of MAP K-10 (Figure 3A). TEM images of MAP K-10 showed characteristic spore features (e.g. condensed core and coat layers) (Figure 3A). Since A–K agar is not a typical method used to induce sporulation from bacterial cultures, we included spore enrichment using potato extract agar (PEA) using a described method (Figure 3B) [34]. Again, year old liquid culture of MAP K-10 was capable of producing spore-like morphotypes and displayed identifiable spore structures (Figure 3B). B. subtilis and E. coli K-12 were included as positive and negative controls, respectively (Figure 3B). MAP K-10 PEA spore-like structures were free of contamination as determined by the absence of growth on blood agar (Figure 3d). Furthermore, we sought to determine if other MAP strains were capable of sporulation. A–K enrichment of and TEM visualization showed that MAP 7565 (sheep strain), Ben (human strain) and Linda (human strain) sporulated (Figure 4). Both Ben and Linda strains were isolated from patients with Crohn's disease [35], [36]. Although controversial, the presence of MAP (either in blood or intestinal tissue) has been shown to be associated with Crohn's disease but an etiological link between the two remains to be established [37], [38], [39].
Figure 3. Sporulation is reproducible on traditional sporulation medium.
Spore enrichment was independently conducted by the National Animal Disease Center (Ames, IA) using a separate MAP K-10 culture inoculated on A–K agar. MAP K-10 year old MB7H9 broth culture, B. subtilis and E. coli K-12 were inoculated on B) Potato extract agar with mycobactin J at 37°C and 39°C. MAP K-10 growth was observed after two weeks of incubation at 39°C in comparison to overnight growth of B. subtilis and E. coli K-12 controls. Biomasses were collected similarly to A–K cultures and processed for TEM (B). MAP K-10 TEM images showed similar structures as those observed in Figure 2 . Furthermore, biomasses were streaked on blood agar and incubated at 37°C and 39°C for 4 weeks to confirm purity (C). MAP K-10 failed to grow for the entire duration of incubation in comparison to B. subitlis and E. coli K-12 controls.
Figure 4. Sporulation occurs in multiple MAP strains.
MAP strains 7565, Ben and Linda were inoculated on A–K agar. Biomasses were collected and processed for TEM. All strains show characteristic spore structures.
MAP spore-like forms survive heat treatment and are positive for MAP 16SrRNA and IS900
To be considered a true spore former, isolated novel MAP morphotypes must survive exposure to heat and be capable to germinate given a nutrient source [26]. We heat treated MAP K-10 spores at 70°C and 90°C for 30 min in addition to 2% lysozyme, proteinase K (PK), kanamycin and anaerobic exposure. Heat treatment served two purposes to: 1) determine temperature threshold for survival and 2) eliminate any remaining vegetative cells such that the re-grown culture only originated from spores. Both lysozyme and PK are typically used as a standard DNA extraction protocol that functions by damaging the cell wall of vegetative MAP cells, which causes bacterial lysis. MAP K-10 spores survived exposure to 70°C but not at 90°C (Figure 5a). Heat exposed spores treated in combination with either lysozyme, PK or kanamycin were capable of re-growth due to the coat layer resistance to these enzymes (Figure 5a). Incubation under anaerobic conditions was included to rule out Clostridium spp. contamination. Exposure to anaerobic conditions post heat treatment failed to produce any visible growth (Figure 5a). Contamination during heat treatment was also eliminated as can be observed by the absence of growth on blood agar plates (Figure 5b). Ten colonies from each heat treated MAP plate (70°C alone/and+lysozyme, +PK, +kanamycin) were selected and submitted to 16SrRNA sequencing. Sequencing results showed that all colonies were positive for MAP 16SrRNA (Figure 5c). These colonies were also positive for MAP-specific IS900 (Figure 5d). MAP spore survival post exposure to 70°C may in fact not be extremely surprising since many studies have shown the presence of MAP as a food contaminant in pasteurized (also treated at 70°C) milk, cheeses and yogurt [40], [41], [42], [43]. It is currently unknown if the MAP found in these dairy products may exist in a spore or spore-like state. It is important to note that MAP is hypothesized to be one potential trigger or causative agent for Crohn's disease (CD) onset [37], [38], [39], [44]. Several studies indicate that the gross pathology of JD and CD are similar, such as the thickened intestinal mucosa and transmural inflammation [36], [45], [46]. It is proposed that MAP survival in pasteurized dairy products may serve as a vehicle for MAP infection in a subset of CD patients [41], [47], [48]. If MAP does survive pasteurization as a spore, this may result in an important finding and further understanding of MAP's potential role in public health.
Figure 5. MAP morphotypes survive 70°C and are positive for MAP 16SrRNA and IS900.
(A) MAP K-10 log phase and spores, B. subtilis and C. perfringens were heat treated at 70°C for 30 min and subsequently treated with 2% lysozyme, PK, or kanamycin. Heat treated samples were plated on MB7H9 or blood agar and incubated at 37°C under aerobic or anaerobic conditions. (B) Heat treated cultures were plated on blood agar to determine growth of any contaminates. (C) 16SrRNA sequences of germinated heat treated MAP spores compared to reference sequences from MAP, Bacillus spp., Streptomyces spp. and Clostridium spp.. Ten colonies from each plate were selected for sequences. Sequences shown are a consensus from the ten colonies. (D) IS900 duplex PCR of germinated heat treated MAP spores.
Dormant MAP cultures express elevated transcript levels of sporulation related genes
In addition to spore visualization, we have identified a number of mycobacterial candidate genes corresponding to those in the sporulation pathway of several Bacillus and Streptomyces species (Figure 6a). We show that MAP 1002c, which has a 57% similarity to the sporulation response regulator spo0A in Bacillus spp., has a 40-fold increase expression in dormant MAP K-10 compared to respective log-phase culture (Figure 6b). Other studies have also noted the presence of spore related genes in M. tuberculosis and other mycobacterial species [49].
Figure 6. Dormant MAP cultures upregulate spore-related transcripts.
(A) A BLAST comparison and reciprocal BLAST searches were conducted between known sporulation genes from Bacillus spp.† and Streptomyces spp.‡ against MAP. Percent similarity was determined by protein alignment. (B) Quantitative real-time PCR was performed on dormant MAP cultures to determine the presence of carD (MAP0475 and MAP0987) and spo0A (MAP1002c). All three genes are upregulated in comparison to log-phase MAP K-10 culture. All samples were conducted in triplicate. C) Multiple sequence alignment of selected CarD proteins. CarD has recently been shown by Stallings et al. to be necessary component of stringency regulation in mycobacteria. Other studies indicate that the stringency response is also necessary for the initiation of sporulation. A multiple sequence alignment of CarD amino acid sequences from mycobacteria and sporulating bacteria was conducted using CLUSTALW.
Research investigating Bacillus spp. and S. coelicolor A3(2) morphology indicate that the stringent response is essential to robust spore production [50], [51]. A recent study by Stallings et al. showed that the mycobacterial gene, carD, is an essential regulator of the stringent response that is also found in a number of sporulating bacteria that downregulates rRNA by binding onto the β-subunit of RNA polymerase (RNAP) in response to nutrient starvation and oxidative stress (Figure 6c) [52]. Transcript levels in dormant MAP culture show a 15 and 2-fold upregulation of MAP 0475 and MAP 0987(carD orthologues), respectively (Fig. 6b). The presence and absence of orthologous genes alone is unlikely to shed any deeper understanding of the sporulation process in mycobacteria but will require the addition of an evolutionary systems biology approach [53]. In other words, functional assays on identified genes are necessary to assess their impact on the generation of this new MAP morphotype. Evolutionary systems biology, which focuses on the changing relationships between genes and gene products, will reveal developmental networks that may include regulatory molecules and feed-forward networks. Ongoing studies in our laboratory are seeking to create knock-out and knock-in mutations of genes identified in Table 1 to determine what role if any they may have in formation of the MAP spore-like morphotype.
Table 1. Primers used in this study.
| Gene and direction | Sequence |
| IS900, L1 ± | CCCGTGACAAGGCCGAAGA |
| IS900, L9 ± | CGGCCCTGGCGTTCCTATG |
| IS900, 900 R ŧ | ACGCTGTCTACCACCCCGCA |
| spoIVA, F | AAATCGGCACACGAAAAGTC |
| spoIVA, R | TGCCAATACCGGGATATCAT |
| Clostridium 16SrDNA, F Ψ | AAAGATGGCATCATCATTCAAC |
| Clostridium 16SrDNA, R Ψ | TACCGTCATTATCTTCCCCAAA |
| Universal 16SrRNA, F | AGAGTTTGATCCTGGCTCAG |
| Universal 16SrRNA, R | GGGTGGATCCTCCTT |
| MAP1002c, F | CGGGTGTGGAACTACGACTT |
| MAP1002c, R | TCTTCTTCCTCAGGTACGAGATGT |
| MAP0475, F | GACAAGGTATTCCAGGTGCTG |
| MAP0475, R | CTCGGCGACCTTGTTGAC |
| MAP0987, F | GCACGACGGCATCGTTAT |
| MAP0987, R | GTCAAGTCCGTCCGTCTCGGTGA |
Motiwala, A.S., et al. Molecular epidemiology of Mycobacterium avium subsp. paratuberculosis: evidence for limited strain diversity, strain sharing, and identification of unique targets for diagnosis. J Clin Micobiol, 2003. 41(5):2015–26.
Bull, T.J., et al. Characterization of IS900 loci in Mycobacterium avium subsp. paratuberculosis and development of multiplex PCR typing. Microbiology, 2000. 146:3285.
Wang, R.F., et al. A 16S rDNA-based PCR method for rapid and specific detection of Clostridium perferingens in food. Mol Cell Probes,m1994. 8(2): 131–7.
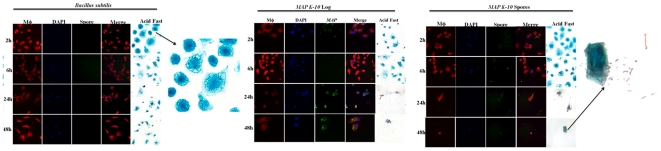
MAP spores retain infectivity and germinate into acid-fast bacilli during exposure to bovine monocyte derived macrophages (MDMs)
Previous studies have reported that MAP can readily be isolated in soil and aquatic environments, which may come into contact with animals and serve as transmission routes [15], [54], [55], [56], [57], [58]. For example, livestock manure stored in liquid lagoons is often applied to agricultural land and MAP may persist within this environment upwards of 175 days [59]. Therefore, we asked the question if MAP spore-like forms could transmit and maintain infection in a well-developed bovine monocyte derived macrophage (MDM) model. Both MAP and B. subtilis spores are readily phagocytosed by MDMs; however, B. subtilis spores are cleared within 6 h p.i. (post infection) (Figure 7). All vegetative cells were lysed as stipulated in materials and methods. MAP spores are maintained within MDMs and germinated by 24 h (Figure 7). Spore germination into developed bacilli show strong acid-fast staining, which is a major diagnostic feature of mycobacteria. Furthermore, the progression of infection with MAP spores was enhanced when compared to the MAP log-phase control and MDMs lysed at 48 h p.i. (Figure 7). These data combined with heat resistance suggest that sporulation in MAP may aid and impact the rate of transmission and consequently establishment of infection in host species.
Figure 7. MAP spores retain infectivity and germinate into acid-fast bacilli in a bovine MDM model.
MAP spores, MAP log-phase and Bacillus subtilis spores were allowed to infect MDMs for 0.5, 2, 6, 24, and 48 h p.i.. MAP spores readily infected MDMs and germinated by 24 h p.i.. Upon 48 h p.i., MDMs were lysed and MAP spores successfully germinated into acid fast bacilli.
Conclusions
This is the first study to identify and describe a new spore-like morphotype in MAP associated with nutrient starvation. We propose that MAP may utilize sporulation as a mechanism to persist in unfavorable conditions such as those encountered in soil and aquatic environments. MAP may also commit itself to a spore-like state to survive the pressures applied by pasteurization and thereby provide one explanation for MAP detection in commercialized dairy products. Although significant strides have been made, especially within the last ten years, in understanding mycobacterial persistence, it continues to be fraught with ambiguities and dissension. The findings by Ghosh et al., which identified spores in M. marinum, and those presented in this paper for MAP are difficult to grapple with as they defy key concepts and change our perceptions of persistence, dormancy and transmission for MAP. This new MAP morphotype or spore readily invaded bovine MDMs, germinated and developed into acid-fast bacilli. More importantly, enrichment and isolation of this new morphotype was independently conducted by a second laboratory (NADC) using a separate MAP culture grown at that facility. Concerns are raised due to the similarities of spore formation in MAP and Bacillus given widely divergent genera. However, 1) certain species of Streptomyces, another genus of the Actinobacteria, are capable of endospore formation at suitable conditions and 2) DPA is also present in Streptomyces despite the lack of spoVF operon. Identification of unique MAP spore coat proteins as well as the cues leading to sporulation may aid in future diagnostics for food and environmental safety. Further studies are needed to examine the role of this newly described MAP morphotype in soil and aquatic environments as well as post pasteurization in dairy products utilizing the above aspects to assess impact in transmission and persistence.
Materials and Methods
Bacterial and MDM Cell Culture
Dormant cultures of MAP strains K-10, 7565, Ben and Linda were grown at 37°C in MB7H9 broth supplemented with oleic acid, albumin, dextrose and catalase (OADC) enrichment and mycobactin J (2.0 mg/L) (Allied Monitor, Fayette, MO) for one year without agitation. MAP cultures were determined to be free of contamination by absence of growth on BHI agar at 37°C and 39°C. Bacillus subtilis and Escherichia coli K-12 were cultured in Luria-Bertani (LB) broth at 37°C with shaking at 150 rpm. Clostridium perfringes was grown on blood agar at 37°C in an anaerobic chamber. Mitsubishi anaeropacks (Fisher Scientific, Pittsburgh, PA) used for the anaerobic chamber were changed every 24 h.
Peripheral blood was collected from the jugular veins of two JD free cattle (542 and 2170) at the University of Minnesota's dairy barn and teaching facility. Monocyte derived macrophages (MDMs) were elutriated using an established protocol [60]. All cattle work performed was in concordance with the institutional guidelines and approved animal care and use protocols at the University of Minnesota.
Spore-like Morphotype Isolation
Approximately 250.0 µL of dormant cultures of MAP (K-10, 7565, Ben and Linda), log-phase B. subtilis (O.D.570 = 0.5) and log-phase E. coli (O.D.600 = 0.5) were cultured separately on Arret-Kirshbaum (A–K) sporulation agar (BD, Franklin Lakes, NJ) at 37°C and 39°C for either 72 (MAP) or 24 h (B. subtilis and E. coli). A time point of 72 h for MAP cultures were selected based on the time required to observe MAP growth on A–K plates. MAP cultures were also grown on Potato Extract Agar (PEA) at 39°C until growth was achieved (2 weeks) [34]. Upon completion of incubation times, A–K and PEA agar plates were allowed to rest at room temperature (RT) for 48 h. Biomasses were collected in autoclaved distilled water (dH2O) and incubated at RT for 72 h.. Spores were washed 3× in 1× phosphate buffer saline (PBS), centrifuged at 13,000 rpm for 10 min to sediment and resuspended in 20.0 mL of autoclaved dH2O. C. perfringes spores were enriched by incubation with Duncan and Strong broth at 37°C for 24 h under anaerobic conditions. All spore samples were heat treated at 70°C for 30 min to lyse any vegetative cells. All spore samples were differentially stained with malachite green/safranin and visualized on an Olympus IX70 inverted fluorescence microscope (Olympus, Center Valley, PA).
DNA Extraction and Polymerase Chain Reaction (PCR)
DNA was extracted from 500.0 mg (wet weight) MAP K-10 log-phase (O.D.600 = 0.5), dormant cultures and spores by incubating samples for 10 min at 37°C with 10% sodium dodecyl sulfate (SDS) followed by homogenization in a mini-bead beater (Roche) using 0.2 mL of 0.1 mm sterile RNase free zirconium beads (Biospec, Bartlesville, OK) for 4 min. DNA samples were cleaned using PE buffer and spin columns (Qiagen, Valencia, CA). B. subtilis DNA was isolated from subcultures grown for 16 h at 37°C using the QIAamp DNA mini kit (Qiagen, Valencia, CA) per manufacturer's instructions. Clostridium perfringens DNA was generously provided by Arpita Ghosh (University of Minnesota). All DNA samples were checked for purity and concentration using the 260/280 ratio provided by the NanoDrop sample retention system (Thermoscientific, Wilmington, DE). All primers with the exception of Clostridium 16SrDNA [61] were designed using Primer3 software (http://frodo.wi.mit.edu/primer3/) (Table 1). PCR was conducted using Hot-Start Taq (Denville, Metuchen, NJ) per manufacturer's instructions. All MAP DNA reactions contained 5% dimethyl sulfoxide (DMSO). The following cycling programs were used for the corresponding genes: IS900 95°C for 15 min., 94°C for 15 s, 58°C for 20 s, 72°C for 20 s and 72°C for 7 min. for 35 cycles, spoIVA 94°C 15 min., 94°C 15 s, 51.5°C for 15 s, 72°C for 30 s, and 72°C for 1 min. for 35 cycles, and Clostridium 16SrDNA 94°C for 15 min., 94°C for 15 s, 52°C for 15 s, 72°C for 30 s and 72°C for 1 min for 35 cycles.
Transmission Electron Microscopy (TEM)
TEM ultrastructural imaging was conducted on MAP log-phase, dormant and isolated spore cultures using the methods described by Ghosh et al. [10]. Dormant cultures and A–K and PEA biomasses of MAP strains K-10, 7565, Linda and Ben were centrifuged at 3,000 rpm and supernatants were discarded. Pellets were fixed with 2.5% glutaraldhyde in 0.1 M sodium cacodylate buffer overnight at 4°C. Samples were washed 3× using 0.1 M sodium cacodylate buffer and post fixed in 1% osmium tetroxide in 0.1 M sodium cacodylate buffer reduced with ferroyanide and washed 3× in autoclaved water. After a series of acetone dehydration, all samples were infiltrated with 1∶2, 2∶1 resin∶acetone mixtures and 3 100% resin. Samples were embedded and cured at 60°C for 48 h and later visualized using TEM.
DPA Assay
DPA was detected using a previously reported colorimetric assay [29]. Wet pellet weight of 250.0 mg was used for each bacterial sample. Autoclaved and “intact” mycobactin J (250 µg/mL) were used as controls. DPA concentrations were calculated based on a DPA (MP Biomedicals, Riverside CA) protein concentration curve. DPA assay was performed six times and all samples were conducted in triplicate with duplicate technical replicates. Figure 2C shows a representative experiment.
Heat Treatment
MAP K-10 spores, B. subtilis spores and C. perfringes spores were heat treated at 70°C and 90°C for 30 min. Additional treatments post heating included exposure to 2% lysozyme at 37°C for 10 min, PK (100 µL of 2 mg/mL) digestion at 37°C for 10 min, kanamycin (50 µg/mL) at 37°C for 2 h and anaerobic exposure at 37°C for 3 weeks. To ensure that MAP cultures were devoid of contaminants post heat treatment, all samples were grown on blood agar at 37°C for 4 weeks. Ten colonies were selected from each MAP heat treatment plate and re-grown in MB7H9 broth at 37°C for 10 weeks. DNA was extracted from re-grown cultures and sequenced for 16SrRNA (Table 1). The universal 16SrRNA cycling program is as follows: 94°C for 15 min., 94°C for 15 s, 54°C for 15 s, 72°C for 30 s and 72°C for 1 min for 30 cycles. 16SrRNA sequences were analyzed using Sequencher (Gene Codes Corporation, Ann Arbor, MI) and aligned using MEGA software (http://www.megasoftware.net/) [62]. Colonies were further submitted to IS900 PCR. All samples were plated twice and heat treatment experiments were conducted three separate times.
RNA Extraction and Qt-RT-PCR
1.0 mL of dormant and log-phase MAP K-10 cultures were centrifuged separately in 1.5 mL eppendorf tubes for 10 min. at 13,000 rpm. Supernatants were decanted and pellets were washed 3× in 1× PBS. 1.0 mL of TRIzol reagent (Invitrogen, Carlsbad, CA) was added to each sample and allowed to incubate at room temperature for 5 min. MAP K-10 samples mixed with TRIZol were homogenized using 0.3 ml of 0.1 mm sterile RNase-free zirconium beads for 4 min. in the MagNa Lyser system (Roche, Basel, Switzerland). RNA was extracted following TRIzol protocol. RNA was subsequently treated with TurboDNase (Ambion, Austin, TX) for 30 min at 37°C. All samples used had a 260/280 ratio of at least 1.9 as measured by NanoDrop sample retention system (Thermoscientific, Wilmington, DE). Primers used for Qt-RT-PCR were designed using Primer3 software (http://frodo.wi.mit.edu/primer3/) (Table 1). Qt-RT-PCR analysis was conducted on 50.0 µg of purified dormant or log-phase MAP K-10 culture combined with the Quantifast one-step RT-PCR reagents (Qiagen, Valencia, CA) using the Lightcycler 480 II (Roche, Basel, Switzerland) programmed for the following: 50°C for 10 min, 95°C for 5 min, 95°C for 10 s, 60°C for 30 s for 40 cycles. Fold change was calculated using the 2−ΔΔCt method. All samples were conducted in triplicate.
Spore Invasion Assay
Bovine MDMs in RPMI containing 2% autologous serum were seeded at 2.0×104 cells/mL in a 24 well plate containing 1.0 No. 1.5 thickness glass coverslips and allowed to adhere for 2 h at 37°C in humidified incubator containing 5% CO2. Following incubation, MDMs were washed 3× in 1× Dulbecco's phosphate buffer saline (D-PBS) to remove non-adherent cells and medium was replaced with fresh serum-free RPMI prior to infection. MAP K-10 subculture and spores and B. subtilis spores were pelleted at 13,000 rpm for 5 min. or 10 min., respectively, washed 2× with warm 1× D-PBS and resuspended in 37°C warmed serum-free RPMI such that a MOI (multiplicity of infection) ratio of 10∶1 was achieved. Serum free RPMI was used to prevent spore germination outside of MDMs. Spores were heat treated at 70°C for 30 min. Subsequently, cultures were vigorously vortexed and allowed to rest for 5 min. at 37°C so that potential clumps would settle to the bottom of the tube. An 18.5 gauge syringe needle was used to repeatedly draw the upper three-fourths of the newly suspended MAP cultures to disperse any remaining clumps. MDMs were separately infected with upper three-fourths of RPMI-MAP/spore cultures for 2 h at 37°C in humidified incubator containing 5% CO2, rinsed 3× with 1× D-PBS and resuspended in RPMI containing 2% autologous serum for the following post infection (p.i.) time points: 0, 0.5, 6, 2, 24, and 48 h. Upon completion of post infection time points, MDMs were rinsed 3× in D-PBS and fluorescently or acid-fast stained for visualization. All time points were conducted in triplicate.
Cell Staining
Fluorescent staining was conducted as stipulated by Lamont et al. [16] with the exception that log-phase and spore cultures were pre-stained for 30 min in 2.0 mg/mL of 5-carboxyfluorescein diacetate (CFDA) (Sigma-Aldrich, St. Louis, MO) at 37°C. Fluorescent images were visualized and collected as a Z-series (step size: 1.0 µm) using DAPI, FITC and Cy5 lasers on an Olympus Fluoview 1000 upright confocal microscope (Olympus, Center Valley, PA). Joint acid-fast images were stained using a modified Zeil-Neelsen protocol (Trend Laboratories Inc., Atlanta, GA) and imaged on an Olympus IX70 inverted fluorescence microscope.
Acknowledgments
This work is dedicated to Dr. Samuel K. Maheswaran, who recently retired after forty-two years of a distinguished academic career at the University of Minnesota. We acknowledge the Veterinary Diagnostic Laboratory and College of Biological Sciences Imaging Center, University of Minnesota for use of microscopy facilities. We thank the University of Minnesota's dairy barn and teaching facility. We also thank Anil Thachil, Arpita Ghosh, and Patricia Goodman, University of Minnesota, for providing Clostridium 16SrDNA primers and C. perfringens culture, Clostridium perfringens DNA, and Bacillus subtilis culture, respectively. We thank Dr. Samuel K. Maheswaran and Dr. Richard Isaacson for their review of this manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by Johne's Disease Integrated Program (USDA-CSREES 2008-55620-18710) awarded to Dr. Sreevatsan. No additional external funding received for this study. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hett EC, Rubin EJ. Bacterial growth and cell division: a mycobacterial perspective. Microbiol Mol Biol Rev. 2008;72:126–156. doi: 10.1128/MMBR.00028-07. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ventura M, Canchaya C, Tauch A, Chandra G, Fitzgerald GF, et al. Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol Mol Biol Rev. 2007;71:495–548. doi: 10.1128/MMBR.00005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bloom BR, McKinney JD. The death and resurrection of tuberculosis. Nat Med. 1999;5:872–874. doi: 10.1038/11309. [DOI] [PubMed] [Google Scholar]
- 4.Gumber S, Taylor DL, Marsh IB, Whittington RJ. Growth pattern and partial proteome of Mycobacterium avium subsp. paratuberculosis during the stress response to hypoxia and nutrient starvation. Vet Microbiol. 2009;133:344–357. doi: 10.1016/j.vetmic.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 5.Russell DG. Mycobacterium tuberculosis: here today, and here tomorrow. Nat Rev Mol Cell Biol. 2001;2:569–577. doi: 10.1038/35085034. [DOI] [PubMed] [Google Scholar]
- 6.Rustad TR, Sherrid AM, Minch KJ, Sherman DR. Hypoxia: a window into Mycobacterium tuberculosis latency. Cell Microbiol. 2009;11:1151–1159. doi: 10.1111/j.1462-5822.2009.01325.x. [DOI] [PubMed] [Google Scholar]
- 7.Cardona PJ. A dynamic reinfection hypothesis of latent tuberculosis infection. Infection. 2009;37:80–86. doi: 10.1007/s15010-008-8087-y. [DOI] [PubMed] [Google Scholar]
- 8.Ehlers S. Lazy, dynamic or minimally recrudescent? On the elusive nature and location of the mycobacterium responsible for latent tuberculosis. Infection. 2009;37:87–95. doi: 10.1007/s15010-009-8450-7. [DOI] [PubMed] [Google Scholar]
- 9.Honer zu Bentrup K, Russell DG. Mycobacterial persistence: adaptation to a changing environment. Trends Microbiol. 2001;9:597–605. doi: 10.1016/s0966-842x(01)02238-7. [DOI] [PubMed] [Google Scholar]
- 10.Ghosh J, Larsson P, Singh B, Pettersson BM, Islam NM, et al. Sporulation in mycobacteria. Proc Natl Acad Sci U S A. 2009;106:10781–10786. doi: 10.1073/pnas.0904104106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Traag BA, Driks A, Stragier P, Bitter W, Broussard G, et al. Do mycobacteria produce endospores? Proc Natl Acad Sci U S A. 2010;107:878–881. doi: 10.1073/pnas.0911299107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anonymous. Johne's disease continues to be the most common cause of bovine enteric disease. Vet Rec. 2008;163:171–174. doi: 10.1136/vr.163.6.171. [DOI] [PubMed] [Google Scholar]
- 13.Whittington RJ, Marshall DJ, Nicholls PJ, Marsh IB, Reddacliff LA. Survival and dormancy of Mycobacterium avium subsp. paratuberculosis in the environment. Appl Environ Microbiol. 2004;70:2989–3004. doi: 10.1128/AEM.70.5.2989-3004.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pribylova R, Slana I, Kaevska M, Lamka J, Babak V, et al. Soil and plant contamination with Mycobacterium avium subsp. paratuberculosis after exposure to naturally contaminated mouflon feces. Curr Microbiol. 2011;62:1405–1410. doi: 10.1007/s00284-011-9875-7. [DOI] [PubMed] [Google Scholar]
- 15.Dhand NK, Toribio JA, Whittington RJ. Adsorption of Mycobacterium avium subsp. paratuberculosis to soil particles. Appl Environ Microbiol. 2009;75:5581–5585. doi: 10.1128/AEM.00557-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lamont EA, Sreevatsan S. Paradigm redux–Mycobacterium avium subspecies paratuberculosis-macrophage interactions show clear variations between bovine and human physiological body temperatures. Microb Pathog. 2010;48:143–149. doi: 10.1016/j.micpath.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 17.Ramakrishnan L, Falkow S. Mycobacterium marinum persists in cultured mammalian cells in a temperature-restricted fashion. Infect Immun. 1994;62:3222–3229. doi: 10.1128/iai.62.8.3222-3229.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamilton HK, Levis WR, Martiniuk F, Cabrera A, Wolf J. The role of the armadillo and sooty mangabey monkey in human leprosy. Int J Dermatol. 2008;47:545–550. doi: 10.1111/j.1365-4632.2008.03722.x. [DOI] [PubMed] [Google Scholar]
- 19.Fukutomi Y, Maeda Y, Matsuoka M, Makino M. Temperature dependency for survival of Mycobacterium leprae in macrophages. Nihon Hansenbyo Gakkai Zasshi. 2009;78:7–16. doi: 10.5025/hansen.78.7. [DOI] [PubMed] [Google Scholar]
- 20.Garcia D, van der Voort M, Abee T. Comparative analysis of Bacillus weihenstephanensis KBAB4 spores obtained at different temperatures. Int J Food Microbiol. 2010;140:146–153. doi: 10.1016/j.ijfoodmicro.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 21.Wekesa VW, Moraes GJ, Ortega EM, Delalibera I., Jr Effect of temperature on sporulation of Neozygites floridana isolates from different climates and their virulence against the tomato red spider mite, Tetranychus evansi. J Invertebr Pathol. 2010;103:36–42. doi: 10.1016/j.jip.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 22.Yang WW, Crow-Willard EN, Ponce A. Production and characterization of pure Clostridium spore suspensions. J Appl Microbiol. 2009;106:27–33. doi: 10.1111/j.1365-2672.2008.03931.x. [DOI] [PubMed] [Google Scholar]
- 23.Kanetsuna F. A Study of Malachite Green Staining of Leprosy Bacilli. Int J Lepr. 1964;32:185–194. [PubMed] [Google Scholar]
- 24.Solari AA, Herrero MM, Painceira MT. Use of malachite green for staining flagella in bacteria. Appl Microbiol. 1968;16:792. doi: 10.1128/am.16.5.792-.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cunningham AF, Spreadbury CL. Mycobacterial stationary phase induced by low oxygen tension: cell wall thickening and localization of the 16-kilodalton alpha-crystallin homolog. J Bacteriol. 1998;180:801–808. doi: 10.1128/jb.180.4.801-808.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Setlow P. I will survive: DNA protection in bacterial spores. Trends Microbiol. 2007;15:172–180. doi: 10.1016/j.tim.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 27.Amaha M, Ordal ZJ, Touba A. Sporulation requirements of Bacillus coagulans var. thermoacidurans in complex media. J Bacteriol. 1956;72:34–41. doi: 10.1128/jb.72.1.34-41.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Daniel RA, Errington J. Cloning, DNA sequence, functional analysis and transcriptional regulation of the genes encoding dipicolinic acid synthetase required for sporulation in Bacillus subtilis. J Mol Biol. 1993;232:468–483. doi: 10.1006/jmbi.1993.1403. [DOI] [PubMed] [Google Scholar]
- 29.Janssen FW, Lund AJ, Anderson LE. Colorimetric assay for dipicolinic acid in bacterial spores. Science. 1958;127:26–27. doi: 10.1126/science.127.3288.26. [DOI] [PubMed] [Google Scholar]
- 30.Ratledge C. Iron, mycobacteria and tuberculosis. Tuberculosis (Edinb) 2004;84:110–130. doi: 10.1016/j.tube.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 31.Filippova SN, Gorbatiuk EV, Poglazova MN, Soina VS, Kuznetsov VD, et al. [Endospore formation by Streptomyces avermitilis in submerged culture]. Mikrobiologiia. 2005;74:204–214. [PubMed] [Google Scholar]
- 32.Stastna J, Goodfellow M, Kristufek V, Novotna J, Jizba J, et al. Characteristics of Streptomyces globisporus strain 0234A forming endospores in submerged cultures. Folia Microbiol (Praha) 1992;37:111–116. doi: 10.1007/BF02836614. [DOI] [PubMed] [Google Scholar]
- 33.Orsburn BC, Melville SB, Popham DL. EtfA catalyses the formation of dipicolinic acid in Clostridium perfringens. Mol Microbiol. 2010;75:178–186. doi: 10.1111/j.1365-2958.2009.06975.x. [DOI] [PubMed] [Google Scholar]
- 34.Robinow CF. Observations on the nucleus of resting and germinating spores of Bacillus megaterium. J Bacteriol. 1953;65:378–382. doi: 10.1128/jb.65.4.378-382.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chiodini RJ, Van Kruiningen HJ, Merkal RS, Thayer WR, Jr, Coutu JA. Characteristics of an unclassified Mycobacterium species isolated from patients with Crohn's disease. J Clin Microbiol. 1984;20:966–971. doi: 10.1128/jcm.20.5.966-971.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chiodini RJ, Van Kruiningen HJ, Thayer WR, Merkal RS, Coutu JA. Possible role of mycobacteria in inflammatory bowel disease. I. An unclassified Mycobacterium species isolated from patients with Crohn's disease. Dig Dis Sci. 1984;29:1073–1079. doi: 10.1007/BF01317078. [DOI] [PubMed] [Google Scholar]
- 37.Sechi LA, Mura M, Tanda E, Lissia A, Fadda G, et al. Mycobacterium avium sub. paratuberculosis in tissue samples of Crohn's disease patients. New Microbiol. 2004;27:75–77. [PubMed] [Google Scholar]
- 38.Di Sabatino A, Paccagnini D, Vidali F, Rosu V, Biancheri P, et al. Detection of Mycobacterium avium subsp. paratuberculosis (MAP)-specific IS900 DNA and antibodies against MAP peptides and lysate in the blood of Crohn's disease patients. Inflamm Bowel Dis. 2011;17:1254–1255. doi: 10.1002/ibd.21461. [DOI] [PubMed] [Google Scholar]
- 39.Naser SA, Ghobrial G, Romero C, Valentine JF. Culture of Mycobacterium avium subspecies paratuberculosis from the blood of patients with Crohn's disease. Lancet. 2004;364:1039–1044. doi: 10.1016/S0140-6736(04)17058-X. [DOI] [PubMed] [Google Scholar]
- 40.Van Brandt L, Coudijzer K, Herman L, Michiels C, Hendrickx M, et al. Survival of Mycobacterium avium ssp. paratuberculosis in yoghurt and in commercial fermented milk products containing probiotic cultures. J Appl Microbiol. 2011;110:1252–1261. doi: 10.1111/j.1365-2672.2011.04979.x. [DOI] [PubMed] [Google Scholar]
- 41.Shankar H, Singh SV, Singh PK, Singh AV, Sohal JS, et al. Presence, characterization, and genotype profiles of Mycobacterium avium subspecies paratuberculosis from unpasteurized individual and pooled milk, commercial pasteurized milk, and milk products in India by culture, PCR, and PCR-REA methods. Int J Infect Dis. 2010;14:e121–126. doi: 10.1016/j.ijid.2009.03.031. [DOI] [PubMed] [Google Scholar]
- 42.Singh SV, Sohal JS, Singh PK, Singh AV. Genotype profiles of Mycobacterium avium subspecies paratuberculosis isolates recovered from animals, commercial milk, and human beings in North India. Int J Infect Dis. 2009;13:e221–227. doi: 10.1016/j.ijid.2008.11.022. [DOI] [PubMed] [Google Scholar]
- 43.Donaghy JA, Totton NL, Rowe MT. Persistence of Mycobacterium paratuberculosis during manufacture and ripening of cheddar cheese. Appl Environ Microbiol. 2004;70:4899–4905. doi: 10.1128/AEM.70.8.4899-4905.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Greenstein RJ. Is Crohn's disease caused by a mycobacterium? Comparisons with leprosy, tuberculosis, and Johne's disease. Lancet Infect Dis. 2003;3:507–514. doi: 10.1016/s1473-3099(03)00724-2. [DOI] [PubMed] [Google Scholar]
- 45.El-Zaatari FA, Osato MS, Graham DY. Etiology of Crohn's disease: the role of Mycobacterium avium paratuberculosis. Trends Mol Med. 2001;7:247–252. doi: 10.1016/s1471-4914(01)01983-9. [DOI] [PubMed] [Google Scholar]
- 46.Tanaka K, Wilks M, Coates PJ, Farthing MJ, Walker-Smith JA, et al. Mycobacterium paratuberculosis and Crohn's disease. Gut. 1991;32:43–45. doi: 10.1136/gut.32.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chacon O, Bermudez LE, Barletta RG. Johne's disease, inflammatory bowel disease, and Mycobacterium paratuberculosis. Annu Rev Microbiol. 2004;58:329–363. doi: 10.1146/annurev.micro.58.030603.123726. [DOI] [PubMed] [Google Scholar]
- 48.Eltholth MM, Marsh VR, Van Winden S, Guitian FJ. Contamination of food products with Mycobacterium avium paratuberculosis: a systematic review. J Appl Microbiol. 2009;107:1061–1071. doi: 10.1111/j.1365-2672.2009.04286.x. [DOI] [PubMed] [Google Scholar]
- 49.Orme IM. The latent tuberculosis bacillus (I'll let you know if I ever meet one). International Journal of Tuberculosis and Lung Disease. 2001;5:589–593. [PubMed] [Google Scholar]
- 50.Hesketh A, Chen WJ, Ryding J, Chang S, Bibb M. The global role of ppGpp synthesis in morphological differentiation and antibiotic production in Streptomyces coelicolor A3(2). Genome Biol. 2007;8:R161. doi: 10.1186/gb-2007-8-8-r161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Schaik W, Prigent J, Fouet A. The stringent response of Bacillus anthracis contributes to sporulation but not to virulence. Microbiology. 2007;153:4234–4239. doi: 10.1099/mic.0.2007/010355-0. [DOI] [PubMed] [Google Scholar]
- 52.Stallings CL, Stephanou NC, Chu L, Hochschild A, Nickels BE, et al. CarD is an essential regulator of rRNA transcription required for Mycobacterium tuberculosis persistence. Cell. 2009;138:146–159. doi: 10.1016/j.cell.2009.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.de Hoon MJ, Eichenberger P, Vitkup D. Hierarchical evolution of the bacterial sporulation network. Curr Biol. 2010;20:R735–745. doi: 10.1016/j.cub.2010.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fischer O, Matlova L, Dvorska L, Svastova P, Bartl J, et al. Diptera as vectors of mycobacterial infections in cattle and pigs. Med Vet Entomol. 2001;15:208–211. doi: 10.1046/j.1365-2915.2001.00292.x. [DOI] [PubMed] [Google Scholar]
- 55.Fischer OA, Matlova L, Bartl J, Dvorska L, Svastova P, et al. Earthworms (Oligochaeta, Lumbricidae) and mycobacteria. Vet Microbiol. 2003;91:325–338. doi: 10.1016/s0378-1135(02)00302-4. [DOI] [PubMed] [Google Scholar]
- 56.Fischer OA, Matlova L, Dvorska L, Svastova P, Peral DL, et al. Beetles as possible vectors of infections caused by Mycobacterium avium species. Vet Microbiol. 2004;102:247–255. doi: 10.1016/j.vetmic.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 57.Pickup RW, Rhodes G, Bull TJ, Arnott S, Sidi-Boumedine K, et al. Mycobacterium avium subsp. paratuberculosis in lake catchments, in river water abstracted for domestic use, and in effluent from domestic sewage treatment works: diverse opportunities for environmental cycling and human exposure. Appl Environ Microbiol. 2006;72:4067–4077. doi: 10.1128/AEM.02490-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Raizman EA, Wells SJ, Jordan PA, DelGiudice GD, Bey RR. Mycobacterium avium subsp. paratuberculosis from free-ranging deer and rabbits surrounding Minnesota dairy herds. Can J Vet Res. 2005;69:32–38. [PMC free article] [PubMed] [Google Scholar]
- 59.Grewal SK, Rajeev S, Sreevatsan S, Michel FC., Jr Persistence of Mycobacterium avium subsp. paratuberculosis and other zoonotic pathogens during simulated composting, manure packing, and liquid storage of dairy manure. Appl Environ Microbiol. 2006;72:565–574. doi: 10.1128/AEM.72.1.565-574.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Coussens PM, Verman N, Coussens MA, Elftman MD, McNulty AM. Cytokine gene expression in peripheral blood mononuclear cells and tissues of cattle infected with Mycobacterium avium subsp. paratuberculosis: evidence for an inherent proinflammatory gene expression pattern. Infect Immun. 2004;72:1409–1422. doi: 10.1128/IAI.72.3.1409-1422.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang MY, Olson BH, Chang JS. Improving PCR and qPCR detection of hydrogenase A (hydA) associated with Clostridia in pure cultures and environmental sludges using bovine serum albumin. Appl Microbiol Biotechnol. 2007;77:645–656. doi: 10.1007/s00253-007-1196-1. [DOI] [PubMed] [Google Scholar]
- 62.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]