Abstract
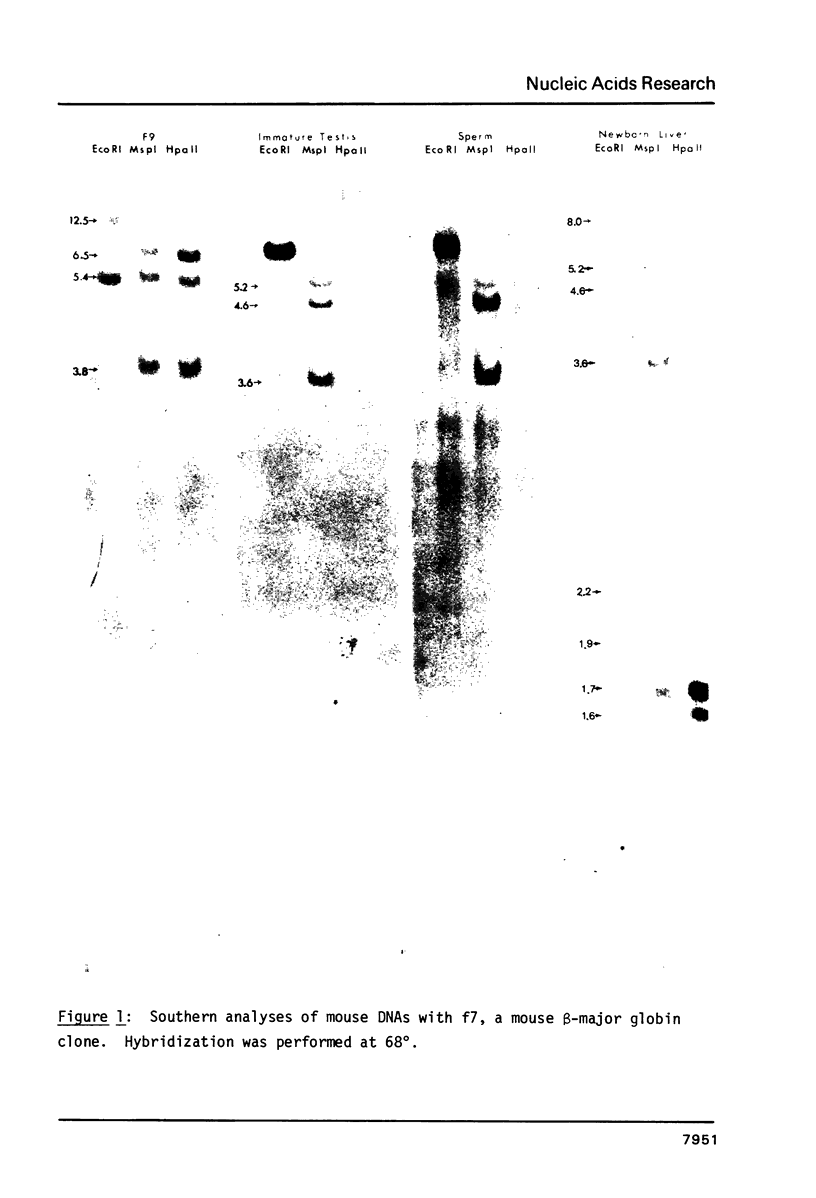
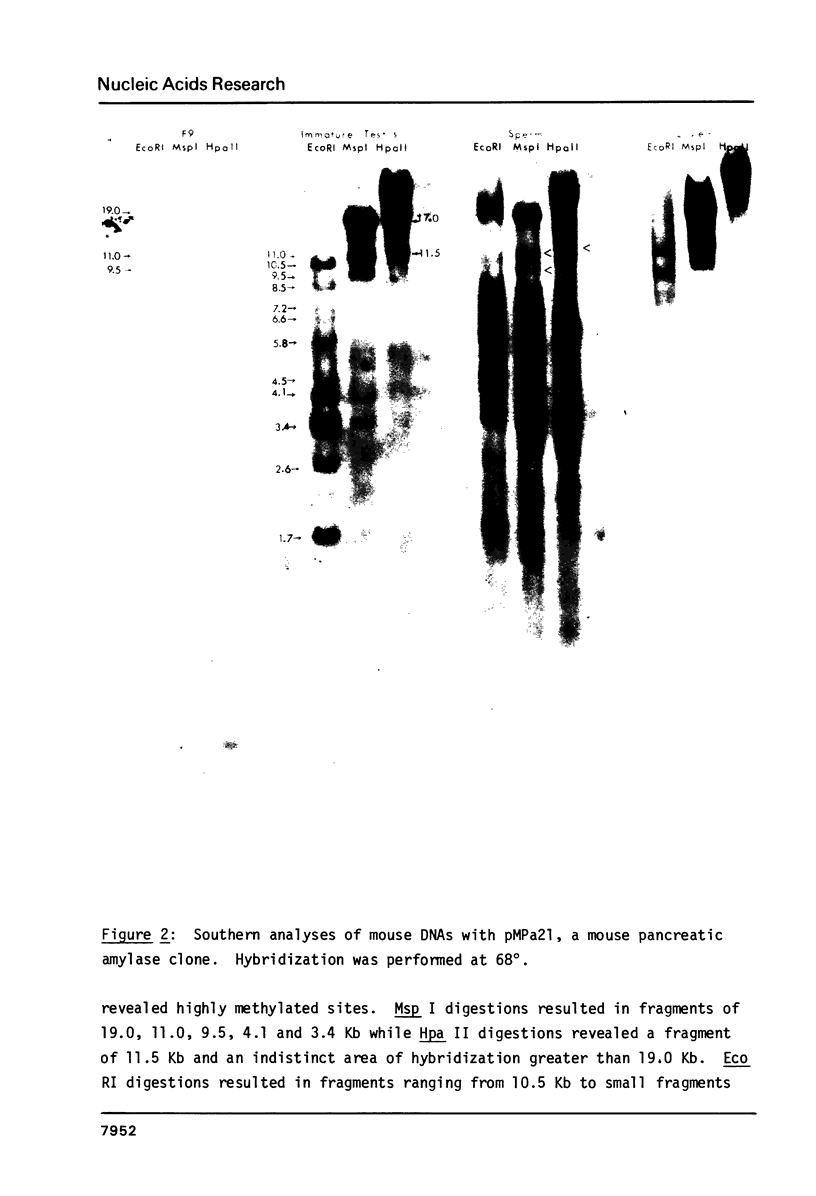
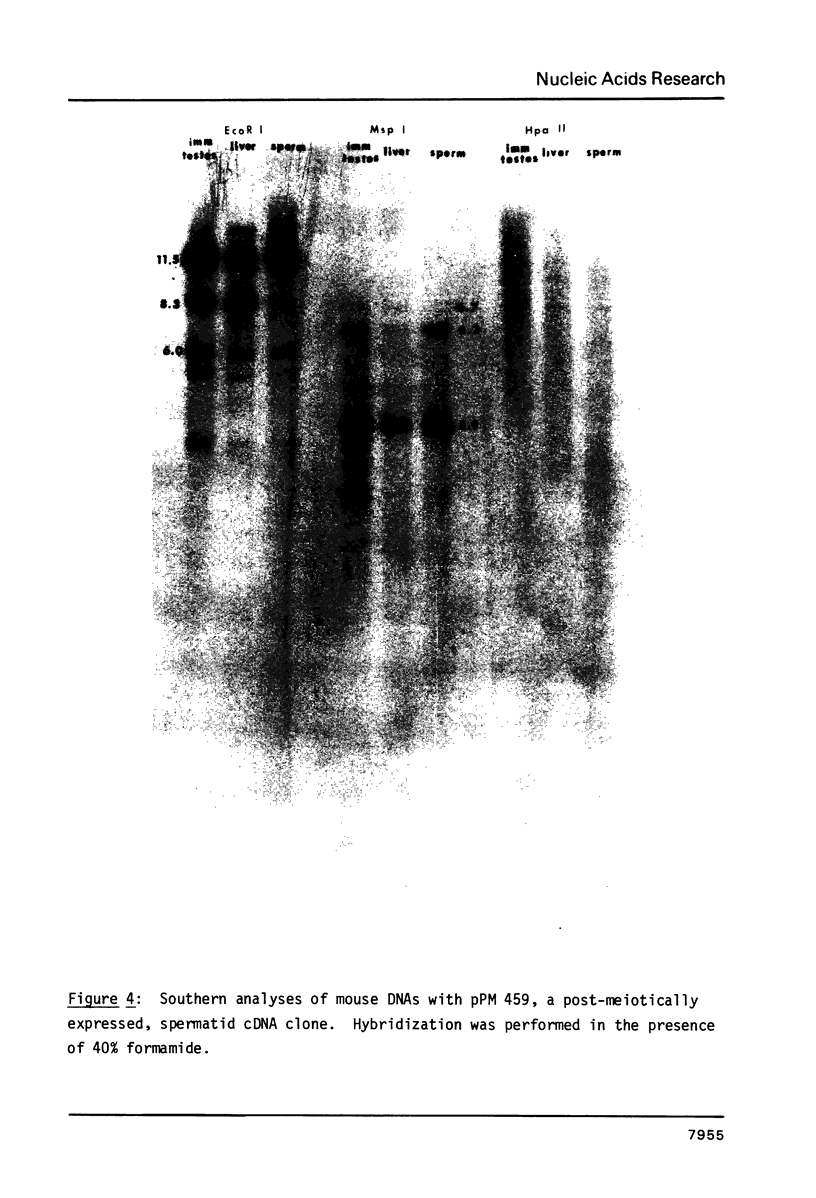
In order to study whether changes in methylation of unique sequence DNA were related to meiosis, DNA was purified from F9 embryonal carcinoma (a "primordial germ cell" equivalent), germ cells from immature testes (containing germ cells up to early spermatocytes), sperm, and appropriate somatic tissues. Restriction was performed with the isoschizomers Msp I and Hpa II, and Eco RI as a control. Electrophoresis and Southern transfers were followed by hybridization to a mouse major beta-globin clone (f7), a mouse pancreatic amylase clone (pMPa21), a type I, histocompatibility-2 clone (pH-2D-4), and a spermatid cDNA clone (pPM 459). The variably methylated sites were all hypomethylated in the embryonal carcinoma DNA and hypermethylated in DNA from immature testes and sperm, irrespective of the transcription state of the gene. The pattern in control tissues generally conformed to an inverse correlation of methylation with transcription. These results suggest that hypermethylation of sperm DNA persists from hypermethylation of these sequences early in testicular development, independent of gene expression.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson D. J., Bach D. L., Yunis E. J., DeWolf W. C. Major histocompatibility antigens are not expressed on human epididymal sperm. J Immunol. 1982 Aug;129(2):452–454. [PubMed] [Google Scholar]
- Artzt K., Dubois P., Bennett D., Condamine H., Babinet C., Jacob F. Surface antigens common to mouse cleavage embryos and primitive teratocarcinoma cells in culture. Proc Natl Acad Sci U S A. 1973 Oct;70(10):2988–2992. doi: 10.1073/pnas.70.10.2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artzt K., Jacob F. Letter: Absence of serologically detectable H-2 on primitive teratocarcinoma cells in culture. Transplantation. 1974 Jun;17(6):632–634. doi: 10.1097/00007890-197406000-00015. [DOI] [PubMed] [Google Scholar]
- DOSKOCIL J., SORM F. Distribution of 5-methylcytosine in pyrimidine sequences of deoxyribonucleic acids. Biochim Biophys Acta. 1962 Jun 11;55:953–959. doi: 10.1016/0006-3002(62)90909-5. [DOI] [PubMed] [Google Scholar]
- Desrosiers R. C., Mulder C., Fleckenstein B. Methylation of Herpesvirus saimiri DNA in lymphoid tumor cell lines. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3839–3843. doi: 10.1073/pnas.76.8.3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerfler W. DNA methylation--a regulatory signal in eukaryotic gene expression. J Gen Virol. 1981 Nov;57(Pt 1):1–20. doi: 10.1099/0022-1317-57-1-1. [DOI] [PubMed] [Google Scholar]
- Ehrlich M., Gama-Sosa M. A., Huang L. H., Midgett R. M., Kuo K. C., McCune R. A., Gehrke C. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res. 1982 Apr 24;10(8):2709–2721. doi: 10.1093/nar/10.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman J., Vitetta E. S. Absence of H-2 antigens capable of reacting with cytotoxic T cells on a teratoma line expressing a T/t locus antigen. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3661–3665. doi: 10.1073/pnas.72.9.3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradin A., Manley J. L., Prives C. L. Methylation of simian virus 40 Hpa II site affects late, but not early, viral gene expression. Proc Natl Acad Sci U S A. 1982 Sep;79(17):5142–5146. doi: 10.1073/pnas.79.17.5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto H., Erickson R. P. Functional assays for mRNA detect many new messages after male meiosis in mice. Biochem Biophys Res Commun. 1982 Oct 29;108(4):1369–1375. doi: 10.1016/s0006-291x(82)80057-0. [DOI] [PubMed] [Google Scholar]
- Gross-Bellard M., Oudet P., Chambon P. Isolation of high-molecular-weight DNA from mammalian cells. Eur J Biochem. 1973 Jul 2;36(1):32–38. doi: 10.1111/j.1432-1033.1973.tb02881.x. [DOI] [PubMed] [Google Scholar]
- Holliday R., Pugh J. E. DNA modification mechanisms and gene activity during development. Science. 1975 Jan 24;187(4173):226–232. [PubMed] [Google Scholar]
- Jagiello G., Tantravahi U., Fang J. S., Erlanger B. F. DNA methylation patterns of human pachytene spermatocytes. Exp Cell Res. 1982 Oct;141(2):253–259. doi: 10.1016/0014-4827(82)90213-0. [DOI] [PubMed] [Google Scholar]
- Jahn C. L., Hutchison C. A., 3rd, Phillips S. J., Weaver S., Haigwood N. L., Voliva C. F., Edgell M. H. DNA sequence organization of the beta-globin complex in the BALB/c mouse. Cell. 1980 Aug;21(1):159–168. doi: 10.1016/0092-8674(80)90123-3. [DOI] [PubMed] [Google Scholar]
- Jeffreys A. J., Flavell R. A. A physical map of the DNA regions flanking the rabbit beta-globin gene. Cell. 1977 Oct;12(2):429–439. doi: 10.1016/0092-8674(77)90119-2. [DOI] [PubMed] [Google Scholar]
- Kaput J., Sneider T. W. Methylation of somatic vs germ cell DNAs analyzed by restriction endonuclease digestions. Nucleic Acids Res. 1979 Dec 20;7(8):2303–2322. doi: 10.1093/nar/7.8.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalanne J. L., Bregegere F., Delarbre C., Abastado J. P., Gachelin G., Kourilsky P. Comparison of nucleotide sequences of mRNAs belonging to the mouse H-2 multigene family. Nucleic Acids Res. 1982 Feb 11;10(3):1039–1049. doi: 10.1093/nar/10.3.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel J. L., Chambon P. DNA methylation: organ specific variations in the methylation pattern within and around ovalbumin and other chicken genes. Nucleic Acids Res. 1979 Dec 20;7(8):2081–2103. doi: 10.1093/nar/7.8.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann M. B., Smith H. O. Specificity of Hpa II and Hae III DNA methylases. Nucleic Acids Res. 1977 Dec;4(12):4211–4221. doi: 10.1093/nar/4.12.4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeady M. L., Jhappan C., Ascione R., Vande Woude G. F. In vitro methylation of specific regions of the cloned Moloney sarcoma virus genome inhibits its transforming activity. Mol Cell Biol. 1983 Mar;3(3):305–314. doi: 10.1128/mcb.3.3.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee J. D., Ginder G. D. Specific DNA methylation sites in the vicinity of the chicken beta-globin genes. Nature. 1979 Aug 2;280(5721):419–420. doi: 10.1038/280419a0. [DOI] [PubMed] [Google Scholar]
- Naveh-Many T., Cedar H. Active gene sequences are undermethylated. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4246–4250. doi: 10.1073/pnas.78.7.4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott M. O., Sperling L., Cassio D., Levilliers J., Sala-Trepat J., Weiss M. C. Undermethylation at the 5' end of the albumin gene is necessary but not sufficient for albumin production by rat hepatoma cells in culture. Cell. 1982 Oct;30(3):825–833. doi: 10.1016/0092-8674(82)90287-2. [DOI] [PubMed] [Google Scholar]
- Pages M., Roizes G. Tissue specificity and organisation of CpG methylation in calf satellite DNA I. Nucleic Acids Res. 1982 Jan 22;10(2):565–576. doi: 10.1093/nar/10.2.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs A. D. X inactivation, differentiation, and DNA methylation. Cytogenet Cell Genet. 1975;14(1):9–25. doi: 10.1159/000130315. [DOI] [PubMed] [Google Scholar]
- Rogers J., Wall R. Immunoglobulin heavy chain genes: demethylation accompanies class switching. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7497–7501. doi: 10.1073/pnas.78.12.7497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schibler U., Tosi M., Pittet A. C., Fabiani L., Wellauer P. K. Tissue-specific expression of mouse alpha-amylase genes. J Mol Biol. 1980 Sep 5;142(1):93–116. doi: 10.1016/0022-2836(80)90208-9. [DOI] [PubMed] [Google Scholar]
- Shen C. K., Maniatis T. Tissue-specific DNA methylation in a cluster of rabbit beta-like globin genes. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6634–6638. doi: 10.1073/pnas.77.11.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman M. I., Miller R. A. F9 embryonal carcinoma cells can differentiate into endoderm-like cells. Dev Biol. 1978 Mar;63(1):27–34. doi: 10.1016/0012-1606(78)90110-0. [DOI] [PubMed] [Google Scholar]
- Shiurba R., Nandi S. Isolation and characterization of germ line DNA from mouse sperm. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3947–3951. doi: 10.1073/pnas.76.8.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmookler Reis R. J., Goldstein S. Variability of DNA methylation patterns during serial passage of human diploid fibroblasts. Proc Natl Acad Sci U S A. 1982 Jul;79(13):3949–3953. doi: 10.1073/pnas.79.13.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer J., Roberts-Ems J., Luthardt F. W., Riggs A. D. Methylation of DNA in mouse early embryos, teratocarcinoma cells and adult tissues of mouse and rabbit. Nucleic Acids Res. 1979 Dec 20;7(8):2369–2385. doi: 10.1093/nar/7.8.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneider T. W. The 5'-cytosine in CCGG1 is methylated in two eukaryotic DNAs and Msp I is sensitive to methylation at this site. Nucleic Acids Res. 1980 Sep 11;8(17):3829–3840. doi: 10.1093/nar/8.17.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stein R., Sciaky-Gallili N., Razin A., Cedar H. Pattern of methylation of two genes coding for housekeeping functions. Proc Natl Acad Sci U S A. 1983 May;80(9):2422–2426. doi: 10.1073/pnas.80.9.2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickland S., Mahdavi V. The induction of differentiation in teratocarcinoma stem cells by retinoic acid. Cell. 1978 Oct;15(2):393–403. doi: 10.1016/0092-8674(78)90008-9. [DOI] [PubMed] [Google Scholar]
- Sturm K. S., Taylor J. H. Distribution of 5-methylcytosine in the DNA of somatic and germline cells from bovine tissues. Nucleic Acids Res. 1981 Sep 25;9(18):4537–4546. doi: 10.1093/nar/9.18.4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanyushin B. F., Tkacheva S. G., Belozersky A. N. Rare bases in animal DNA. Nature. 1970 Mar 7;225(5236):948–949. doi: 10.1038/225948a0. [DOI] [PubMed] [Google Scholar]
- Vardimon L., Kressmann A., Cedar H., Maechler M., Doerfler W. Expression of a cloned adenovirus gene is inhibited by in vitro methylation. Proc Natl Acad Sci U S A. 1982 Feb;79(4):1073–1077. doi: 10.1073/pnas.79.4.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waalwijk C., Flavell R. A. DNA methylation at a CCGG sequence in the large intron of the rabbit beta-globin gene: tissue-specific variations. Nucleic Acids Res. 1978 Dec;5(12):4631–4634. doi: 10.1093/nar/5.12.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H., Larsen A., Groudine M. Alpha-Globin-gene switching during the development of chicken embryos: expression and chromosome structure. Cell. 1981 May;24(2):333–344. doi: 10.1016/0092-8674(81)90323-8. [DOI] [PubMed] [Google Scholar]
- Yagi M., Koshland M. E. Expression of the J chain gene during B cell differentiation is inversely correlated with DNA methylation. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4907–4911. doi: 10.1073/pnas.78.8.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Ploeg L. H., Flavell R. A. DNA methylation in the human gamma delta beta-globin locus in erythroid and nonerythroid tissues. Cell. 1980 Apr;19(4):947–958. doi: 10.1016/0092-8674(80)90086-0. [DOI] [PubMed] [Google Scholar]