Abstract
Activation of many multi-domain signaling proteins requires rearrangement of autoinhibitory interdomain interactions that occlude activator binding sites. In one model for activation, the major inactive conformation exists in equilibrium with activated-like conformations that can be stabilized by ligand binding or post-translational modifications. The molecular basis for this model is established for the archetypal signaling adapter protein Crk-II by measuring the thermodynamics and kinetics of the equilibrium between autoinhibited and activated-like states using fluorescence and NMR spectroscopies, together with segmental isotopic labeling via expressed protein ligation. The results demonstrate that intramolecular domain-domain interactions both stabilize the autoinhibited state and induce the activated-like conformation. A combination of favorable interdomain interactions and unfavorable intradomain structural changes fine-tunes the population of the activated-like conformation and allows facile response to activators. This mechanism suggests a general strategy for optimization of autoinhibitory interactions of multi-domain proteins.
Many autoinhibited signaling proteins are responsive to small changes in the populations of cognate binding partners1–5. The dynamic equilibrium between autoinhibited and activated-like conformations of signaling proteins in the absence of activators is believed to play a key role in achieving the seemingly contradictory tasks of preventing uncontrolled activation and enabling prompt response to cellular stimuli6,7. This model implies that the interdomain interface responsible for autoinhibition must be sufficiently stable to prevent constitutive activation, but sufficiently labile to allow facile kinetic exchange between autoinhibited and activation-competent states.
The conformation of a protein in complex with a target molecule must, in the absence of the target, have a free energy equal to or greater than that of the experimentally observed apo structure. Consequently, the binding affinity for the target must be sufficient to overcome the energetic penalty associated with forming higher energy holo conformations8. The thermodynamics and kinetics of such conformational changes are critical for understanding the physical principles controlling binding affinity; however, detection of low populations of high energy states in the absence of binding partners represents a considerable experimental challenge.
A full description of the molecular mechanisms responsible for controlling the dynamic equilibrium between autoinhibited (closed) and activated-like (open) states requires comparison of the structures and thermodynamics of individual domains in isolation and in the context of full-length molecules, as well as knowledge of the kinetics of interconversion between the various populated conformational states. Moreover, unique spectroscopic probes are needed for studying the individual domains within the full-length multi-domain protein9,10. Consequently, complete descriptions are not yet available of the molecular basis for control of autoinhibition of multi-domain signaling proteins.
Archetypical signaling adaptor proteins of the Crk family are comprised of Src homology 2 (SH2) and 3 (SH3) domains and have been implicated in many cellular processes including cell motility, proliferation and adhesion11–14 (Fig. 1a). Crk proteins mediate diverse protein-protein interactions, through their SH2 and SH3 domains, in downstream transduction of growth and differentiation signals11. Crk-II consists of an N-terminal SH2 domain and two SH3 domains (SH2-nSH3-cSH3). The binding of the N-terminal SH3 (nSH3) domain to C3G, a guanine-nucleotide exchange factor for small molecular weight GTPase15, Abl16 and DOCK18017 contributes to the integrin signaling18 and the regulation of fibroblast migration11. The biological activity of the nSH3 domain of Crk-II is negatively regulated by the C-terminal SH3 (cSH3) domain via intramolecular domain-domain interactions11,19. The solution structure of Crk-II shows that the cSH3 domain stabilizes a closed autoinhibited state of Crk-II in which the ligand binding site of the nSH3 domain is blocked by interdomain interactions with the SH2 domain; however, the active site of the nSH3 domain is not directly occluded by the cSH3 domain19 (Fig. 1b). Alternative splicing of the c-crk gene yields Crk-I (SH2-nSH3), which lacks the C-terminal SH3 domain and is constitutively active, causing transformation of hematopoietic and fibroblast cells12,20. The NMR solution structure of Crk-I indicates that the active site of the nSH3 domain is exposed to solvent19, consistent with the inability of the SH2 domain alone to exert autoinhibitory control of nSH3 activity9. Various models have been proposed for control of autoinhibition of Crk-II19,21,22; however, a complete biophysical analysis of this process, which necessarily involves analysis of the full-length protein and individual domains, has not been reported. Herein, we present a thermodynamic, kinetic and structural analysis that reveals a critical role for selective domain destabilization in tuning the responsiveness of Crk-II to activation.
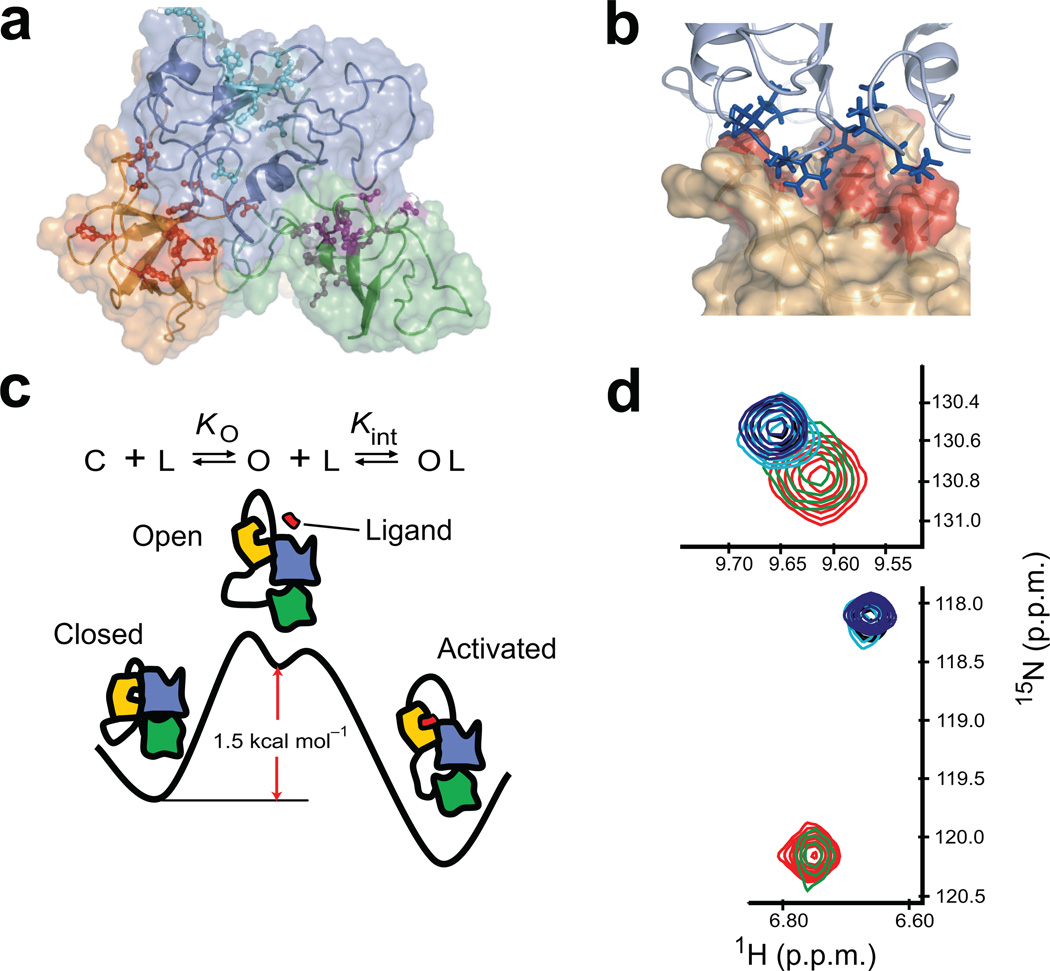
Figure 1. Scheme for the autoinhibition and activation of Crk-II.
(a) Solution structure of Crk-II (SH2; blue, nSH3; yellow, cSH3; green) (pdb id; 2EYZ19). Active site residues of each domain are shown in stick-and-ball format. (b) Expanded view of the interface between the active site of the nSH3 and SH2 domain in Crk-II. Active site residues are shown in red on the surface representation of the nSH3 domain. Interface residues of SH2 domain are shown in stick format. (c) Schematic representation of a three-state equilibrium of ligand binding to the nSH3 domain in Crk-II (SH2; blue, nSH3; yellow, cSH3; green, ligand; red). KO is the equilibrium constant between the closed and open state, and Kint is the intrinsic binding affinity of the ligand to the nSH3 domain in the open state. C and O represent the closed and open state of Crk-II, respectively. L represents the ligand that binds to the nSH3 domain. (d) Change in the chemical shift of W169 (upper panel) and A172 (lower panel) as a function of C3G-ligand concentration. The ratios of protein and ligand are 1:0 (red), 1:0.22 (green), 1:0.63 (cyan), 1:0.79 (black) and 1:1.19 (blue).
Results
Thermodynamics and kinetics of a dynamic equilibrium of Crk-II
Crk-II exists as an equilibrium mixture of the autoinhibited state and the open state, in which the binding site of the nSH3 domain is exposed to solvent (Fig. 1c). Target proteins can bind to the nSH3 domain of Crk-II only when the protein adopts an open state. The free energy of the closed autoinhibited state relative to the open activated-like state of Crk-II was determined from the binding affinity of a C3G-derived23 ligand to the nSH3 domain in Crk-II and Crk-I. The binding affinity (Kint) of the ligand to the nSH3 domain in the open state was estimated using Crk-I (Methods). The free energy difference, ΔG° = G°open − G°closed = 1.5 ± 0.1 kcal mol−1, corresponds to closed and open state populations of pclosed = 92.6% and popen = 7.4%, respectively, for Crk-II (Table 1 and Supplementary Fig. 1).
Table 1.
Binding constants of the C3G-ligand to the nSH3 domain of various states of Crk
| Proteins | KD (nM) |
1Stability (referenced to Crk-I) (kcal mol−1) |
|---|---|---|
| 2Crk-I | 22.5 ± 1.3 | - |
| Crk-II | 285.5 ± 19.0 | 1.5 ± 0.1 |
Stabilities were calculated from KO (Methods)
ref. 8
The kinetics of ligand binding to the nSH3 domain in the context of full-length Crk-II were estimated from an NMR-detected titration experiment using uniformly 15N-labeled Crk-II and fluorescein-labeled C3G-derived ligand. Two 15N resonances in the nSH3 domain, W169 (ΔωN = ωbound − ωfree = 790 s−1) and A172 (ΔωN = 165 s−1), undergo slow and fast chemical exchange, respectively, relative to the NMR chemical shift timescale, upon addition of ligand (Fig. 1d). Therefore, the chemical exchange rate constant is 165 s−1 < kex < 790 s−1, corresponding to a bimolecular on-rate constant of 5.8 × 109 M−1 s−1 < kon < 2.8 × 1010 M−1 s−1 for ligand binding to the nSH3 domain in the open state of Crk-II. The observed KD is 1.9 orders of magnitude lower for fluorescein-labeled C3G ligand than for the unlabeled ligand24. Assuming that koff is unaffected by the presence of the label, 6.5 × 107 M−1 s−1 < kon < 3.1 × 108 M−1 s−1 for an unlabeled ligand is estimated (Methods). These results are in excellent agreement with measured on-rates for ligand binding to isolated SH3 domains25 and thus support use of Crk-I to estimate the intrinsic ligand binding affinity, Kint, for the open state of full-length Crk-II (Fig. 1c and Table 1).
Collectively, these results demonstrate that regulation of the interconversion between the open and closed autoinhibited conformations plays a central role in controlling Crk-II function. These data also show that the stability of the interface with the cSH3 domain limits the activation rate of Crk-II, because autoinhibition must be relieved to populate the open state.
Intra- and inter-domain interactions in the closed state
Considerable basal levels (~7%) of the open states of Crk-II persist, even though a large solvent accessible surface area (~1360 Å2) is buried between the cSH3 and other domains in the structure of autoinhibited Crk-II (Fig. 1a). We quantified the energetics of the interdomain interactions by comparing the stability of the cSH3 domain in isolation and in full-length Crk-II. The unfolded cSH3 domain cannot bind to the other domains of Crk-II; consequently, unfolding and dissociation of the cSH3 domain are thermodynamically linked. The effect of ligand binding on global unfolding, by monitoring the change in Tm or H/D-exchange, has been reported for other isolated SH3 domains26,27.
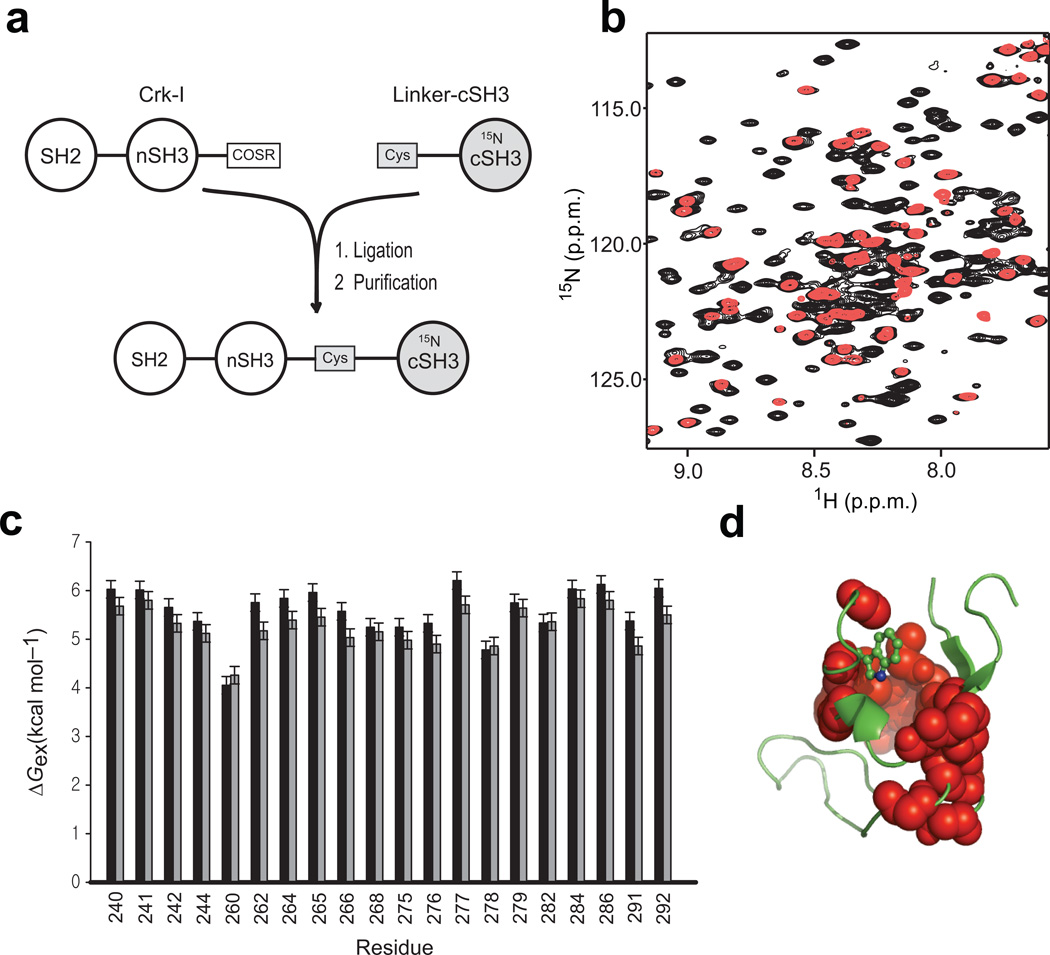
NMR-detected backbone amide H/D-exchange experiments are routinely used to measure the unfolding free energy of small to intermediate sized proteins (<25 kDa); however, peak overlap makes the application of the H/D-exchange experiment difficult for large (>30 kDa) proteins. We used expressed protein ligation (EPL)28 to produce segmentally labeled full-length Crk-II, in which only the cSH3 domain is labeled with 15N (SH214N-nSH314N-cSH315N) (Fig. 2a). Owing to the simplified NMR spectrum, all of the expected 1H-15N resonances in the cSH3 domain are detected in segmentally labeled full-length Crk-II (~34 kDa) (Fig. 2b and Supplementary Fig. 2).
Figure 2. NMR analysis of the cSH3 domain within full-length Crk-II.
(a) Schematic diagram outlining preparation of segmentally labeled Crk-II (residues from 208 to 304 are 15N-labeled). (b) Overlay of the 1H-15N HSQC spectrum of uniformly 15N-labeled Crk-II (black) and segmentally labeled Crk-II (red). (c) NMR-detected H/D-exchange free energy of unfolding of the cSH3 domain in isolated (black), and Crk-II (grey) The error bars represent standard deviation of three repeated experiments. (d) Residues detected in the H/D-exchange experiment are shown as spheres on the structure of the cSH3 domain.
H/D-exchange rates were measured at 290 K for 20 residues (Fig. 2c), located in and around the hydrophobic core of the cSH3 domain (Fig. 2d). Strikingly, the protection factors are nearly equal or larger in the isolated cSH3 domain (average ΔGex = 5.58 ± 0.52 kcal mol−1) relative to the values measured for the cSH3 domain within full-length Crk-II (average ΔGex = 5.29 ± 0.41 kcal mol−1) (Fig. 2c). 1H and 15N chemical shifts (Fig. 3a and 3b) and 15N transverse relaxation rate measurements (Fig. 3c) demonstrate that the similarity in the protection factors of the cSH3 domain in isolation and in the full-length Crk-II are not due to dissociation of the cSH3 domain from the other domains in Crk-II. These results indicate that the cSH3 domain does not experience any net favorable interdomain binding free energy, even though a large surface area is buried in the autoinhibited state.
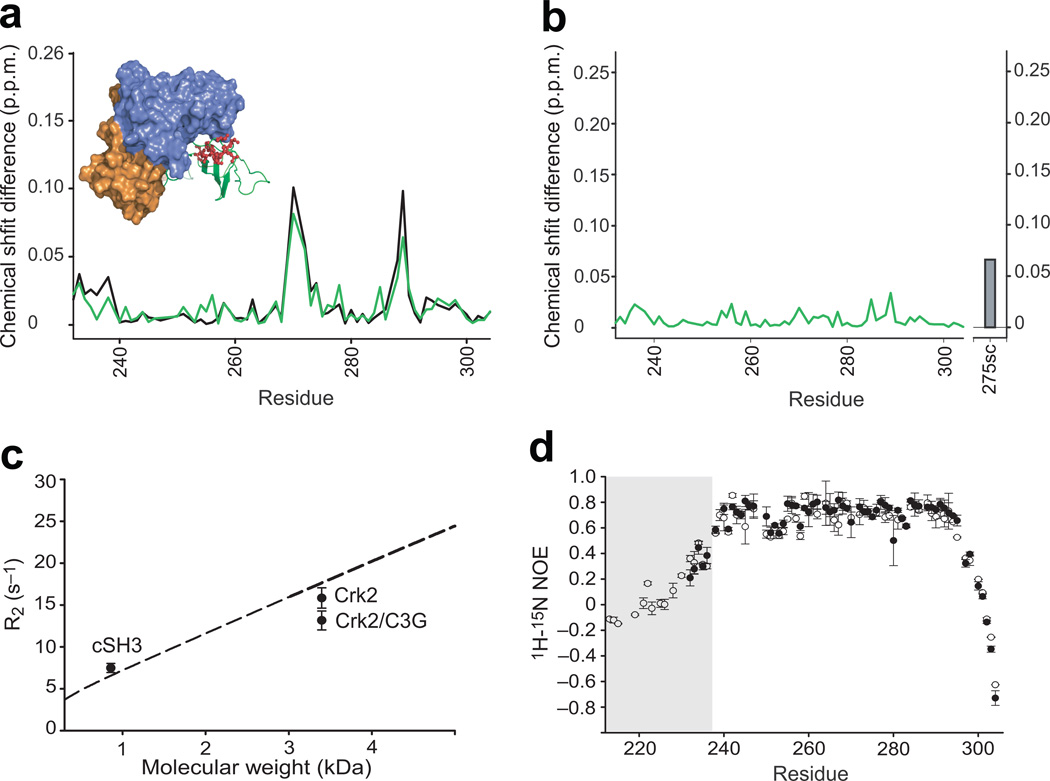
Figure 3. Structural and dynamic analysis of the cSH3 domain within Crk-II.
(a) Chemical shift differences (Δδ) of the cSH3 domain in Crk-II (black), and C3G-ligand bound Crk-II (green), relative to the isolated cSH3 domain. The inset shows the locations of the residues whose resonances changed markedly between isolated cSH3 and Crk-II (top 25% in the deviation plot). (b) Chemical shift differences of the cSH3 domain in C3G-ligand bound Crk-II, compared to those within Crk-II. The difference for the 15Nε1 resonance of Trp275 is shown as a bar. (c) Comparison of average R2 rates of the cSH3 domain in isolation (Mw=8,569 Da), in Crk-II, and in Crk-II/C3G (~33,830 Da). The dashed line represents the molecular weight dependent R2 rates calculated using isotropic rotational correlation times calculated from Stokes’ law. Error bars represent the standard deviation of average R2. (d) {1H}-15N heteronuclear NOE measurements for the cSH3 domain in isolation, residues 232–304, (closed circles) and within Crk-II, residues 208–304, (open circles). The residues corresponding to the linker region, residues 208–236, are shaded in the plot. Error bars represent the propagated uncertainties of two repeated experiments. The uncertainty was estimated using background noise of the spectrum.
We characterized the conformational properties of the linker region between the nSH3 and cSH3 domains, residues 224–237, in full-length Crk-II. Heteronuclear {1H}-15N NOE values are < 0.6 for residues in the linker, compared with values >0.6 for residues in elements of secondary structure, in full-length Crk-II and are similar to values measured for the isolated cSH3 domain (Fig. 3d). We did not observe any resonance cross-peaks in the first NMR spectrum in H/D exchange experiments for residues in the linker region in full-length Crk-II, indicating that amide proton exchange protection factors are small. We note that segmentally labeled protein must be used to measure accurate NMR H/D exchange and spin relaxation parameters for the linker region owing to the serious spectral overlap in uniformly labeled Crk-II, which prevents quantitative peak intensity measurements (Supplementary Fig. 3). Thus, thermodynamically unfavorable interactions must occur simultaneously in the cSH3 domain, and not the linker, upon formation of the thermodynamically favorable domain-domain interface of the autoinhibited state.
Conformational change of Trp275 within the cSH3 domain
We investigated the structural changes in the cSH3 domain that may offset favorable interdomain interactions, and thereby allow facile activation of Crk-II. The backbone 15N and 1H chemical shifts of the cSH3 domain in the context of autoinhibited Crk-II and activated Crk-II (Crk-II/C3G) are broadly similar (Fig. 3b), indicating that modest conformational changes around the interdomain interface of the cSH3 domain are sufficient to induce the open conformation of Crk-II. The 15Nε1 resonance for the indole ring of Trp275 of the cSH3 domain has the largest change upon activation (Fig. 3b and Supplementary Fig. 2). Trp275 is highly conserved in Crk-II (Supplementary Fig. 4), and is critical for stabilizing the cSH3 domain and the autoinhibited state of Crk-II19,29. H/D exchange experiments show that side-chain indole 1Hε1 of Trp275 is well protected from solvent in the isolated cSH3 domain (Supplementary Fig. 5a and 5c), but not in full-length Crk-II (Supplementary Fig. 5b and 5d). We performed fluorescence experiments to confirm that Trp275 is protected from solvent exposure in the isolated cSH3 domain30,31. The Trp fluorescence emission maximum is blue-shifted compared with free indole (Table 2). We also compared the Stern-Volmer constant (Ksv) of the isolated cSH3-WT domain to free indole (Table 2). The fluorescence results show that Trp275 has a single conformation in which the side chain is buried into the hydrophobic core. These results imply that domain-domain interactions in full-length Crk-II perturb the hydrophobic core of the cSH3 domain by switching Trp275 to a more exposed conformation.
Table 2.
Thermodynamic and fluorescence quenching parameters of the wild type cSH3 domain and the cSH3 domain which contains 7AW at position 275.
| Proteins | ΔG° (kcal mol−1) |
m (kcal mol−1M−1) |
Ksv (M−1) |
λmax (nm) |
|---|---|---|---|---|
| cSH3-WT | 3.3 ± 0.3 | 0.7 ± 0.1 | 6.7 ± 0.2 | 325 |
| Indole | - | - | 34.2 ± 1.0 | 340 |
| cSH3-7AW | 2.5 ± 0.4 | 0.5 ± 0.1 | 4.7 ± 1.31 0.001 ± 0.0052 |
373 |
| 7-azaindole | - | - | 5.0 ± 0.1 | 382 |
Stern -Volmer constant of exposed species deduced from the fit of the data.
Stern -Volmer constant of buried species deduced from the fit of the data.
The solvent-protection of the side chain of Trp275 is consistent with solution structure of the isolated cSH3 domain (PDB code; 2GGR). The indole ring of Trp275 is packed against several hydrophobic residues, resulting in the burial of 1117 Å2 of hydrophobic surface area32. In contrast, Trp275 populates a different χ1 and χ2 rotamer in full-length Crk-II and the indole ring is flipped toward the binding interface with the SH2 domain (Fig. 4a).
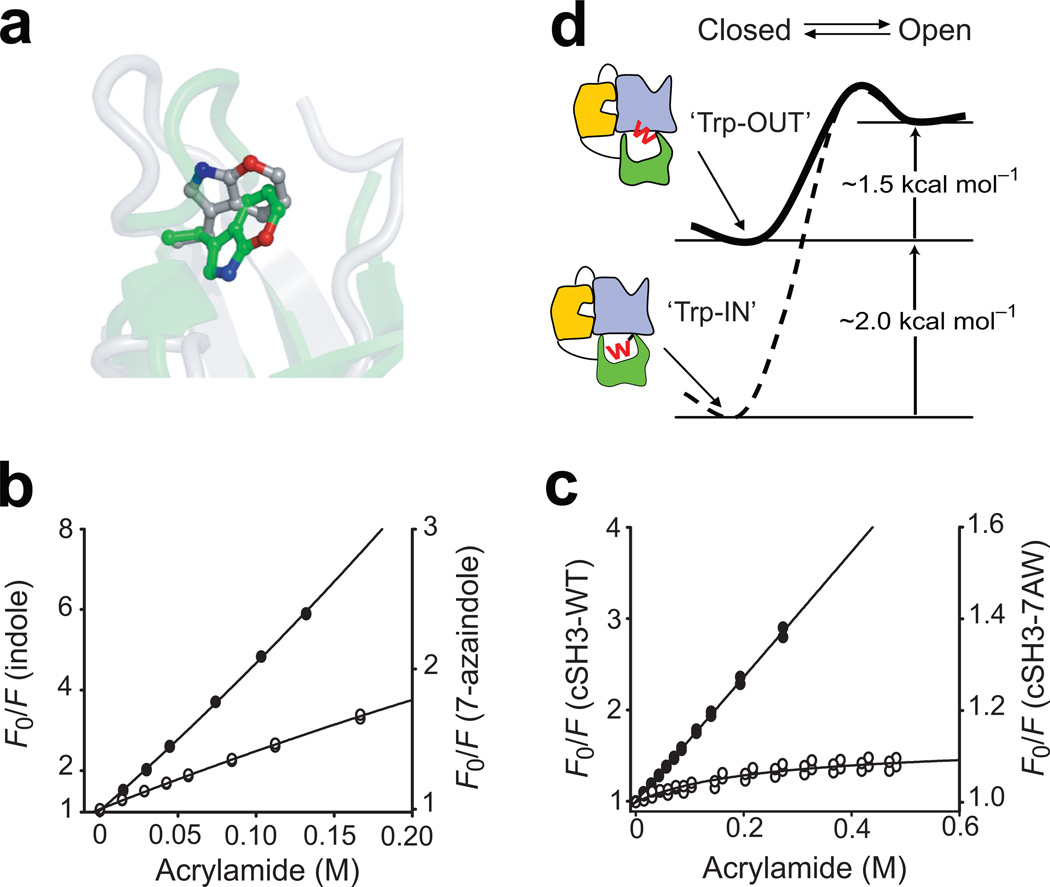
Figure 4. Conformation of Trp275 modulates the stability of the cSH3 domain.
(a) Comparison of the position of the indole ring of Trp275 in the structures of isolated cSH3 domain (green, pdb id: 2GGR32) and full-length Crk-II (silver, pdb id: 2EYZ19). The 7-position of the Trp residue is highlighted in red. Steady-state fluorescence quenching experiments for (b) free indole (closed circles) and 7-azaindole (open circles) and (c) isolated cSH3-WT (closed circles) and isolated cSH3-7AW (open circles). The solid lines represent the best fit model using the Stern-Volmer relationship (Methods). (d) Schematic representation of the effects of the conformational change of Trp275 on the equilibrium between the open and closed states of Crk-II (W represents Trp275). Reduced stability of the cSH3 upon interdomain interactions reduces the activation barrier between the open and closed states of Crk-II.
Rotameric mutation of Trp275 using non-coded amino acid
We sought to design a mutant of cSH3 that would mimic the conformation of Trp275 observed in full-length Crk-II. The energetic consequences of local conformational changes of a single Trp residue are difficult to evaluate using traditional mutagenesis, because substitutions conserving size and molecular structure cannot be made using natural amino acids. We selectively replaced Trp275 in the isolated cSH3 domain by the non-natural amino acid 7-aza Trp (7AW), in which the carbon atom in the 7-position of indole ring is replaced by nitrogen. This mutation does not perturb the structure of the side chain, except for the increased polarity of the ring. The 7-position of the Trp275 side chain is buried in the hydrophobic core of the isolated cSH3 and we reasoned that the side chain of 7AW would reorient to expose the 7-position to solvent, mimicking the effects of changing the χ1 rotamer of Trp275 in Crk-II (Fig. 4a). Thus the 7AW substitution can be used as a “rotameric mutation”. The effect of 7AW on the stability of the cSH3 domain was measured using urea-induced denaturation. The stability of the isolated cSH3-7AW domain was decreased by almost ~1 kcal mol−1 relative to wild type isolated cSH3 (Table 2 and Supplementary Fig. 6). The m-value also decreased, indicating that the hydrophobic core is slightly perturbed in cSH37AW, compared to wild-type cSH3.
We performed fluorescence quenching experiments to confirm that the 7AW substitution perturbed the rotameric conformational state of Trp27530,31. We compared the Ksv of the isolated cSH3-7AW domain to free 7-azaindole, because of the difference in intrinsic quenching efficiencies of W and 7AW (Table 2 and Fig. 4b and 4c). The result for the cSH3-7AW domain shows that the protein populates major (~89%) and minor (~11%) conformations corresponding to the buried and exposed conformation of the side chain of 7AW, respectively. In addition, the 1H-15N HSQC spectrum of the cSH3-7AW showed that several resonances from residues near 7ATrp275 are split into two peaks, further confirming the presence of two different conformations (data not shown). In contrast, the spectrum of the isolated wild type cSH3 domain showed single resonance peaks for each residue. These results demonstrate that the incorporation of 7AW induces the rotation of the side chain toward the surface of the cSH3 domain (Fig. 4a). We used these data in combination with the unfolding free energy of the cSH3-7AW ensemble to estimate an energetic penalty of 2.04 ± 0.38 kcal mol−1 for rotation of Trp275 in the autoinhibited form of Crk-II (for details see Supplementary Methods). The substantial energetic penalty incurred when the side chain of Trp275 flips out to enable favorable interdomain interactions provides an explanation for the absence of stabilization of the cSH3 domain in the context of full-length Crk-II.
Discussion
Quantifying the energetics of the structural changes upon domain-domain interactions is central to understanding signaling processes at the molecular level. We have developed a self-consistent thermodynamic and kinetic model for the control of the autoinhibition of Crk-II in which favorable domain-domain interactions are balanced by structural changes, including perturbations of Trp275, that reduce the stability of the cSH3 domain, in order to avoid over-stabilization of the autoinhibited state (Fig. 4d). The moderate conformational change of the side chain of Trp275 destabilizes the cSH3 domain by a surprisingly large ~2 kcal mol−1. However, packing interactions in the hydrophobic core of SH3 domains are highly conserved and domain stability is very sensitive to perturbations of the hydrophobic core33.
The hypothetical binding energy between the cSH3 and SH2 domains of Crk-II that would be expected if exposure of the Trp275 side chain were not energetically unfavorable is important in assessing whether additional conformational changes contribute to the free energy difference between the closed and open states. The intramolecular binding energy between the two domains can be estimated using the relationship Ka,intra = Ka,inter*Ceffective, in which Ceffective is the effective local concentration of the binding domain and Ka,inter is the intermolecular binding affinity34. The intermolecular binding constant between the cSH3 and SH2 domains of Crk-II is not known; however, the Abl SH3 domain binds to a proline-rich peptide sequence derived from the DE-loop of the Crk-II SH2 domain with Ka,inter ~360 µM35. A theoretical investigation of the intramolecular interaction of SH3 domains showed that the effective local concentration is ~ 80 mM when two domains are separated by the optimum linker length34. Thus, the hypothetical intramolecular binding energy, in which the unfavorable interaction of Trp275 is not considered, would be ~ −3.2 kcal mol−1. This result is consistent with our estimation of free energy difference between the closed state and open states in which the Trp275 side chain is buried (Fig. 4d). Thus, the offset of the favorable interdomain interaction (~ −3.2 kcal mol−1) by unfavorable conformational change from buried to exposed conformations of Trp275 (~2 kcal mol−1) reduces the free energy difference between the closed and open states to ~1.2 kcal mol−1. The self-consistency between this latter quantity and the difference measured independently (Table 1) indicates that the conformational change associated with Trp275 is the dominant contribution to tuning the free energy difference between autoinhibited and activated-like states of Crk-II. The intramolecular binding free energy of the cSH3 domain to other regions of Crk-II is relatively small, compared to other SH3-ligand interactions, perhaps because the intramolecular binding interface of Crk-II is not optimized for ideal binding36.
Two other models have been proposed for control of autoinhibition of Crk-II. In the first model, autoinhibition of chicken Crk-II is controlled by interactions between the nSH3 and cSH3 domain mediated via isomerization of Pro23737. However, that work made use of a truncated Crk construct (nSH3-cSH3) that lacked the SH2 domain. The nSH3 and cSH3 domains do not directly interact in the structure of the full-length human protein19. In addition, the amino acid sequence adjacent to Pro237 in the chicken Crk-II sequence differs from the corresponding human and mouse sequences (Supplementary Fig. 4). These differences suggest that the regulatory mechanism of chicken Crk-II may be different from that of human and mouse Crk-II. In the second model, the autoinhibited state of human Crk-II is stabilized by rigidification of the linker region (residues 224–236) tethering the nSH3 and cSH3 domains19. However, heteronuclear NOE data (Fig. 3d) indicate that the linker region experiences extensive conformational fluctuations on picosecond-nanosecond time scales. In addition, the H/D exchange data indicate that the linker region undergoes slower interconversion between associated (exchange non-competent) and dissociated (exchange competent) states with minor free energy differences.
In conclusion, intramolecular domain-domain interactions induce differential effects on the thermodynamic stabilities of individual domains to finely adjust the overall functional response of Crk-II. The interplay of these competing effects acts as the driving force to induce the basal level of the open (activated-like) state required for activation of Crk-II. The thermodynamically unfavorable conformational change of the cSH3 domain upon autoinhibition contributes to timely activation in response to cellular stimuli. The mode of regulation revealed by the present study is likely to be found in other multi-domain proteins. In the limiting case, order-to-disorder transitions may maximally offset favorable domain-domain interactions38.
Methods
Protein Expression and Purification
Detailed procedures for preparation of protein samples are described in Supplementary Methods. Intactness of segmentally labeled Crk-II was confirmed by comparing the NMR chemical shifts and R2 relaxation rate constants with those of uniformly 15N labeled Crk-II which was prepared in conventional protein preparation method (Supplementary Fig. 7). Isolated cSH3 domain was confirmed to be monomeric by measuring the concentration dependent NMR transverse relaxation rate constants (Supplementary Fig. 7).
Ligand Binding Assays
A fluorescence-based titration assay was used to measure the affinity constants of both C3G-ligand15 and a phosphopeptide-ligand39. All ligand binding measurements were performed at 25°C in a stirred 1 cm pathlength cuvette. The excitation wavelength was 295 nm. C3G-ligand, HPPPPLPPKRRK-ε-fluorescein-G-NH2, was prepared by solid-phase peptide synthesis. Dissociation constants were calculated by assuming the formation of a 1:1 complex and by fitting corrected fluorescence intensities to the equation;
| (1) |
where F and ΔF are the change and maximum change in protein fluorescence, respectively. P is the total protein concentration, L is the total ligand concentration, and KD is the equilibrium dissociation constant. Experiments were performed in triplicate.
The free energy difference between the open and closed states of Crk-II was calculated using eq (2). In our three state binding model (Fig. 1c), the equilibrium constant (KO) for the opening and closing process between the closed and open states of Crk-II was calculated from the apparent binding affinity (KD) of the C3G ligand and the binding affinity (Kint) of C3G ligand:
| (2) |
where, and
| (3) |
Determination of the bimolecular rate constant, kon
The bi-molecular rate constant, kon, was determined from the exchange rate constant, kex, estimated from a NMR-titration experiment. The kex for the ligand binding to the Crk-II is defined by the equation:
| (4) |
where kon and koff represent bimolecular on-rate and unimolecular off-rate constants, respectively, between the open and ligand-bound states (Fig. 1c), [L] represents the free ligand concentration, and Kint represents the dissociation constant of C3G-ligand to Crk-I. The reported kon was calculated with KO of 0.074 (popen = 7.4% in Crk-II) and [L]free = 0.077 µM (protein:ligand = 1:0.22). The NMR titration experiment (Fig. 3c) showed that the exchange rate is 165s−1 < kex < 790 s−1 at this protein:ligand ratio resulting in 5.8 × 109 M−1 s−1 < kon < 2.8 × 1010 M−1 s−1. The observed KD for fluorescein-labeled C3G-ligand is 85-fold lower than fluorescein-free C3G-ligand24. Despite this effect of fluorescein, the label is necessary to quantify the concentration of ligand in solution because the ligand sequence lacks spectroscopic probes and measurements of peptide concentrations using amino acid analysis have large errors. Thus, the observed kon rate was corrected by the ratio of KD for label-free and fluorescein-labeled ligand. The correction yields estimates of 6.5 × 107 M−1 s−1 < kon < 3.1 × 108 M−1 s−1.
NMR spectroscopy
A Bruker DRX600 NMR spectrometer equipped with a CryoProbe was used. Chemical shifts for different protein states and complexes were measured in a buffer containing 10 mM sodium phosphate (pH 7.0), 100 mM NaCl, 2 mM DTT at 298 K. Chemical shift changes were calculated as:
| (5) |
The 1H-15N NOE spectra were collected for segmentally labeled Crk-II (SH214N-nSH314N-cSH315N) and for the isolated cSH3 domain with and without 1H saturation for 4.0 s at 600 MHz equipped with a CryoProbe. Recycle delays for NOE experiment and control experiments were 3.0 s and 9.0 s, respectively. The delay between NOE and control experiments was 18.0 s. 1H-15N HSQC, 3D-HNCO, HNCACB, CBCA(CO)NH, and HN(CA)CO experiments were used for backbone assignment.
Equilibrium Unfolding Measurements
Urea-denaturation experiments were performed at 298 K by monitoring the change in fluorescence signal intensity recorded at 340 nm and 398 nm for wild type cSH3 and cSH3-7AW, respectively. Details of experimental conditions are described in Supplementary Methods.
H/D Exchange Rate Measurements
H/D exchange experiments were performed at 290 K on a Bruker DRX 600 MHz NMR spectrometer equipped with a CryoProbe. Samples for H/D exchange experiments were exchanged into a buffer containing 10 mM sodium phosphate (pD 6.6), 200 mM NaCl, 2 mM DTT (d11) in 100% D2O, using Micro Bio-spin columns (BIO-RAD). H/D-exchange rates were determined from peak intensity changes in a series of 1H-15N HSQC experiments. Reported exchange rates are the average of two or three repeated experiments. Exchange rates for residues whose exchange was dominated by local conformational fluctuations could not be determined, because the intrinsic rate of exchange is too high at the experimental pH (pD = 6.6). The folding rate for the cSH3 domain has previously been measured to be 82.6 ± 2.9 s−1,32; thus the system is within the EX2 regime.
Fluorescence Quenching Experiments
Steady state quenching experiments were performed at 298K. Samples were prepared in a buffer containing 20mM Na-phosphate (pH 7.2), 100mM NaCl, 3mM TCEP. The concentration of each sample was adjusted to ensure that the OD was 0.1 or less at the excitation wavelength to prevent inner filter effects. Acrylamide was used as a quencher. Observed signal intensity was corrected for both dilution effects and for the inner filter effect upon adding acrylamide using OD295nm = 0.23 M−1 cm−1. The excitation wavelengths for cSH3-WT (and free indole) and cSH3-7AW (and 7-azaindole) were 295 nm and 310 nm, respectively. Quenching data for homogenously emitting samples (free indole, 7-azaindole and cSH3-WT) were analyzed using the Stern-Volmer equation30:
| (6) |
For cSH3-7AW, the Stern-Volmer equation was re-written to take into account heterogeneous fluorescent components31:
| (7) |
where F0 and F represent fluorescence intensity in the absence and presence of quencher, Q, respectively. Ksv,1 and f1 is the Stern-Volmer constant and population fraction of the exposed conformation and Ksv,2 is the Stern-Volmer constant of the buried conformation. V represents the sphere of action. Application of equation 7 to the quenching data of the cSH3-WT domain yielded unrealistic fitting, indicating that equation 6 is adequate for fitting the data.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health (NIH) grants GM070941 (D.P.R.), EB001991 (T.W.M.), GM55843 (T.W.M.), and GM59273 (A.G.P.). D.P.R., T.W.M., and A.G.P. are members of the New York Structural Biology Center (NYSBC) supported by NIH grant GM66354. We thank Drs. Kaushik Dutta and Shibani Bhattacharya for their help in collecting NMR data at NYSBC.
Footnotes
Author Contributions J.H.C. designed and performed all experiments, analyzed the data, and wrote paper. V.M. and M.V.P. contributed to synthesis of the ligands and preparation of the segmentally labeled protein. D.P.R., T.W.M., and A.G.P. designed the experiments, analyzed the data, and wrote paper.
References
- 1.Schlessinger J. Autoinhibition Control. Science. 2000;300:750–752. doi: 10.1126/science.1082024. [DOI] [PubMed] [Google Scholar]
- 2.Ferguson KM, Berger MB, Mendrola JM, Cho HS, Leahy DJ, Lemmon MA. EGF activates its receptor by removing interactions that autoinhibit ectodomain dimerization. Mol. Cell. 2003;11:507–517. doi: 10.1016/s1097-2765(03)00047-9. [DOI] [PubMed] [Google Scholar]
- 3.Smock RG, Gierasch LM. Sending Signals dynamically. Science. 2009;324:198–203. doi: 10.1126/science.1169377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen L, et al. Structural insight into the autoinhibition mechanism of AMP-activated protein kinase. Nature. 2009;459:1146–1149. doi: 10.1038/nature08075. [DOI] [PubMed] [Google Scholar]
- 5.Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–452. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- 6.Yao X, Rosen MK, Gardner KH. Estimation of the available free energy in a LOV2-Jα photoswitch. Nat. Chem. Biol. 2008;4:491–497. doi: 10.1038/nchembio.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li P, Martins IRS, Amarasinghe GK, Rosen MK. Internal dynamics control activation and activity of the autoinhibited Vav DH domain. Nat. Struct. Biol. 2008;15:613–618. doi: 10.1038/nsmb.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miloushev VZ, et al. Dynamic properties of a type II cadherin adhesive domain: implications for the mechanism of strand-swapping of classical cadherins. Structure. 2008;16:1195–1205. doi: 10.1016/j.str.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muralidharan V, et al. Domain-specific incorporation of noninvasive optical probes into recombinant proteins. J. Am. Chem. Soc. 2004;126:14004–14012. doi: 10.1021/ja0466199. [DOI] [PubMed] [Google Scholar]
- 10.Tang L, Roulhac PL, Fitzgerald MC. H/D exchange and mass spectrometry-based method for biophysical analysis of multidomain proteins at the domain level. Anal. Chem. 2007;79:8728–8739. doi: 10.1021/ac071380a. [DOI] [PubMed] [Google Scholar]
- 11.Feller SM. Crk family adaptors-signalling complex formation and biological roles. Oncogene. 2001;20:6348–6371. doi: 10.1038/sj.onc.1204779. [DOI] [PubMed] [Google Scholar]
- 12.Nojima Y, et al. Integrin-mediated cell adhesion promotes tyrosine phosphorylation of p130Cas, a Src homology 3-containing molecule having multiple Src homology 2-binding motifs. J. Biol. Chem. 1995;270:15398–15402. doi: 10.1074/jbc.270.25.15398. [DOI] [PubMed] [Google Scholar]
- 13.Schaller MD, Parsons JT. pp125FAK-dependent tyrosine phosphorylation of paxillin creates a high-affinity binding site for Crk. Mol. Cell. Biol. 1995;15:2635–2645. doi: 10.1128/mcb.15.5.2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parsons JT, Schaller MD. Focal adhesion kinase and associated proteins. Curr. Opin. Cell. Biol. 1994;6:705–710. doi: 10.1016/0955-0674(94)90097-3. [DOI] [PubMed] [Google Scholar]
- 15.Knudsen BS, Feller SM, Hanafusa H. Four proline-rich sequences of the guanine-nucleotide exchange factor C3G bind with unique specificity to the first Src homology 3 domain of Crk. J. Biol. Chem. 1994;269:32781–32787. [PubMed] [Google Scholar]
- 16.Ren R, Ye ZS, Baltimore D. Abl protein-tyrosin kinase selects the Crk adapter as a substrate using SH3-binding sites. Genes Dev. 1994;8:783–795. doi: 10.1101/gad.8.7.783. [DOI] [PubMed] [Google Scholar]
- 17.Hasegawa H, et al. DOCK180, a major CRK-binding protein, alters cell morphology upon translocation to the cell membrane. Mol. Cell. Biol. 1996;16:1770–1776. doi: 10.1128/mcb.16.4.1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiyokawa E, et al. Activation of Rac1 by a Crk SH3-binding protein, DOCK180. Genes Dev. 1998;12:3331–3336. doi: 10.1101/gad.12.21.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kobashigawa Y, et al. Structural basis for the transforming activity of human cancer-related signaling adaptor protein CRK. Nat. Struct. Mol. Biol. 2007;14:503–510. doi: 10.1038/nsmb1241. [DOI] [PubMed] [Google Scholar]
- 20.Feller SM, Knudsen B, Hanafusa H. c-Abl kinase regulates the protein binding activity of c-Crk. EMBO J. 1994;13:2341–2351. doi: 10.1002/j.1460-2075.1994.tb06518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sakar P, Reichman C, Saleh T, Birge RB, Kalodimos CG. Proline cis-trans isomerization controls autoinhibition of a signaling protein. Mol. Cell. 2007;25:413–426. doi: 10.1016/j.molcel.2007.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cowburn D. Moving parts: how the adaptor protein CRK is regulated, and regulates. Nat. Struct. Mol. Biol. 2007;14:465–466. doi: 10.1038/nsmb0607-465. [DOI] [PubMed] [Google Scholar]
- 23.Mochizuki N, et al. Crk activation of JNK via C3G and R-Ras. J. Biol. Chem. 2000;275:12667–12671. doi: 10.1074/jbc.275.17.12667. [DOI] [PubMed] [Google Scholar]
- 24.Wu X, et al. Structural basis for the specific interaction of lysine-containing proline-rich peptides with the N-terminal SH3 domain of c-Crk. Structure. 1995;3:215–226. doi: 10.1016/s0969-2126(01)00151-4. [DOI] [PubMed] [Google Scholar]
- 25.Demers J-P, Mittermaier A. Binding mechanism of an SH3 domain studied by NMR and ITC. J. Am. Chem. Soc. 2009;131:4355–4367. doi: 10.1021/ja808255d. [DOI] [PubMed] [Google Scholar]
- 26.Wildes D, Marqusee S. Hydrogen exchange and ligand binding: Ligand-dependent and ligand-independent protection in the Src SH3 domain. Protein Sci. 2005;14:81–88. doi: 10.1110/ps.04990205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang C, Pawley NH, Nicholson LK. The role of backbone motions in ligand binding to the c-Src SH3 domain. J. Mol. Biol. 2001;313:873–887. doi: 10.1006/jmbi.2001.5083. [DOI] [PubMed] [Google Scholar]
- 28.Cowburn D, Muir TW. Segmental isotopic labeling using expressed protein ligation. Methods Enzymol. 2001;339:41–54. doi: 10.1016/s0076-6879(01)39308-4. [DOI] [PubMed] [Google Scholar]
- 29.Zvara A, et al. Activation of the focal adhesion kinase signaling pathway by structural alterations in the carboxyl-terminal region of c-CrkII. Oncogene. 2001;20:951–961. doi: 10.1038/sj.onc.1204173. [DOI] [PubMed] [Google Scholar]
- 30.Lakowicz JR. Principles of fluorescence spectroscopy. New York: Springer; 2006. [Google Scholar]
- 31.Eftink MR, Ghiron CA. Exposure of tryptophanyl residues in proteins. Quantitative determination by fluorescence quenching studies. Biochemistry. 1976;15:672–680. doi: 10.1021/bi00648a035. [DOI] [PubMed] [Google Scholar]
- 32.Muralidharan V, et al. Solution structure and folding characteristics of the C-terminal SH3 domain of c-Crk-II. Biochemistry. 2006;45:8874–8884. doi: 10.1021/bi060590z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Di Nardo AA, Larson SM, Davidson AR. The relationship between conservation, thermodynamic stability, and function in the SH3 domain hydrophobic core. J. Mol. Biol. 2003;333:641–655. doi: 10.1016/j.jmb.2003.08.035. [DOI] [PubMed] [Google Scholar]
- 34.Zhou HX. Quantitative relation between intermolecular and intramolecular binding of pro-rich peptides to SH3 domains. Biophys J. 2006;91:3170–3181. doi: 10.1529/biophysj.106.090258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Donaldson LW, Gish G, Pawson T, Kay LE, Forman-Kay JD. Structure of a regulatory complex involving the Abl SH3 domain, the Crk SH2 domain, and a Crk-derived phosphopeptide. Proc. Natl. Acad. Sci. USA. 2002;99:14053–14058. doi: 10.1073/pnas.212518799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Porter M, Schindler T, Kuriyan J, Miller WT. Reciprocal regulation of Hck activity by phosphorylation of Tyr527 and Tyr416. J. Biol. Chem. 2000;275:2721–2726. doi: 10.1074/jbc.275.4.2721. [DOI] [PubMed] [Google Scholar]
- 37.Sarkar P, Reichman C, Saleh T, Birge RB, Kalodimos CG. Proline cis-trans isomerization controls autoinhibtion of a signaling protein. Mol. Cell. 2007;25:413–426. doi: 10.1016/j.molcel.2007.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swain JF, et al. Hsp70 chaperone ligands control domain association via an allosteric mechanism mediated by the interdomain linker. Mol. Cell. 2007;26:27–39. doi: 10.1016/j.molcel.2007.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blaschke UK, Cotton GJ, Muir TW. Synthesis of multi-domain proteins using expressed protein ligation: Strategies for segmental isotopic labeling of internal regions. Tetrahedron. 2000;56:9461–9470. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.