Abstract
Pre-steady state kinetic studies on Y-family DNA polymerase η (Polη) have suggested that the polymerase undergoes a rate-limiting conformational change step before the phosphoryl transfer of the incoming nucleotide to the primer terminus. However, the nature of this rate-limiting conformational change step has been unclear, due in part to the lack of structural information on the Polη binary complex. We present here for the first time a crystal structure of human Polη (hPolη) in binary complex with its DNA substrate. We show that the hPolη domains move only slightly on dNTP binding and that the polymerase by and large is pre-aligned for dNTP binding and catalysis. We also show that there is no major reorientation of the DNA from a nonproductive to a productive configuration and that the active site is devoid of metals in the absence of dNTP. Together, these observations lead us to suggest that the rate-limiting conformational change step in the Polη replication cycle likely corresponds to a rate-limiting entry of catalytic metals in the active site.
Introduction
A comparison of the binary and ternary crystal structures of replicative DNA polymerases has revealed many of the hallmarks of classical induced-fit mechanism wherein the binding of dNTP causes a large conformational change in the enzyme 1; 2; 3. That is, when the polymerase binds the DNA template/primer to form the binary complex, it adopts an open conformation in which the fingers domain is oriented away from the active site. On subsequent dNTP binding to form the ternary complex, the fingers domain rotates inward toward the active site to form the closed conformation 1; 2; 3. This serves to both orient the functional groups and helps to discriminate the correct from the incorrect incoming nucleotide, and thereby achieve error rates as low as 1 in 105 nucleotide incorporation events.
The Y-family DNA polymerases lack the fidelity of replicative polymerases but they maintain the continuity of the replication fork by replicating through distorting DNA lesions 4; 5. Humans have four Y-family polymerases – Polη, Polι, Polκ, and Rev1 – each with a unique DNA damage bypass profile. Polη is unique in its ability to replicate through an ultraviolet (UV)-induced cis-syn thymine-thymine (T-T) dimer by inserting two As opposite the two Ts of the dimer with the same efficiency and accuracy as opposite undamaged Ts 6. And, recent genetic studies have indicated that Polη replicates through UV-induced cyclobutane pyrimidine dimers (CPDs) in a highly error-free manner in mouse and human cells 7. Because of the involvement of Polη in promoting error-free replication through CPDs, its inactivation in humans causes the variant form of xeroderma pigmentosum 8; 9, a genetic disorder characterized by a greatly enhanced predisposition to sun induced skin cancers 10. Polη is the only DNA polymerase demonstrated to act as a tumor suppressor in humans.
Does Polη, like the classical replicative polymerases, undergo a major conformational transition on dNTP binding ? Pre-steady state kinetic studies on yeast Polη (yPolη) have suggested that the polymerase undergoes a rate-limiting conformational change step before the phosphoryl transfer of the incoming nucleotide to the primer terminus 11. However, the nature of this rate-limiting conformational change step has been unclear, due in part to the lack of structural information on the Polη binary complex. From the crystal structures of apo yPolη 12 and that of more recent ternary complexes of yeast and human Polη 13; 14 the rate-limiting step does not appear to correspond to a conformational change in the fingers domain, which occupies almost the same position with respect to the palm (and the active site residues) as in the apo-enzyme structure. Alternatively, based on ternary structures of yPolη with cisplatin adducts, it has been suggested that the DNA “rotates” from a nonproductive to a productive configuration on dNTP binding 15. To help resolve these various models, we present here for the first time a crystal structure of human Polη (hPolη) in binary complex with its DNA substrate. The structure shows that the fingers and other domains in hPolη are generally pre-oriented for dNTP binding and catalysis. The most significant change in the polymerase corresponds to the local reorientation of some side chains and to the binding of catalytic metals to the active site We conclude from the structure that the rate-limiting conformational change step observed in pre-steady state kinetic studies on Polη does not correspond to any major conformational transition in the protein or the DNA but, instead, likely corresponds to the rate-limiting binding of catalytic metals to the active site.
Results
Structure determination
The catalytic core of hPolη (residues 1 to 432) was crystallized with a 9-nt/13-nt primer/template from a mix containing Mg2+, but lacking dNTPs. Diffraction quality crystals of the binary complex were obtained after multiple rounds of micro seeding from solutions containing 30% PEG 3350 (w/v). Like the ternary complex 13, the binary complex belongs to space group P6(1) and has similar unit cell dimensions (a=98.9Å, b=98.9Å, c=81.2Å). The structure was solved by molecular replacement (MR) using the structure of the hPolη ternary complex of (with DNA and dATP omitted) as a search model 13. Clear electron density was observed for the primer and template strands in the initial maps, and after subsequent refinement, no electron density was observed for any metal ion in the active site despite the inclusion of Mg2+ in the protein-DNA mix used for crystallization.
The refined hPolη binary complex (Rfree of 24.8%; Rwork of 18.6%) contains protein residues 1 to 432 (residues 154–159 of the palm domain, and residues 410–412 of the PAD domain could not be modeled due to lack of electron density), template strand nucleotides 2–13, primer strand nucleotides 2–9, and 76 waters. The structure has good stereochemistry, with 96.9% of the residues in the most favored regions of the Ramachandran plot, based on MolProbity Ramachandran analysis 16.
Overall arrangement
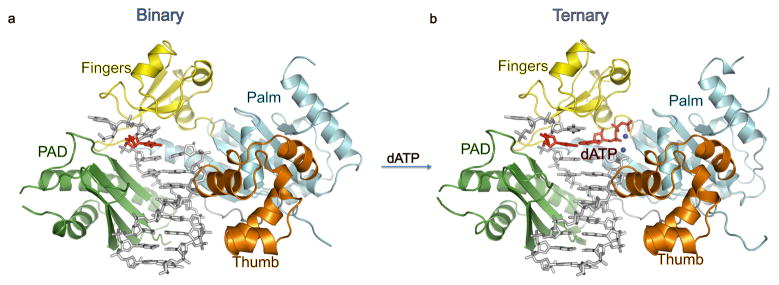
As in the ternary complex 13, hPolη embraces the template-primer with its palm (residues 1–13 and 90–238), fingers (residues 17–87), and thumb (residues 241–301) domains, as well as the polymerase associated domain (PAD (residues 319–432; also known as the little finger domain) unique to Y-family polymerases (Fig. 1). The palm carries the active site residues, Asp13, Asp115 and Glu116, which catalyze the nucleotidyl transfer reaction upon dNTP binding. The fingers domain lies above the templating base, while the thumb and the PAD straddle the duplex portion of the template-primer (Fig. 1). The thumb skims the minor groove surface of the substrate DNA, while the PAD is anchored in the major groove. The majority of DNA interactions are mediated by the PAD, wherein the main chain amides on the “outer” β-strands of the PAD β-sheet make a series of hydrogen bonds with the template and primer strands.
Fig. 1.
Human Polη–DNA binary and ternary complexes. (A) structure of hPolη in binary complex with undamaged DNA. The palm, fingers, thumb domains and the PAD are shown in cyan, yellow, orange and green, respectively. The DNA is in grey and the templating 3′T is in red. (B) structure of hPolη in ternary complex with undamaged DNA and incoming dATP. The templating 3′T and incoming dATP are shown in red. The putative Mg2+ ions are shown in dark blue.
Conformational changes
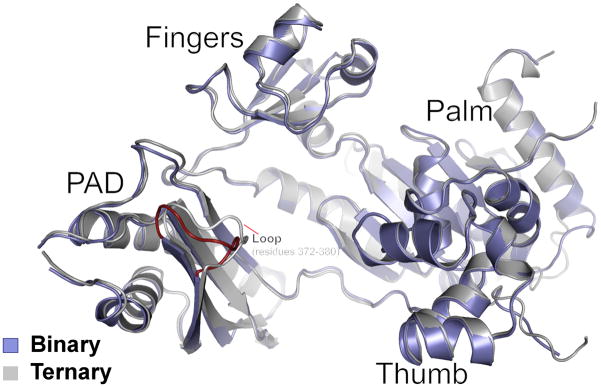
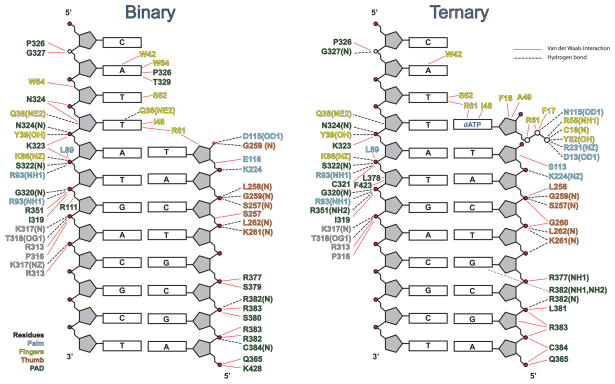
To assess the conformational changes in hPolη on dNTP binding, we superimposed the binary and ternary complexes via their palm domains. The superposition reveals a small (~1.5 Å) outward movement of the fingers domain on dNTP binding (Fig. 2), which slightly enlarges the active site to bind dNTP more comfortably. That is, without this slight outward movement, the Ile48 main chain carbonyl and the Ala49 side chain would partially overlap with the sugar of dNTP. Also, Arg61 in the binary complex extends into the space that would normally be occupied by the base of incoming dNTP and makes van der Waals contacts with the templating base (Fig. 2A). By contrast, Arg61 adopts three different rotamers in the ternary complex 13, making contacts with the base 5′ to the templating base as well as to the incoming nucleotide (Fig. 2B). The PAD is the only other domain that shows significant movement on dNTP binding (~2.3Å) (Fig. 3). However, the contacts that the PAD makes with the template and primer strands are essentially the same as in the ternary complex (Fig. 4). Curiously, the Cα of K376 in the loop between the residues 372 to 380 in the PAD moves outward by ~4Å as compared to the ternary complex (Fig. 3). This loop contains residues such as L378 that are involved in direct interactions with the DNA in the ternary complex (Fig. 4). By contrast, L378 is disordered in the binary complex in accordance with the local rearrangement of the loop in the absence of dNTP (Fig. 3).
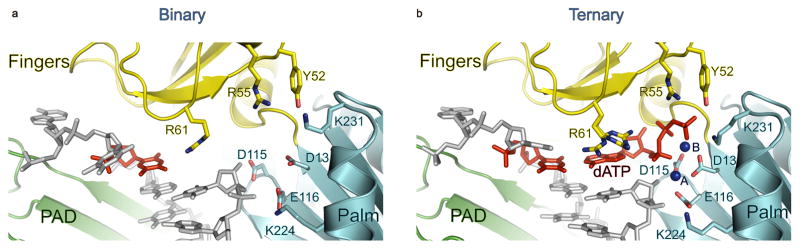
Fig. 2.
Close-up views of the active site regions of human Polη–DNA binary and ternary complexes. (A) the active site region in the binary complex, and (B) the active site region in the ternary complex. The palm and fingers domains and the PAD are shown in cyan, yellow and green, respectively. The DNA is colored grey, and the putative Mg2+ ions (A and B) are dark blue. The undamaged templating T is shown in red in both of the structures and incoming dATP is in red for the ternary structure. Highlighted and labeled are the catalytic residues (D13, D115 and E116), and residues that interact with the triphosphate moiety of incoming dATP (Y52, R55 and K231). R61 has one conformation in the binary complex where it interacts with the templating T, in contrast to multiple conformations in the ternary complex. The catalytic residues are configured slightly different when compared to the ternary complex and do not bind Mg2+ in the absence of dNTP. The residues are colored to match the color of their respective domains.
Fig. 3.
Comparison of human Polη in binary and ternary complexes. The two complexes are superimposed based on the hPolη palm domain. hPolη in the binary and ternary complexes is shown in dark blue and gray, respectively. The superposition reveals subtle conformational changes in the fingers and the PAD domain. The loop region encompassing residues 372 to 380 in the PAD domain of the binary complex is shown in red.
Fig. 4.
A schematic comparison of hydrogen bonds and van der Waals interactions between human Polη and DNA in the binary and ternary complexes. Hydrogen bonds are indicated by broken black lines (determined by interaction distance < 3.2Å) and van der Waals interactions are indicated by continuous red lines (determined by interaction distance < 4.2Å). The residues are colored to match the domain they belong to: cyan for the palm domain, yellow for the fingers domain, orange for the thumb domain, and green for the PAD.
In the hPolη ternary complex 13, incoming dNTP is bound with its triphosphate moiety interlaced between the fingers and palm domains, making hydrogen bonds with Tyr52 and Arg55 from the fingers domain and Lys231 from the palm domain (Fig. 2B). In the binary complex, Tyr52 and Arg55 are in almost the same position as in the ternary complex and Lys231 requires only a change in rotamer to interact with dNTP (Fig. 2A). The catalytic residues Asp13, Asp115, and Glu116 also require only a change in rotamer to coordinate to the two Mg2+ (Mg2+A and Mg2+B) observed in the active site of the ternary complex (Fig. 2). Significantly, Glu116 is buttressed in the binary complex via hydrogen bonds with a conserved Lys224.
There is no indication of metals in the active site in the binary complex (despite the presence of 1 mM MgCl2 in the crystallization mix) (Figs. 1 & 2). In the hPolη ternary complex 13, Mg2+B is coordinated by the unesterified oxygens of dNTP α-, β- and γ phosphates, the carboxylates of Asp13 and Asp115, and the main chain carbonyl of Met14, while Mg2+A is coordinated by the α-phosphate, the carboxylates of Asp13, Asp 115, and Glu116 and the 3′OH at the primer terminus (Fig. 2B). Thus, even though the three catalytic residues are positioned similarly to those in the ternary complex (Fig. 2A), they are insufficient by themselves to bind the catalytic metals in the absence of dNTP.
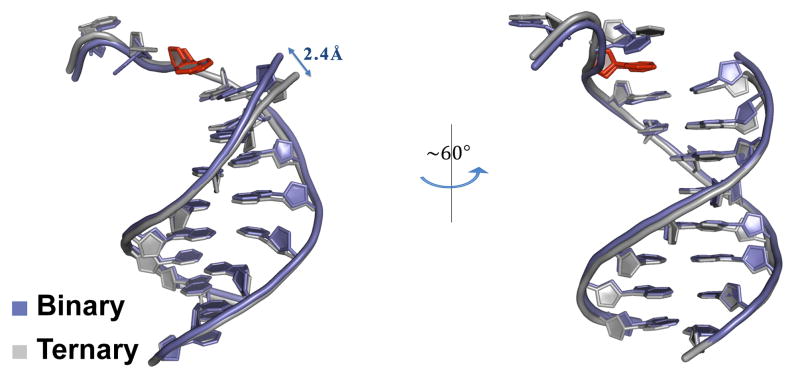
The duplex portion of the DNA has a similar B-DNA-like conformation in both the binary and ternary complexes (Figs. 1 & 5). The average helical twist and rise values in the binary complex are ~31.5° and 3.31Å, respectively, as compared to ~31.2° and 3.34Å in the ternary complex. The most significant local conformational change in the DNA corresponds to an ~2.4Å shift in the primer 3′OH - towards the “empty” dNTP binding site in the binary complex (Fig. 5). The base 5′ to the templating T also changes in conformation. Thus, like the polymerase, changes in the DNA are limited to local adjustments rather to any gross re-orientation of the DNA. The shift in the primer terminus on dNTP (and metal) binding appears to be important in aligning the 3′ OH for its in-line nucleophilic attack on the α-phosphate of incoming dNTP.
Fig. 5.
Comparison of the DNA template and primer strands in the human Polη binary and ternary complexes. The two complexes are again superimposed based on the hPolη palm domain. The DNA is shown in blue and gray in the binary and ternary complexes, respectively. The templating base is shown in red in both binary and ternary template strands in both the complexes. The view on the right is obtained by ~60° rotation along the DNA axis.
Discussion
Despite differences in sequence, structure, and function, all DNA polymerases follow essentially the same reaction pathway 1; 2; 3; 4; 5; 17; 18. That is, preceding phosphoryl transfer, DNA polymerases first bind the template-primer to form the preinsertion polymerase-DNA binary complex, and then bind incoming dNTP to form the preinsertion polymerase-DNA-dNTP ternary complex. This is then followed by a step leading to the formation of a phosphodiester bond between the α-phosphate of dNTP and the 3′OH at the primer terminus. Together, these transitions preceding phosphoryl transfer serve as kinetic checkpoints for the rejection of incorrect nucleotides early in the reaction pathway 2. From pre-steady state kinetic studies it has been inferred that classical polymerases as well as Polη undergo a rate-limiting conformational change step before the phosphoryl transfer reaction 2; 3; 11. In the case of classical polymerases, this rate-limiting step was considered to be the large “open-to-closed” conformational transition in the fingers domain observed in crystal structures, which brings the domain closer to the active site residues 1; 2; 3. However, from recent kinetic studies, the motion of the fingers appears to be relatively fast 19; 20; 21; 22; 23, and it has been suggested that the basis of the rate limiting step in the catalytic cycles of these classical polymerases may correspond to a change in metal ion occupancy in the active site 24; 25; 26. A similar mechanism may operate in Polη. From a comparison of the binary and ternary hPolη stuctures presented here, the fingers domain occupies almost the same position with respect to the palm domain (and the active site residues) in the presence or absence of dNTP. The residues that normally interact with the triphosphate moiety of dNTP (Tyr52, Arg55 and Lys231) and the active site residues Asp13, Asp115, and Glu116) are similarly disposed in the binary and ternary complexes, suggesting that Polη is, for the most part, pre-configured for dNTP binding and catalysis upon binding the template-primer. A notable difference between the structures is the absence of catalytic metals in the binary complex. Analogous to the more classical polymerases, we suggest that the rate- limiting conformational change step in Polη may correspond to the rate-limiting entry of catalytic metals, Mg2+A and Mg2+B, in the active site.
The mechanism we propose here for Polη differs from that suggested previously 15. Based on the observation of more than one DNA orientation in the ternary structures of yPolη with cisplatin adducts, it has been suggested, for example, that the DNA “rotates” from a nonproductive to a productive configuration on dNTP binding 15. However, there is no indication from our comparison of the hPolη binary and ternary structures that the DNA binds in more than one configuration, with the PAD maintaining a tight and almost identical grip on the DNA in both complexes. Interestingly, when Dpo4, an archaeal homolog of Polκ, binds 8-oxoG modified template-primer, the PAD slides or translocates by one nucleotide step in the presence of dNTP, while the thumb domain-DNA contacts remain fixed 27. By contrast, the hPolη PAD maintains the same register with respect to template-primer and other domains in the absence or presence of dNTP. Overall, the hPolη structures are more reminiscent of binary and ternary of complexes human Polι 28, wherein the catalytic pocket is essentially preformed and the binding of dNTP imposes a local rather than any large conformational change in the inter-domain positions.
In summary, we show here that the hPolη domains move only slightly on dNTP binding and that the polymerase by and large is pre-aligned for dNTP binding and catalysis. We also show that there is no major reorientation of the DNA from a nonproductive to a productive configuration, as previously suggested15. Instead, as with the classical polymerases, we suggest that the rate- limiting conformational change step in the Polη replication cycle likely corresponds to a rate-limiting entry of catalytic metals in the active site. A similar conclusion was reached for Dpo4 based on structures with matched or mismatched incoming dNTP 29, suggesting that metal ion coordination may be the key rate-limiting step for most DNA polymerases.
Materials and Methods
Crystallization
The catalytic core of hPolη (residues 1–432) harboring an N-terminal hexa-histadine (6XHis) tag and a C406M mutation was overexpressed in E.coli and purified as previously described by Biertumpfel et al 13. Briefly, the 6XHis tag was removed via overnight incubation with PreScission protease and the protein subsequently purified via ion-exchange (MonoS) and size-exclusion (Superdex75) chromatography, prior to concentration of the protein for crystallization experiments. A 13–nt template strand (5′-TCATTATGACGCT–3′) was annealed to a 9-nt primer strand (5′ – TAGCGTCAT – 3′) to yield the 9-nt/13-nt primer/template. The binary complex was prepared by mixing hPolη with the primer/template in a 1:1.1 molar ratio. 1 mM MgCl2 was added to the mixture and the complex incubated for at least an hour and then concentrated to ~6 mg/ml. Initial crystallization trials performed using various commercially available crystallization screens. Thin needle like crystals were obtained in 0.1M Bis-Tris (pH 5.5) and 20%–35% PEG 3350 (w/v) by using the sitting-drop vapor diffusion method. Several rounds of micro-seeding were required to produce good quality crystals. For data collection, the crystals were cryoprotected by soaks for 5 min in mother liquor solutions containing 5%, 10%, and 15% glycerol (v/v), respectively, and then flash frozen in liquid nitrogen. X-ray diffraction data were collected on beamline X25 at Brookhaven National Laboratory (BNL). The crystals diffract to 2.55 Å resolution with synchrotron radiation and belong to space group P6(1) with unit cell dimensions a = 98.9 Å, b = 98.9 Å and c = 81.2 Å. Matthew’s coefficient calculation indicates one protein-DNA complex in the asymmetric unit. The ternary complex crystals also belong space group P6(1) and have similar unit cell dimensions as the binary complex 13.
Structure determination and refinement
The structure of hPolη binary complex was solved by molecular replacement (MR), using the hPolη ternary structure (PDB Id. 3MR2) 13, with the DNA, incoming nucleotide, and water molecules omitted, as the starting model. The program PHASER gave a unique MR solution 30. The first round of refinement and map calculation was carried out without the DNA using PHENIX 31. The electron density maps (2Fo-Fc and Fo-Fc) showed unambiguous densities for the DNA, which was then built into the map using COOT 32. Iterative rounds of refinement and water picking were performed with PHENIX and REFMAC 33, and model building with COOT. The final model has good stereochemistry, as shown by Molprobity with 96.9% of the residues in the most favored regions of the Ramachandran plot 16. Figures were prepared using PyMol 34
First structure of tumor suppressor human Polη in binary complex with DNA
Surprisingly, the polymerase is pre-aligned for dNTP binding and catalysis
No reorientation of the DNA from a nonproductive to a productive configuration
The active site is devoid of metals in the absence of dNTP
Replication cycle likely limited by rate-limiting entry of metals in the active site
Table 1.
Data collection and refinement statistics
| Data collection | |
|---|---|
| Space group | P61 |
| Cell dimensions | |
| a, b, c (Å) | 98.9, 98.9, 81.2 |
| α, β, γ (°) | 90.0, 90.0, 120.0 |
| Resolution (Å)a | 42.27-2.55 (2.64-2.55) |
| Rsym (%)b | 10.1 (44.1) |
| I/σ(I) | 14.7 (2.6) |
| Completeness (%) | 99.8 (99.1) |
| Redundancy | 4.4 (3.9) |
|
| |
| Refinement | |
|
| |
| Resolution (Å) | 42.27-2.56 (2.62–2.56) |
| Number of reflections | 13,569 |
| Rwork/Rfreec, d | 0.186/0.248 |
| Number of atoms | |
| Protein | 3191 |
| DNA | 389 |
| Water | 76 |
| B-factors (Å2) | |
| Protein | 58.8 |
| DNA | 74.7 |
| Water | 35.4 |
| R.m.s deviations | |
| Bond lengths (Å) | 0.007 |
| Bond angles (°) | 1.034 |
| Ramachandran Plot Quality | |
| Most favored (%) | 96.9% |
| Generously allowed | 2.9% |
| Disallowed (%) | 0.2% |
Values for outermost shells are given in parentheses
Rsym=Σ|I -<I>|/ΣI, where I is the integrated intensity of a given reflection.
Rwork= Σ ||Fobserved| – |Fcalculated||/Σ|Fobserved|
For Rfree calculations 7.5% of the data excluded from refinement were used.
Acknowledgments
We thank the staff at BNL (beamline X25) for facilitating X-ray data collection, and O. Rechkoblit for discussions. This work was supported by grant ES017767 from the National Institutes of Health.
Footnotes
Coordinates
Coordinates and structure factors have been deposited in the PDB with an accession code 3TQ1.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Doublie S, Sawaya MR, Ellenberger T. An open and closed case for all polymerases. Structure Fold Des. 1999;7:R31–5. doi: 10.1016/S0969-2126(99)80017-3. [DOI] [PubMed] [Google Scholar]
- 2.Joyce CM, Benkovic SJ. DNA polymerase fidelity: kinetics, structure, and checkpoints. Biochemistry. 2004;43:14317–24. doi: 10.1021/bi048422z. [DOI] [PubMed] [Google Scholar]
- 3.Rothwell PJ, Waksman G. Structure and mechanism of DNA polymerases. Adv Protein Chem. 2005;71:401–40. doi: 10.1016/S0065-3233(04)71011-6. [DOI] [PubMed] [Google Scholar]
- 4.Prakash S, Johnson RE, Prakash L. Eukaryotic Translesion Synthesis DNA Polymerases: Specificity of Structure and Function. Annu Rev Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 5.Washington MT, Carlson KD, Freudenthal BD, Pryor JM. Variations on a theme: eukaryotic Y-family DNA polymerases. Biochim Biophys Acta. 2010;1804:1113–23. doi: 10.1016/j.bbapap.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson RE, Prakash S, Prakash L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Poleta. Science. 1999;283:1001–4. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 7.Yoon JH, Prakash L, Prakash S. Highly error-free role of DNA polymerase eta in the replicative bypass of UV-induced pyrimidine dimers in mouse and human cells. Proc Natl Acad Sci U S A. 2009;106:18219–24. doi: 10.1073/pnas.0910121106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson RE, Kondratick CM, Prakash S, Prakash L. hRAD30 mutations in the variant form of xeroderma pigmentosum [see comments] Science. 1999;285:263–5. doi: 10.1126/science.285.5425.263. [DOI] [PubMed] [Google Scholar]
- 9.Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta [see comments] Nature. 1999;399:700–4. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 10.Inui H, Oh KS, Nadem C, Ueda T, Khan SG, Metin A, Gozukara E, Emmert S, Slor H, Busch DB, Baker CC, DiGiovanna JJ, Tamura D, Seitz CS, Gratchev A, Wu WH, Chung KY, Chung HJ, Azizi E, Woodgate R, Schneider TD, Kraemer KH. Xeroderma pigmentosum-variant patients from America, Europe, and Asia. J Invest Dermatol. 2008;128:2055–68. doi: 10.1038/jid.2008.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Washington MT, Prakash L, Prakash S. Yeast DNA polymerase eta utilizes an induced-fit mechanism of nucleotide incorporation. Cell. 2001;107:917–27. doi: 10.1016/s0092-8674(01)00613-4. [DOI] [PubMed] [Google Scholar]
- 12.Trincao J, Johnson RE, Escalante CR, Prakash S, Prakash L, Aggarwal AK. Structure of the catalytic core of S. cerevisiae DNA polymerase eta: implications for translesion DNA synthesis. Mol Cell. 2001;8:417–26. doi: 10.1016/s1097-2765(01)00306-9. [DOI] [PubMed] [Google Scholar]
- 13.Biertumpfel C, Zhao Y, Kondo Y, Ramon-Maiques S, Gregory M, Lee JY, Masutani C, Lehmann AR, Hanaoka F, Yang W. Structure and mechanism of human DNA polymerase eta. Nature. 2010;465:1044–8. doi: 10.1038/nature09196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Silverstein TD, Johnson RE, Jain R, Prakash L, Prakash S, Aggarwal AK. Structural basis for the suppression of skin cancers by DNA polymerase eta. Nature. 2010;465:1039–43. doi: 10.1038/nature09104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alt A, Lammens K, Chiocchini C, Lammens A, Pieck JC, Kuch D, Hopfner KP, Carell T. Bypass of DNA lesions generated during anticancer treatment with cisplatin by DNA polymerase eta. Science. 2007;318:967–70. doi: 10.1126/science.1148242. [DOI] [PubMed] [Google Scholar]
- 16.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–83. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steitz TA. DNA polymerases: structural diversity and common mechanisms. J Biol Chem. 1999;274:17395–8. doi: 10.1074/jbc.274.25.17395. [DOI] [PubMed] [Google Scholar]
- 18.McCulloch SD, Kunkel TA. The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res. 2008;18:148–61. doi: 10.1038/cr.2008.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim SJ, Beard WA, Harvey J, Shock DD, Knutson JR, Wilson SH. Rapid segmental and subdomain motions of DNA polymerase beta. J Biol Chem. 2003;278:5072–81. doi: 10.1074/jbc.M208472200. [DOI] [PubMed] [Google Scholar]
- 20.Purohit V, Grindley ND, Joyce CM. Use of 2-aminopurine fluorescence to examine conformational changes during nucleotide incorporation by DNA polymerase I (Klenow fragment) Biochemistry. 2003;42:10200–11. doi: 10.1021/bi0341206. [DOI] [PubMed] [Google Scholar]
- 21.Rothwell PJ, Mitaksov V, Waksman G. Motions of the fingers subdomain of klentaq1 are fast and not rate limiting: implications for the molecular basis of fidelity in DNA polymerases. Mol Cell. 2005;19:345–55. doi: 10.1016/j.molcel.2005.06.032. [DOI] [PubMed] [Google Scholar]
- 22.Luo G, Wang M, Konigsberg WH, Xie XS. Single-molecule and ensemble fluorescence assays for a functionally important conformational change in T7 DNA polymerase. Proc Natl Acad Sci U S A. 2007;104:12610–5. doi: 10.1073/pnas.0700920104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Santoso Y, Joyce CM, Potapova O, Le Reste L, Hohlbein J, Torella JP, Grindley ND, Kapanidis AN. Conformational transitions in DNA polymerase I revealed by single-molecule FRET. Proc Natl Acad Sci U S A. 2010;107:715–20. doi: 10.1073/pnas.0910909107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arndt JW, Gong W, Zhong X, Showalter AK, Liu J, Dunlap CA, Lin Z, Paxson C, Tsai MD, Chan MK. Insight into the catalytic mechanism of DNA polymerase beta: structures of intermediate complexes. Biochemistry. 2001;40:5368–75. doi: 10.1021/bi002176j. [DOI] [PubMed] [Google Scholar]
- 25.Yang L, Arora K, Beard WA, Wilson SH, Schlick T. Critical role of magnesium ions in DNA polymerase beta’s closing and active site assembly. J Am Chem Soc. 2004;126:8441–53. doi: 10.1021/ja049412o. [DOI] [PubMed] [Google Scholar]
- 26.Joyce CM, Potapova O, Delucia AM, Huang X, Basu VP, Grindley ND. Fingers-closing and other rapid conformational changes in DNA polymerase I (Klenow fragment) and their role in nucleotide selectivity. Biochemistry. 2008;47:6103–16. doi: 10.1021/bi7021848. [DOI] [PubMed] [Google Scholar]
- 27.Rechkoblit O, Malinina L, Cheng Y, Kuryavyi V, Broyde S, Geacintov NE, Patel DJ. Stepwise translocation of Dpo4 polymerase during error-free bypass of an oxoG lesion. PLoS Biol. 2006;4:e11. doi: 10.1371/journal.pbio.0040011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. An incoming nucleotide imposes an anti to syn conformational change on the templating purine in the human DNA polymerase-iota active site. Structure. 2006;14:749–55. doi: 10.1016/j.str.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 29.Vaisman A, Ling H, Woodgate R, Yang W. Fidelity of Dpo4: effect of metal ions, nucleotide selection and pyrophosphorolysis. Embo J. 2005;24:2957–67. doi: 10.1038/sj.emboj.7600786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr. 2005;61:458–64. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 31.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 33.Winn MD, Murshudov GN, Papiz MZ. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 2003;374:300–21. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
- 34.Delano WL. The PyMol Molecular Graphics System. Delano Scientific LLC; San Carlos, USA: 2002. [Google Scholar]