Abstract
Background
Exacerbation of migraine with menses is common in adolescent girls and women with migraine, occurring in up to 60% of females with migraine. These migraines are oftentimes longer and more disabling and may be related to estrogen levels and hormonal fluctuations.
Objective
This study identifies the unique genomic expression pattern of menstrually-related migraine (MRM) in comparison to migraine occurring outside the menstrual period and headache free controls.
Methods
Whole blood samples were obtained from female subjects having an acute migraine during their menstrual period (MRM) or outside of their menstrual period (nonMRM) and controls (C) – females having a menstrual period without any history of headache. The mRNA was isolated from these samples and genomic profile was assessed. Affymetrix Human Exon ST 1.0 arrays were used to examine the genomic expression pattern differences between these three groups.
Results
Blood genomic expression patterns were obtained on 56 subjects (MRM = 18, nonMRM = 18 and C = 20). Unique genomic expression patterns were observed for both MRM and nonMRM. For MRM, 77 genes were identified that were unique to MRM, while 61 genes were commonly expressed for MRM and nonMRM and 127 genes appeared to have a unique expression pattern for nonMRM. In addition, there were 279 genes that differentially expressed for MRM compared to nonMRM that were not differentially expressed for nonMRM. Gene ontology of these samples indicated many of these groups of genes were functionally related and included categories of immunomodulation/inflammation, mitochondrial function and DNA homeostasis.
Conclusions
Blood genomic patterns can accurately differentiate MRM from nonMRM. These results indicate that MRM involves a unique molecular biology pathway that can be identified with a specific biomarker and suggest that individuals with MRM have a different underlying genetic etiology.
Keywords: Migraine, Pediatric Headache, Adolescent Headaches, Menstrual Migraine, Gene Expression, Microarray, Personalized Medicine
Introduction
Headache and migraine are significant health problems creating disability and socioeconomic impact.1, 2 A female predominance of migraine has long been recognized for adults with a recent delineation of the specifics for adolescents.3 This pattern only begins to develop once children start to enter puberty and is established once girls start menstruation This differential presentation has been partially attributed to hormonal effects on migraine, although the exact pathophysiological etiology is only beginning to be elucidated (for review see Martin and Behbehani).4, 5 One of the key effectors in this pathophysiology is the fluctuation of estrogen throughout a woman’s lifetime and within a menstrual cycle.
Menstrually-related migraine (MRM) is defined in the appendix of the International Classification of Headache Disorders, 2nd Edition (ICHD-II),6 with the criteria separating the relationship into 3 separate disorders – pure menstrual migraine without aura (PMM), menstrually-related migraine without aura (MRM), and non-menstrual migraine without aura. The first two diagnoses have a temporal relationship with the migraine starting 2 days before to 3 days after the start of menses in two-thirds of the menstrual period with the only difference being that patients with MRM also have additional attacks at other times of the cycle. These two are differentiated from the third in that there is no relationship to the menstrual cycle.
The development and pathophysiology of the menstrual relationship to migraine is beginning to be clarified (reviewed).4, 5, 7 One of the key underlying components of this pathophysiology is the estrogen levels with the migraine appearing to be triggered when the estrogen level drops prior to the start of menses. What remains to be answered is why this results in the only time that some women have migraine (PMM), while in others it occurs both during their menses as well as other times of their cycle (MRM) and furthermore in ~40% of women there is no consistent relationship with their menstrual cycle. These differences suggest that there is an underlying pathophysiological difference among these three groups of women
New molecular biology techniques can assist in identifying the genetic and environmental influences on disease. Gene expression profiling using microarray technology is a powerful technique that can quickly and efficiently screen expression levels in the entire human genome8 and has helped in diagnosing and classifying cancers.9–11 Blood cells inherit the same genetic information as brain cells and blood genomic profiling patterns for neurological diseases have been described for both rats and humans.12–24 Recently, we have demonstrated the use of this technique in migraine by differentiating chronic migraine without mediation overuse from those patients with medication overuse.25 The ability to detect disease-specific gene expression changes in the blood of patients with neurological diseases greatly increases the likelihood of identifying biochemical pathways involved in the pathophysiology of polygenetic, neurological diseases such as migraine where brain tissue samples are not readily available.
Based on the observation of differences in MRM and nonMRM and the ability to assess differences at the gene expression level, we speculated that blood genomic profiling could be used to identify underlying molecular differences and potential biomarkers for the identification of MRM. In the future, this genomic fingerprint may allow for unique diagnosis and treatment of MRM, resulting in a personalized approach to the management with improved response and outcome.
Materials and Methods
Subjects
As part of their standard, multidisciplinary headache care, patients complete a detailed questionnaire with confirmatory history, physical examination, neurological examination, and comprehensive headache examination that is maintained in an extensive, relational database, allowing for screening of complex phenotypes.26 In addition, patients with acute headache exacerbations during the clinic visit or patients with acute headaches that are unresponsive to at-home therapies requiring infusion therapies complete an additional questionnaire that details the features of the acute headache. One component of this acute headache questionnaire is the relationship to a girl’s menstrual period. Female patients with an established menstrual cycle and an acute headache were asked to participate in genomic expression analysis. This group was representative of the entire female adolescent population seen based on analysis of the larger clinic population3 and no additional selection criteria were applied. These subjects were recruited in a balanced manner to obtain an equal number of females with an acute migraine during their menstrual period with those having an acute migraine, but not having a menstrual period and outside the defined risk period for MRM. Inclusion criteria for females with MRM were 1) females with MRM must be actively having or recovering from their menstrual period; 2) must have an acute migraine attack during a menstrual cycle; and 3) age greater than 11 years old and less than 18 years old. Inclusion criteria for non-MRM were 1) females with non-MRM must not be actively having or recovering from or have their menstrual period start within 5 days of sample; 2) must have an acute migraine attack; and 3) age greater than 11 years old and less than 18 years old.
Age and race matched controls were obtained from the Cincinnati Children’s Hospital Genomic Cohort of females that did not have a history of headaches based on completion of a questionnaire and validated by direct interview, but were having an menstrual period at the time of the sampling. The Cincinnati Children’s Hospital Genomic Cohort is a population based control that is representative of the general population and the healthy controls (i.e., lacking recurrent illness) from this study were selected by age and race matching. Whole blood was obtained prior to acute treatment and processed for mRNA isolation as previously described.25 Patients and their parents gave informed consent/assent, based on age and institutional policy. Patients and parents also authorized the use of medical information for research purposes including data collection and analysis as approved by the Institutional Review Board at Cincinnati Children’s Hospital Medical Center.
Blood sample collection, RNA isolation, and Microarray hybridization
Blood sample collection, RNA isolation, microarray hybridization, and normalization were performed as previously described.25, 27 Whole blood was collected into 6 Paxgene Blood RNA tubes, RNA isolated using Paxgene Blood RNA Kit and concentrated using RNeasy MinElute Cleanup Kit (Qiagen, Valencia, CA, USA, http://www.preanalytix.com/RNA-Instr.asp). mRNA was assessed for concentration by spectrophotometry (1 µg/µl total RNA) and for quality using the ratio of 28S:18S ribosomal RNA with Agilent Bioanalyzer 2100 (http://www.chem.agilent.com).
RNA was prepared and hybridized to Affymetrix Human Exon ST 1.0 microarrays28 (http://www.affymetrix,com) using standard Affymetrix labeling protocol (http://www.affymetrix.com/support/technical/manual/expression_manual.affx) in batches of 12 samples with 4 samples representing MRM, 4 samples matched by age and race representing nonMRM and 4 samples of controls matched by age and race. This generated cell intensity (CEL) files containing the raw data. The quality and identification of array outliers in the CEL files were assessed using dChip, version. 2005 (DNA-Chip analyzer, http://www.dchip.org). The perfect match-only (PM-only) model was used in dChip. All microarrays found to have greater than 5% array outliers were excluded from further analysis. These were due to technical issues due to microarray binding of the labeled samples. Microarrays that passed quality control criteria were then normalized using RMA.29
Microarray Analysis
Samples were grouped by MRM, nonMRM, and controls. Differences between all three groups as well as individual comparison between each group were assessed using Genespring GX version 11 (Agilent Technologies, Santa Clara, CA). Minimum average fold change cutoffs were established, and lists of probesets generated for difference. Group analysis was performed to minimize individual variation including variation due to age or timing of cycle. On the Affymetrix microarray each gene’s probeset is represented by approximately 40 probes across the exon regions of each gene with approximately 4 probes per exon. Thus, these probesets can be correlated as gene expression. A t-test with Benjamini and Hochberg False Discovery Rate multiple testing correction was performed on each fold change list. Further analysis was performed on all probesets that were significantly different (p < 0.05).
Identified gene lists were analyzed for overrepresentation in tissue expression, biological pathway and gene ontology using DAVID 2008 (Database for Annotation, Visualization and Integrated Discovery, NIAID / NIH, http://david.abcc.ncifcrf.gov/). DAVID compares the experimental list with databases of probesets shown to be expressed within specific tissues, pathways, or ontology categories, and determines whether the experimental lists contains more probesets than would statistically be expected by chance alone using an EASE score (a modified Fishers Exact Test).
RESULTS
Genomic patient demographics
A total of 56 subjects were enrolled (18 with MRM, 18 with non-MRM, and 20 controls). Demographic features of subjects did not significantly vary. All were female, mean age for the entire group was 15.2 ± 1.7 years (MRM – 15.3 ± 1.6, nonMRM – 15.0 ± 2.0, and control 15.3 ± 1.6) and racial distribution was proportional to the regional population (white – 87.5%, black – 10.7%, and biracial – 1.8%). All subjects with headache met the ICHD-II for migraine without aura, while the controls did not have a history of headache. Of note, none of the MRM subjectgs had pure MRM.
After mRNA isolation, 13 samples had either insufficient or poor quality nRNA (MRM – 5, nonMRM – 6, and controls – 2). The remainder of the samples were matched for age and race for all three parameters to generate 3 balanced batches for Affymetrix microarray processing (4 of MRM, 4 of non-MRM and 4 controls). This resulted in 1 MRM and 6 controls not being processed. After GeneSpring analysis, 4 probesets were determined to be outliers and restricted from further analysis.
Genomic expression patterns
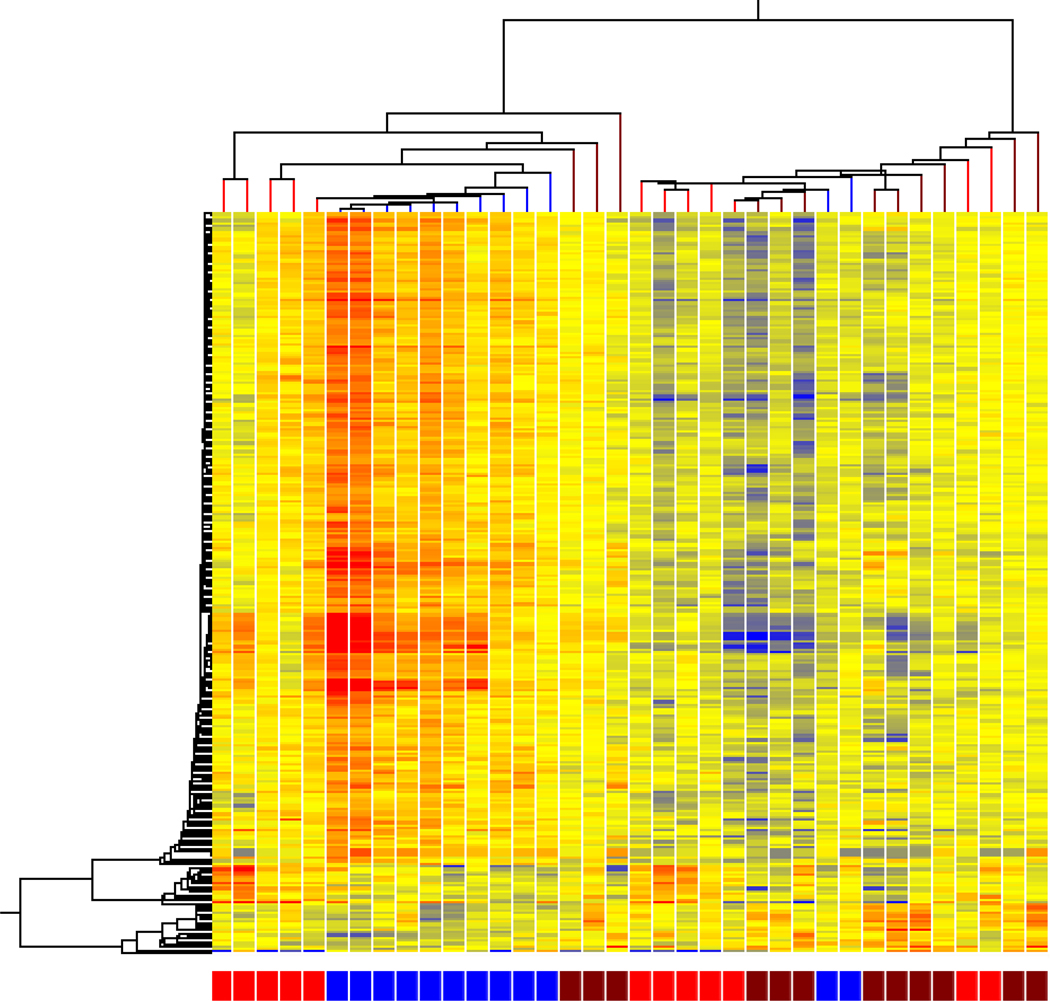
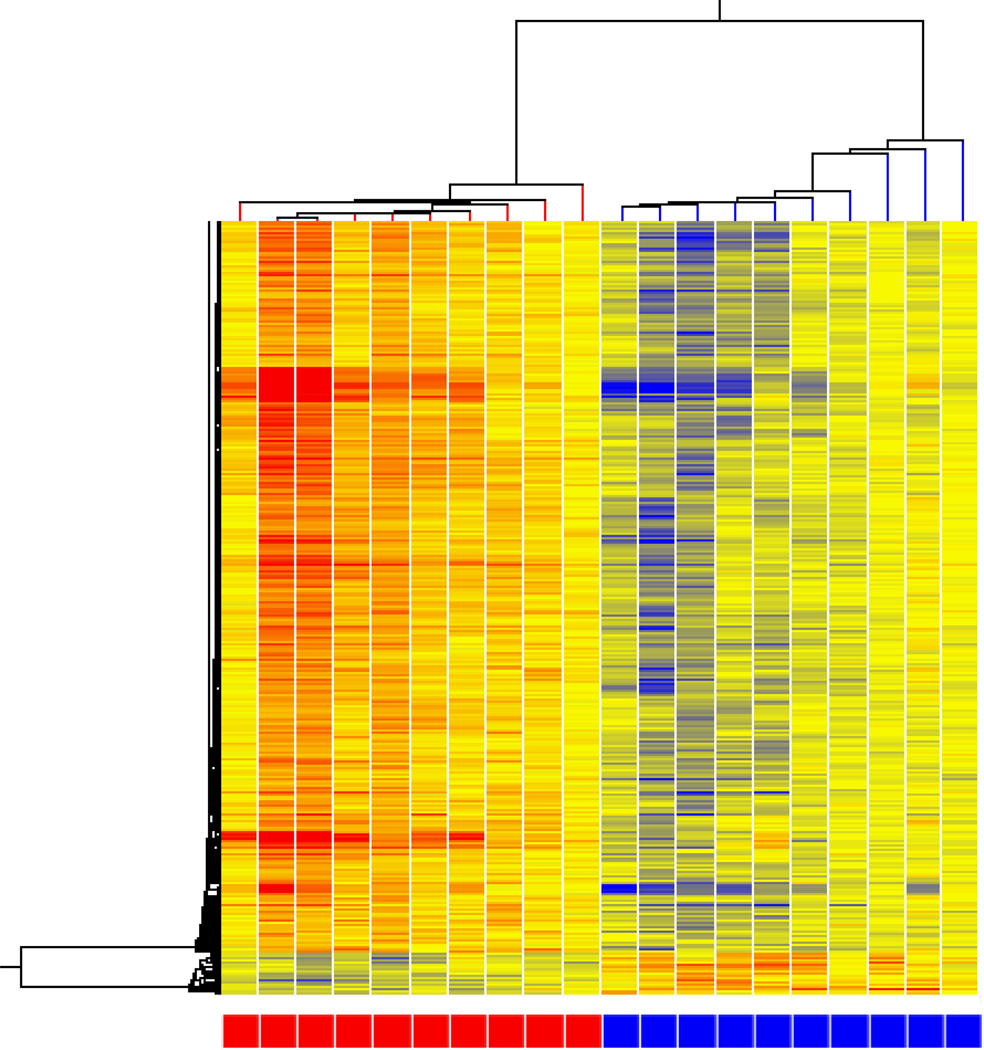
Statistically significant gene expression differences between MRM, nonMRM, and controls were determined by filtering all of microarrays probesets by average fold change of greater than 1.3. When all three conditions were compared, 270 genes were identified with a p <0.05. The hierarchal distribution demonstrated groups of subjects with the largest grouping consisting of all but 2 MRM subjects clustered together (Figure 1).
Figure 1. Cluster analysis of MRM (blue) vs nonMRM (brown) vs controls (red).
Hierarchical cluster analysis of expression patterns of all genes expressed at a significantly different level in at three way comparison between MRM, nonMRM and controls. Individual subjects are represented in each column. Individual probesets are represented by each row. Hierarchical cluster analysis groups subjects and probesets that are most alike together and presented as a branching pattern of subjects (top) and probesets (left). Subjects within the same branch are the most similar. For individual probesets, red indicates that the probeset is expressed at a higher level of expression than the average expression for that probeset, while blue represents a lower level of expression. MRM subjects are represented by blue boxes at the bottom with 10 subjects clustered together with nonMRM indicated by brown boxes and controls by red boxes.
Genomic expression patterns – MRM vs. controls
Females with MRM were compared to female controls that were having a menstrual period to determine the migraine specific genes during a menstrual period. At a 1.3 fold change or higher with a p <0.05, 431 genes differed between MRM and controls – 340 expressed at a higher level in MRM compared to controls. Hierarchal analysis demonstrated a clear separation (Figure 2). As all of these subjects had a menstrual period at the time of the blood draw, the genes expression differences represent those genes that are differentially expressed due to migraine (whether MRM or nonMRM). This gene set is represented by the lower circle in the Venn diagram (Figure 5).
Figure 2. MRM vs controls.
Hierarchical cluster analysis of expression patterns of MRM (blue boxes at bottom) compared to controls (red boxes at bottom).
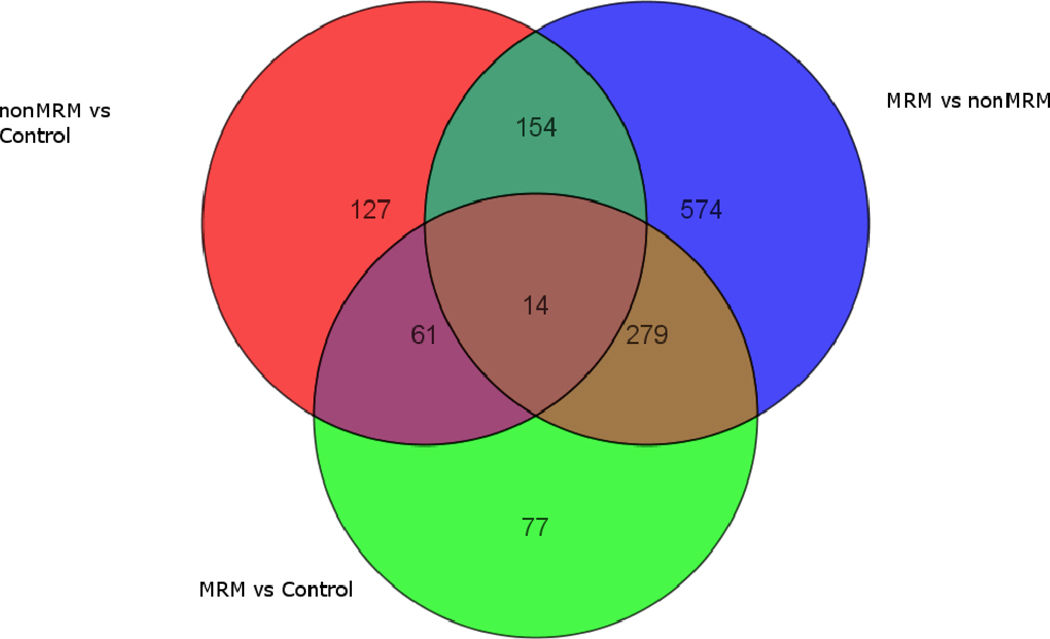
Figure 5. Composite comparison of gene expression profiles.
The top Venn diagrams represents three groups of genes that are differentially expressed with a Fold Change ≥ 1.3 between pairs of groups. The top left circle contains the genes that are differentially expressed between the Control and the Non-MMR groups, the top right circle contains those between the Non-MMR and MMR groups, and the bottom circle contains those between the Controls and the MMR groups.
The bottom Venn diagram represents three groups of genes that are differentially expressed with a Fold Change ≥ 1.3 for each group vs. the other two groups combined. The top left circle contains the genes that are differentially expressed between the MMR group and the combined Non-MMR and Control groups, the top right circle contains those for the Non-MMR group and the combined MMR and Control groups, and the bottom circle contains those for the Control group and the combined MMR and Non-MMR groups.
Genomic expression patterns – nonMRM vs. controls
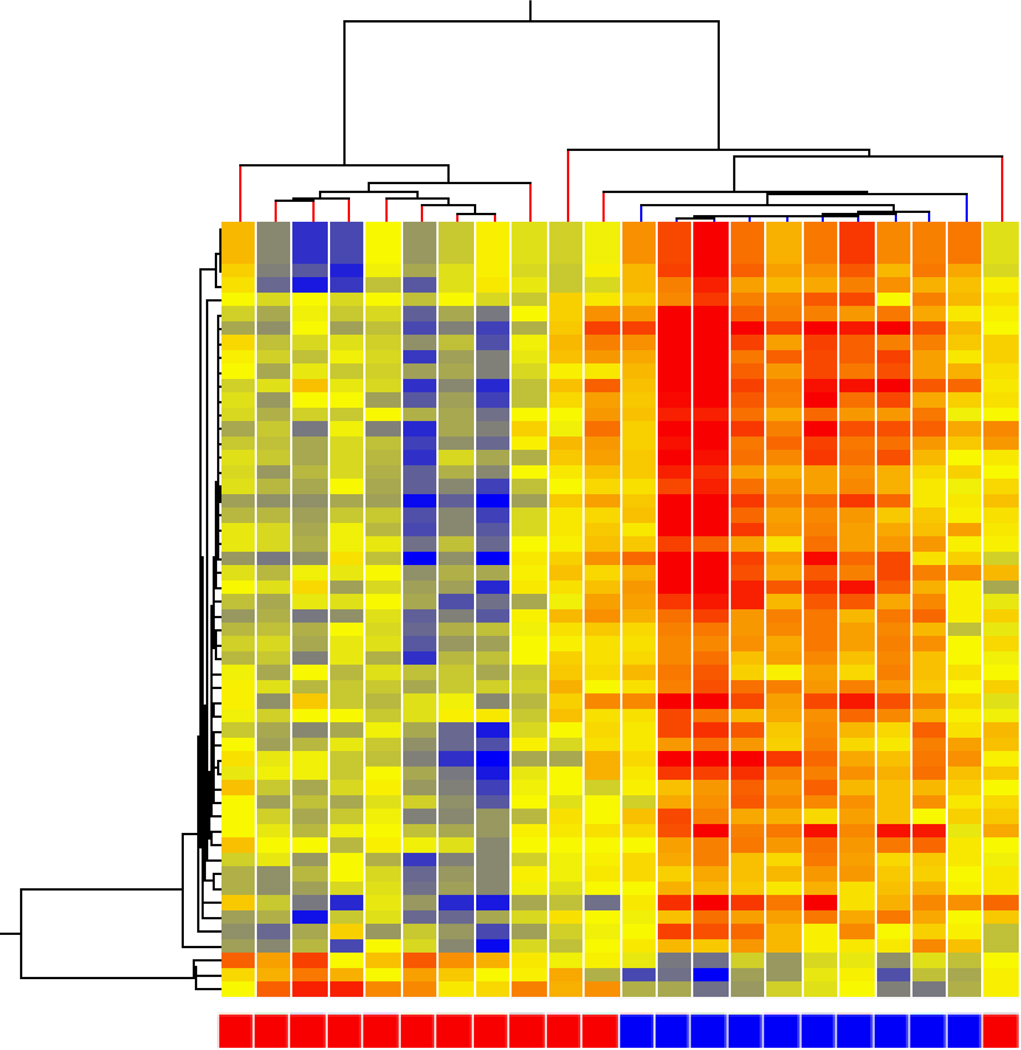
Females with an acute migraine not occurring during their menstrual were compared with females having their menstrual period to determine both gene expression related to migraine (nonMRM expressed) and menstrual (controls). At a 1.3 fold change or higher with a p <0.05, 356 genes differed between nonMRM and controls – 238 expressed at a higher level in nonMRM compared to controls. Hierarchal analysis demonstrated a clear separation (Figure 3). As this comparison includes the presence (nonMRM) or absence (controls) of migraine and the presence (controls) or absence (nonMRM) of a menstrual period, these gene expression alterations represent those genes due to both migraine not occurring during a menstrual period and the genes due to a menstrual period. This is represented by the upper left circle in the Venn diagram (Figure 5).
Figure 3. nonMRM vs controls.
Hierarchical cluster analysis of expression patterns of nonMRM (blue boxes at bottom) compared to controls (red boxes at bottom).
Genomic expression patterns – MRM vs. nonMRM
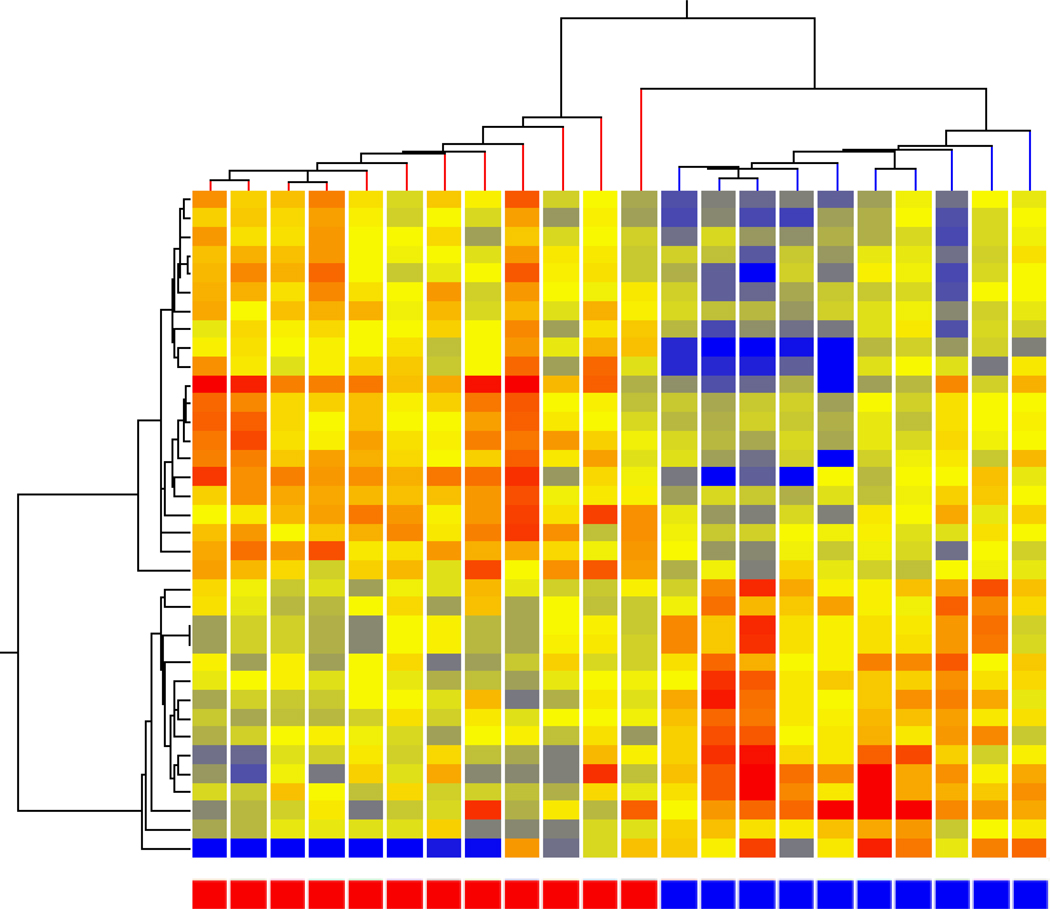
Females with acute migraine either during their menstrual period (MRM) or outside of their menstrual period (nonMRM) were compared to determine both the contribution of the menstrual period (MRM) as well as any unique difference in the two types of migraine. At a 1.3 fold change or higher with a p <0.05, 1021 genes differed between nonMRM and controls – 739 expressed at a higher level in MRM compared to nonMRM. Hierarchal analysis demonstrated a clear separation (Figure 4). This comparison includes the effects of comparing MRM with nonMRM, as well as menstrual period effects (MRM) with those subjects without a menstrual period. The former comparison would be anticipated to identify the differential expression of genes due to a difference between MRM and nonMRM, while the latter would detect those gene expression patterns due to menstrual cycling. This is represented by the upper right circle in the Venn diagram (Figure 5).
Figure 4. MRM vs nonMRM.
Hierarchical cluster analysis of expression patterns of MRM (red boxes at bottom) compared to nonMRM (blue boxes at bottom).
Genomic expression patterns – Composite
In order to determine the gene expression contribution that were unique to each of the potential features (i.e. pure menstrual-related migraine effects, pure non-menstrual migraine effects, combined migraine effects, and pure menstrual effects), the three sets of gene expression patterns (pairwise comparison) were compared for common genes using a Venn diagram (Figure 5).
In the overlap between the gene list for non-MRM/controls (upper left circle) and the MRM/controls (lower circle), 75 genes were identified including 14 common to the entire population of genes. As the MRM/controls neutralize all of the effects of menstrual period, the overlapping genes must represent those genes commonly altered for both MRM and nonMRM (migraine non-specific genes). This group of genes represents the genes commonly expressed for migraine independent of the menstrual effect, while the remaining genes in the non-MRM/control represent genes due to both a difference in migraine and menstrual effects. In contrast, in the MRM/control the remaining genes represent MRM related unique genes and thus are the pure MRM genes.
For the nonMRM/control (upper left circle) comparison to the nonMRM/MRM (upper right circle), there were 168 overlapping genes that had altered expression in common between these two comparisons. The common feature between these two comparisons was changes due to both menstrual effects and non-MRM changes. This is due to both the nonMRM/control and nonMRM/MRM having half the subjects having a menstrual period (MRM and control) and half the subjects in both comparisons having nonMRM. However, the migraine related genes in these overlapping areas only represent those genes that are not expressed in MRM. The remaining genes in the nonMRM/controls thus represent nonMRM genes (127 unique to nonMRM and 61 common for both MRM and nonMRM), while the remaining genes in the nonMRM/MRM represent those genes due to either menstrual periods or MRM.
In the comparison between nonMRM/MRM (upper right circle) and MRM/control (lower circle), both groups compared MRM to either nonMRM or controls, while the nonMRM/MRM also compared menstrual to non-menstrual effects. Thus, the overlapping region represents those genes due to MRM differentially expressed from nonMRM, yet do not include those genes that are uniquely expressed in MRM from controls (i.e., the 77 genes uniquely represented in the lower circle). The remaining genes in the nonMRM/MRM thus represent genes with altered expression due to menstrual effects or due to genes differentially expressed in nonMRM.
Putting the all three comparison together (Figure 5) identifies 7 distinct areas that can serve as potential biomarkers for separating MRM from nonMRM and controls. The 77 genes in the lower circle represent the MRM specific genes that are either not expressed in nonMRM or do not differentiate from nonMRM. The 61 genes in the convergence between MRM/control and nonMRM/control represent the genes that are commonly expressed in migraine compared to control. The 127 genes expressed in the nonMRM/control most likely represent nonMRM genes that are not expressed in MRM. The 279 genes in the convergence between nonMRM/MRM and MRM/controls are most likely those genes that are expressed differentially in MRM from nonMRM are the genes that are most likely to lead to a separation of the two conditions. The remaining sections are affected by the influences of menstruation, with the 574 genes in the upper right most likely to represent this effect.
Functional Annotation Clustering
The probeset identified in three way comparison were analyzed to identify genes expressed at a higher level than would be expected by chance alone with DAVID. Using the Functional Annotation Clustering tool in David for each unique set of genes, distinct clusters of gene functions were identified. This tool analyzes all of the genes that were differentially expressed and groups them by reported function including gene pathways, tissue localization, diseases described and gene similarity.
For the MRM related genes sets, in the 77 genes that were identified to be unique to MRM, 17 functional clusters were identified with an overall enrichment score ranging from 2.18 to 0.02. The enrichment score for the group is based on the EASE score for each member of the group with the EASE score representing a more conservative measure than the Fisher Exact test to determine the degree of gene enrichment in the functional group. The group with the highest enrichment were expressed in epithelium and related to phosphoproteins. There were multiple groups related to DNA and RNA metabolism. Of particular interest to migraine pathophysiology was a functional group related to mitochondrial functioning, oxidative phosphorylation and metal ion binding.
In the 61 genes that had altered expression for both MRM and nonMRM (i.e., migraine genes that were not specific to MRM), only 5 annotation groups were identified (enrichment score between 2.11 and 0.44). The highest group was related to immune function with genes that have been demonstrated in infections and diabetic processes, including mitochondrial involvement, adenyl nucleotide and ribonucleotide binding and acetylation. Two of the additional groups were also involved with immune function.
For the 127 genes that appeared to represent nonMRM gene expression, 27 functional annotation groups were identified (enrichment score – 2.38 to 0.04). The highest functional group was focused on cell to cell signaling including signaling peptides, glycophosphorylation and disulfide bonding. The next highest group were consistent with adaptive immunity responses with many of the genes involved immunoglobin presentation and function. Many of the remaining groups were involved with membrane signaling processes. Of particular interest to migraine, several of the groups were related to neurological function including mitochondrial functioning, cell signaling including G-protein signaling and cell death/apoptosis, all of which we have previously observed for gene processes involved in migraine.
In the 279 genes that appeared to be differentially expressed in MRM, but not in nonMRM, 59 functional annotation groups were identified (enrichment score – 9.65 to <0.01(3 groups)). The highest group was related to zinc finger processing in the nucleus and related to gene expression. This also influenced many of the other groups as they were related to neoplasms and high nuclear turnover and ribosomal processing.
In the 154 gene expression group that represented the nonMRM genes, there were also a large number of functional groups with 47 groups identified (enrichment score – 3.22 to 0.01). As was seen for the unique nonMRM genes, the highest group here was in immune response genes followed by glycoprotein signaling and disulfide bonding. In addition, several of the groups were related to cellular signaling especially in relationship to phosphorylation and G-protein coupled binding. In addition, apoptosis genes were also noted.
Finally, in the 574 genes that appeared to represent menstrual only effects, only 5 annotation groups were identified (enrichment score – 1.84 to 0.44). The highest group here was related to genes expressed in the uterus and included genes involved with nuclear turnover and nucleotide binding. The remaining groups were related to membrane formation and catabolic processes.
DISCUSSION
MRM occurs commonly in adolescent females with migraine. The ICHD-II defines MRM in the appendix with the need for further study. Although the age of our subjects was predominantly in the adolescent age range, the patterns of MRM we observed is similar to that noted for adults based on our previous reports of adolescent menstrual migraine patterns.3 Therefore, the potential biomarkers in terms of gene expression alterations that we have identified have the potential to be broadly applied and lead to investigation to identify specific biomarkers that may identify disease risk or progression.
Clinically, patients with MRM describe the migraines that occur during their menstrual period tend to be more severe and of longer duration with more notable migraine associated symptoms.3 This perceived difference may represent a unique or differential phenotype for MRM as compared to nonMRM. These patients often require more aggressive treatment for these particular migraines and may progress to the need of intermittent prophylaxis. 30–44
From epidemiology studies in adults, it is well recognized that females are more prone to migraine than males. Based on the American Migraine Prevalence and Prevention study,45, 46 17.1% of females 12 years of age and older and 5.6% of men have migraine. Approximately 50% of these females have a menstrual pattern to their migraine.7 Subtracting this percentage from the total results in approximately the same number of males and females have headaches that are not related to menstrual periods. This further supports the hypothesis that females with a menstrual pattern to their migraines have a different underlying pathophysiology from migraines that do not occur with menstrual periods. Further analysis into the phenotypic and molecular expression of male and females with or without MRM needs be to done to confirm this speculation.
The results of this gene expression study clearly demonstrate that there are unique genes expressed during MRM that are not expressed during nonMRM. Further studies are needed to replicate these findings both in adolescents and adults, as well as address the changes in individual genes using alternative methods such as quantitative PCR. In addition, there are also a group of genes that are uniquely expressed in nonMRM that are not expressed in MRM. This observation provides support not only for a unique pathophysiological mechanism for MRM that differentiates it from typical migraine. The observation that there is overlapping gene expression also demonstrates the common pathophysiology between MRM and nonMRM.
This observation is clearly influenced by the gene expression effects related to menstruation. Adolescents may lack regular menstrual cycles and potentially limit the generalizability of these finding, the differential gene expression patterns, suggest that the biological changes are due to the acute effects of the menstrual period. These gene effects may represent the pathways that are regulated that lead to the sensitivity for MRM on those females prone to MRM. This is limited by the lack of a comparison of females with nonMRM with controls outside of their menstrual period. This comparison would detect migraine only gene expression.
In addition to these limitations, gene expression profiling itself is inherently limited by the potential of other influences on gene expression that may reflect difference in sample attainment and processing. To minimize this, group analysis was utilized rather than individual sample comparison and all samples were processed in balanced batches.
The functional annotation analysis begins to reveal some of the underlying pathophysiological. The gene expression patterns that are unique for MRM demonstrate that there are alterations in the phosphorylation state of several proteins. Changes in phosphorylation are often used to alter cell signaling sensitivity and protein functioning. This observation suggests that there may be alterations in particular signaling pathways that are sensitized during an acute MRM that leads to its progression. The involvement of mitochondrial genes combines with earlier observations that migraine may be related to energy dysfunction and this may be exacerbated in MRM.
In the nonMRM unique gene expression patterns, the underlying functional annotation clustering also appears to be related to signaling processes, but in a different underlying mechanism that includes alterations in disulfide bonds and changes in glycophosphorylation. This observation demonstrates that for both MRM and nonMRM, cell signaling is integrally involved in the pathogenesis of acute migraine attacks and may be responsible for the progression of an attack.
Of particular interest in the nonMRM is the involvement of immune related pathways. Inflammatory processes have long been noted for migraine and the observation that these pathways are expressed at distinctly differently levels from controls demonstrates that the immune system is altered during an acute migraine. This immune response may lead to the neurovascular inflammation and platelet responses that have been historically reported for migraine.
Cell signaling appears to be a critical component of the migraine pathophysiology. The involvement of G-protein coupled activation and adenyl cyclase compounds is intriguing given the responsiveness of migraine to triptans with the serotonin receptors being G-protein coupled receptors, as well as the evolution of migraine treatment to new compounds including CGRP and adenosine agonists.
For the group of genes that are differentially expressed from controls for both nonMRM and MRM, inflammatory and mitochondrial processing appears to be the common pathophysiological features. These changes may be regulated at the nuclear level with alterations in ribonucleotide binding and thus mRNA production that leads to the common pathophysiology of energy and inflammatory processes. This ultimately leads to a common phenotype expression.
This study raises the issue of differential gene expression patterns as a method to assess the differences that are occurring during acute migraine with our without menstrual periods. Further studies are needed to validate this in adolescents as well as adults. In addition, the genes with altered expression may lead to the identification of biomarkers in the future. Additional groups that would be of interest to compare include within subject comparison (a migraine during menses in comparison to another part of the cycle) as well as in subjects with pure menstrual migraine.
Supplementary Material
Table 1.
Demographics of Subjects with Medication Overuse Headaches
| Genomic Sample | ||||
|---|---|---|---|---|
| MRM | nonMRM | controls | ||
| n | 18 | 18 | 20 | |
| Age | 15.3 ± 1.6 | 15.0 ± 2.0 | 15.3 ± 1.6 | |
| Race | ||||
| White | 15 | 16 | 18 | |
| Black | 3 | 1 | 2 | |
| Mixed | 1 | |||
Acknowledgements
This work was support by a NIH grant (NS045752) and an investigator initiated, independent grant from Endo Pharmaceuticals. The authors would like to thank Frank Sharp for his original support of this project and his consultation in the original NIH grant. The authors would also like to thank Susan LeCates, Shannon Cherney, Polly Vaughan, Ann Segers, Paula Manning, and Judy Bush for their assistance in clinical recruitment and patient management.
Abbreviations
- MRM
Menstrual-Related Migraine
- nonMRM
Migraine not related to menstrual periods
- ICHD-II
International Classification of Headache Disorders, 2nd Edition
- DAVID
Database for Annotation, Visualization and Integrated Discovery
Footnotes
Conflict of Interest: None
References
- 1.Lipton RB, Scher AI, Steiner TJ, et al. Patterns of health care utilization for migraine in England and in the United States. Neurology. 2003;60:441–448. doi: 10.1212/wnl.60.3.441. [DOI] [PubMed] [Google Scholar]
- 2.Lipton RB, Bigal ME, Kolodner K, Stewart WF, Liberman JN, Steiner TJ. The family impact of migraine: population-based studies in the USA and UK. Cephalalgia. 2003;23:429–440. doi: 10.1046/j.1468-2982.2003.00543.x. [DOI] [PubMed] [Google Scholar]
- 3.Crawford MJ, Lehman L, Slater S, et al. Menstrual Migraine in Adolescents. Headache. 2009 doi: 10.1111/j.1526-4610.2009.01347.x. [DOI] [PubMed] [Google Scholar]
- 4.Martin VT, Behbehani M. Ovarian hormones and migraine headache: understanding mechanisms and pathogenesis--part 2. Headache. 2006;46:365–386. doi: 10.1111/j.1526-4610.2006.00370.x. [DOI] [PubMed] [Google Scholar]
- 5.Martin VT, Behbehani M. Ovarian hormones and migraine headache: understanding mechanisms and pathogenesis--part I. Headache. 2006;46:3–23. doi: 10.1111/j.1526-4610.2006.00309.x. [DOI] [PubMed] [Google Scholar]
- 6.Headache Classification Subcommittee of the International Headache Society. The International Classification of Headache Disorders. Cephalagia. 2004;24:1–160. [Google Scholar]
- 7.Martin VT, Lipton RB. Epidemiology and biology of menstrual migraine. Headache. 2008;48 Suppl 3:S124–S130. doi: 10.1111/j.1526-4610.2008.01310.x. [DOI] [PubMed] [Google Scholar]
- 8.Cobb JP, Mindrinos MN, Miller-Graziano C, et al. Application of genome-wide expression analysis to human health and disease. Proc Natl Acad Sci U S A. 2005;102:4801–4806. doi: 10.1073/pnas.0409768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alizadeh AA, Staudt LM. Genomic-scale gene expression profiling of normal and malignant immune cells. Curr Opin Immunol. 2000;12:219–225. doi: 10.1016/s0952-7915(99)00078-3. [DOI] [PubMed] [Google Scholar]
- 10.Golub TR, Slonim DK, Tamayo P, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 11.Yeoh EJ, Ross ME, Shurtleff SA, et al. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell. 2002;1:133–143. doi: 10.1016/s1535-6108(02)00032-6. [DOI] [PubMed] [Google Scholar]
- 12.Tang Y, Xu H, Du X, et al. Gene expression in blood changes rapidly in neutrophils and monocytes after ischemic stroke in humans: a microarray study. J Cereb Blood Flow Metab. 2006 doi: 10.1038/sj.jcbfm.9600264. [DOI] [PubMed] [Google Scholar]
- 13.Lu A, Tang Y, Ran R, Ardizzone TL, Wagner KR, Sharp FR. Brain genomics of intracerebral hemorrhage. J Cereb Blood Flow Metab. 2006;26:230–252. doi: 10.1038/sj.jcbfm.9600183. [DOI] [PubMed] [Google Scholar]
- 14.Du X, Tang Y, Xu H, et al. Genomic profiles for human peripheral blood T cells, B cells, natural killer cells, monocytes, and polymorphonuclear cells: comparisons to ischemic stroke, migraine, and Tourette syndrome. Genomics. 2006;87:693–703. doi: 10.1016/j.ygeno.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 15.Tang Y, Gilbert DL, Glauser TA, Hershey AD, Sharp FR. Blood gene expression profiling of neurologic diseases: a pilot microarray study. Arch Neurol. 2005;62:210–215. doi: 10.1001/archneur.62.2.210. [DOI] [PubMed] [Google Scholar]
- 16.Tang Y, Schapiro MB, Franz DN, et al. Blood expression profiles for tuberous sclerosis complex 2, neurofibromatosis type 1, and Down's syndrome. Ann Neurol. 2004;56:808–814. doi: 10.1002/ana.20291. [DOI] [PubMed] [Google Scholar]
- 17.Tang Y, Lu A, Ran R, et al. Human blood genomics: distinct profiles for gender, age and neurofibromatosis type 1. Brain Res Mol Brain Res. 2004;132:155–167. doi: 10.1016/j.molbrainres.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 18.Tang Y, Hershey AD, Powers SW, et al. Genomic abnormalities in patients with migraine and chronic migraine: preliminary blood gene expression suggests platelet abnormalities. Headache. 2004;44:994–1004. doi: 10.1111/j.1526-4610.2004.04193.x. [DOI] [PubMed] [Google Scholar]
- 19.Tang Y, Glauser TA, Gilbert DL, et al. Valproic acid blood genomic expression patterns in children with epilepsy - a pilot study. Acta Neurol Scand. 2004;109:159–168. doi: 10.1046/j.1600-0404.2003.00253.x. [DOI] [PubMed] [Google Scholar]
- 20.Tang Y, Nee AC, Lu A, Ran R, Sharp FR. Blood genomic expression profile for neuronal injury. J Cereb Blood Flow Metab. 2003;23:310–319. doi: 10.1097/01.WCB.0000048518.34839.DE. [DOI] [PubMed] [Google Scholar]
- 21.Tang Y, Lu A, Aronow BJ, Wagner KR, Sharp FR. Genomic responses of the brain to ischemic stroke, intracerebral haemorrhage, kainate seizures, hypoglycemia, and hypoxia. Eur J Neurosci. 2002;15:1937–1952. doi: 10.1046/j.1460-9568.2002.02030.x. [DOI] [PubMed] [Google Scholar]
- 22.Bernaudin M, Tang Y, Reilly M, Petit E, Sharp FR. Brain genomic response following hypoxia and re-oxygenation in the neonatal rat. Identification of genes that might contribute to hypoxia-induced ischemic tolerance. J Biol Chem. 2002;277:39728–39738. doi: 10.1074/jbc.M204619200. [DOI] [PubMed] [Google Scholar]
- 23.Tang Y, Lu A, Aronow BJ, Sharp FR. Blood genomic responses differ after stroke, seizures, hypoglycemia, and hypoxia: blood genomic fingerprints of disease. Ann Neurol. 2001;50:699–707. doi: 10.1002/ana.10042. [DOI] [PubMed] [Google Scholar]
- 24.Whitney AR, Diehn M, Popper SJ, et al. Individuality and variation in gene expression patterns in human blood. Proc Natl Acad Sci U S A. 2003;100:1896–1901. doi: 10.1073/pnas.252784499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hershey AD, Burdine D, Kabbouche MA, Powers SW. Genomic expression patterns in medication overuse headaches. Cephalalgia. 2010 doi: 10.1177/0333102410373155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hershey AD, Powers SW, Bentti AL, Degrauw TJ. Effectiveness of amitriptyline in the prophylactic management of childhood headaches. Headache. 2000;40:539–549. doi: 10.1046/j.1526-4610.2000.00085.x. [DOI] [PubMed] [Google Scholar]
- 27.Hershey AD, Burdine D, Liu C, Nick TG, Gilbert DL, Glauser TA. Assessing quality and normalization of microarrays: case studies using neurological genomic data. Acta Neurol Scand. 2008;118:29–41. doi: 10.1111/j.1600-0404.2007.00979.x. [DOI] [PubMed] [Google Scholar]
- 28.Affymetrix I. Data Sheet: GeneChip Exon Array System for Human, Mouse, and Rat. http://media.affymetrix.com/support/technical/datasheets/exon_arraydesign_datasheet.pdf [online]. Available. [Google Scholar]
- 29.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.MacGregor EA. Prevention and treatment of menstrual migraine. Drugs. 2010;70:1799–1818. doi: 10.2165/11538090-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 31.MacGregor EA, Pawsey SP, Campbell JC, Hu X. Safety and tolerability of frovatriptan in the acute treatment of migraine and prevention of menstrual migraine: Results of a new analysis of data from five previously published studies. Gend Med. 2010;7:88–108. doi: 10.1016/j.genm.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 32.Brandes JL, Poole A, Kallela M, et al. Short-term frovatriptan for the prevention of difficult-to-treat menstrual migraine attacks. Cephalalgia. 2009;29:1133–1148. doi: 10.1111/j.1468-2982.2009.01840.x. [DOI] [PubMed] [Google Scholar]
- 33.MacGregor EA, Brandes JL, Silberstein S, et al. Safety and tolerability of short-term preventive frovatriptan: a combined analysis. Headache. 2009;49:1298–1314. doi: 10.1111/j.1526-4610.2009.01513.x. [DOI] [PubMed] [Google Scholar]
- 34.Silberstein SD, Berner T, Tobin J, Xiang Q, Campbell JC. Scheduled short-term prevention with frovatriptan for migraine occurring exclusively in association with menstruation. Headache. 2009;49:1283–1297. doi: 10.1111/j.1526-4610.2009.01509.x. [DOI] [PubMed] [Google Scholar]
- 35.Newman LC, Harper S, Jones BA, Campbell J. Frovatriptan for acute treatment of migraine associated with menstruation: results from an open-label postmarketing surveillance study. J Womens Health (Larchmt) 2009;18:1265–1273. doi: 10.1089/jwh.2008.1031. [DOI] [PubMed] [Google Scholar]
- 36.Wade A, Pawsey S, Whale H, Boyce M, Warrington S. Pharmacokinetics of two 6-day frovatriptan dosing regimens used for the short-term prevention of menstrual migraine: A phase I, randomized, double-blind, placebo-controlled, two-period crossover, single-centre study in healthy female volunteers. Clin Drug Investig. 2009;29:325–337. doi: 10.2165/00044011-200929050-00005. [DOI] [PubMed] [Google Scholar]
- 37.Guidotti M, Mauri M, Barrila C, Guidotti F, Belloni C. Frovatriptan vs. transdermal oestrogens or naproxen sodium for the prophylaxis of menstrual migraine. J Headache Pain. 2007;8:283–288. doi: 10.1007/s10194-007-0417-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adelman JU, Calhoun A. A randomized trial of frovatriptan for the intermittent prevention of menstrual migraine. Neurology. 2005;64:931. doi: 10.1212/wnl.64.5.931. author reply 931. [DOI] [PubMed] [Google Scholar]
- 39.Pringsheim T, Davenport WJ, Dodick D. Acute treatment and prevention of menstrually related migraine headache: evidence-based review. Neurology. 2008;70:1555–1563. doi: 10.1212/01.wnl.0000310638.54698.36. [DOI] [PubMed] [Google Scholar]
- 40.Mannix LK, Savani N, Landy S, et al. Efficacy and tolerability of naratriptan for short-term prevention of menstrually related migraine: data from two randomized, double-blind, placebo-controlled studies. Headache. 2007;47:1037–1049. doi: 10.1111/j.1526-4610.2007.00855.x. [DOI] [PubMed] [Google Scholar]
- 41.Brandes JL, Smith T, Diamond M, Ames MH. Open-label, long-term tolerability of naratriptan for short-term prevention of menstrually related migraine. Headache. 2007;47:886–894. doi: 10.1111/j.1526-4610.2007.00809.x. [DOI] [PubMed] [Google Scholar]
- 42.Massiou H, Jamin C, Hinzelin G, Bidaut-Mazel C. Efficacy of oral naratriptan in the treatment of menstrually related migraine. Eur J Neurol. 2005;12:774–781. doi: 10.1111/j.1468-1331.2005.01076.x. [DOI] [PubMed] [Google Scholar]
- 43.Moschiano F, Allais G, Grazzi L, et al. Naratriptan in the short-term prophylaxis of pure menstrual migraine. Neurol Sci. 2005;26 Suppl 2:s162–s166. doi: 10.1007/s10072-005-0435-4. [DOI] [PubMed] [Google Scholar]
- 44.Newman L, Mannix LK, Landy S, et al. Naratriptan as short-term prophylaxis of menstrually associated migraine: a randomized, double-blind, placebo-controlled study. Headache. 2001;41:248–256. doi: 10.1046/j.1526-4610.2001.111006248.x. [DOI] [PubMed] [Google Scholar]
- 45.Stewart WF, Wood C, Reed ML, Roy J, Lipton RB. Cumulative lifetime migraine incidence in women and men. Cephalalgia. 2008;28:1170–1178. doi: 10.1111/j.1468-2982.2008.01666.x. [DOI] [PubMed] [Google Scholar]
- 46.Lipton RB, Bigal ME, Diamond M, Freitag F, Reed ML, Stewart WF. Migraine prevalence, disease burden, and the need for preventive therapy. Neurology. 2007;68:343–349. doi: 10.1212/01.wnl.0000252808.97649.21. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.