Abstract
Background
The emergence of integrase strand-transfer inhibitor (INSTI) resistance-associated mutations was examined in patients with low-level viremia after switching from enfuvirtide to raltegravir in the ANRS 138-Easier trial.
Methods
Integrase genes of plasma virus from raltegravir-treated patients in the Easier trial with low-level viremia (50–500 copies/ml) were sequenced to determine INSTI resistance-associated mutations. Baseline viral load, baseline and nadir CD4 cell count, antiretroviral treatment, genotypic susceptibility score, level of viremia and degree of treatment adherence during the study period were also analyzed.
Results
Forty-nine patients experienced at least one episode of low-level viremia while receiving raltegravir; integrase genotyping was successful in samples from 39 individuals (80%). Among them, three [7.7%, 95% confidence interval (CI) 1.6–20.9%] had significant INSTI resistance mutations consisting of N155H in two and P145S in one.
Absence of these mutations from proviral DNA at baseline suggested selection of INSTI resistance during episodes of low-level viremia. No specific factors significantly associated with emergence of INSTI resistance mutations during low-level viremia were identified.
Conclusion
Emergence of INSTI resistance mutations can occur during episodes of low-level viremia in patients receiving raltegravir-containing regimens.
Keywords: HIV-1, integrase inhibitor, low-level viremia, raltegravir, resistance mutation, treatment experienced
Background
Raltegravir is an integrase strand-transfer inhibitor (INSTI) approved for treatment of HIV-1-infection in antiretroviral-naïve and treatment-experienced patients [1,2]. Raltegravir resistance is conferred by mutations at integrase codons 143, 148 and 155 together with associated secondary mutations [1,3,4]. Mutations associated with raltegravir resistance have been found in 44% of treatment-naïve and 68% of treatment-experienced patients at the time of virologic failure of a raltegravir-containing regimen [1,3]. As most clinical studies of raltegravir resistance have performed integrase genotyping only on samples with plasma HIV-1 RNA levels of at least 1000 copies/ml, few data are available on the emergence of such mutations in patients with low viremia.
The Agence Nationale de Recherche sur le Sida et les Hepatites Virales (ANRS) 138-Easier trialwas a 48-week study in which virologically suppressed treatment-experienced patients receiving a stable enfuvirtide-containing regimen were randomized to switch to raltegravir or remain on enfuvirtide [5,6]. Similar rates of virologic suppression were maintained in both arms. Integrase inhibitor resistance was not detected in the single patient in the raltegravir arm who experienced protocol-defined virologic failure (confirmed virus load of at least 400 copies/ml). However, many patients experienced episodes of low-level viremia, defined as at least one viral load measurement of 50–500 copies/ml while receiving raltegravir-containing antiretroviral therapy. In the present study, we performed integrase genotyping on plasma samples from patients who experienced low-level viremia while receiving raltegravir.
Materials and methods
The Easier trial (ANRS 138) was a multicenter, randomized, comparative, 48-week open-label trial with a primary end point at week 24. Eligible patients had a prior history of triple-class [protease inhibitor, nucleoside reverse transcriptase inhibitor (NRTI), and non-nucleoside reverse transcriptase inhibitor (NNRTI)] antiretroviral drug failure or intolerance, no previous therapy with an INSTI and had received the same enfuvirtide-based treatment and with plasma HIV RNA levels below 400 copies/ml for at least 3 months. Patients were randomized to maintain their enfuvirtide-based regimen or a switch to raltegravir in combination with the same background regimen. Patients in the enfuvirtide continuation arm could switch to raltegravir at week 24. The present study included patients enrolled in Easier trial who experienced at least one episode of low-level viremia (plasma HIV-1 RNA 50–500 copies/ml) while receiving raltegravir, but who did not experience protocol-defined virologic failure. Plasma HIV-1 RNA was quantified using either Cobas TaqMan HIV-1 Roche or M2000RT Abbott or NucliSens EasyQ bioMérieux (threshold of quantification, 50 copies/ml).
Stored plasma samples (1.5 ml) with low-level viremia were subjected to ultracentrifugation at 28 100g for 2 h at 4°C to pellet virus particles prior to RNA extraction using the QIAamp viral RNA minikit (QIAGEN Inc., Valencia, CA). The integrase coding region of pol (codons 48–252; HXB2 nucleotides 4371–4962) was amplified by reverse transcription-coupled polymerase chain reaction (RT-PCR), followed by a nested-PCR using forward and reverse primers IN1 (nucleotides 4336 to 4359) and IN2 (nucleotides 5053 to 5027), respectively, for the RT-PCR and forward and reverse primers IN3 (nucleotides 4371 to 4389) and IN4 (nucleotides 4984 to 4962), respectively, for the nested PCR primer positions are numbered according to HXB2R pol (http://www.hiv.lanl.gov/content/sequence/HIV/REVIEWS/HXB2.html).
Each sample was amplified in duplicate and the amplicons pooled for analysis. Population sequencing was performed on purified amplicons (QIAquick PCR Purification Kit, QIAGEN Inc.) with the Taq Dye Deoxy Terminator cycle sequencing kits (Applied Biosytems, Carlsbad, CA) and resolved on an ABI 3730 automated DNA sequencer. Sequences were processed using Sequencher (Genecodes, Ann Arbor, MI) and aligned to the HIV-1 subtype B reference strain HXB2 (GenBank accession no. K03455). Phylogenetic analysis using PhyML was used to confirm sequence identity and exclude PCR contamination. Primary and secondary INSTI resistance mutations were identified according to the 2009 International AIDS Society (IAS)-USA list (www.iasusa.org).
Population sequencing of proviral DNA from peripheral blood mononuclear cells (PBMCs) was performed on samples obtained at study entry from those patients in whom INSTI resistance mutations were identified during episodes of low-level viremia. DNA was extracted from whole blood (MagnaPure, Roche, Meylan, France) using standard procedures (www.hivfrenchresistance.org). In one patient, clonal analysis of integrase from a week 4 plasma sample was performed in cloning integrase gene PCR products into a pCR4-TOPO vector (InVitrogen, Carlsbad, CA) and sequencing the purified clonal fragments.
Raltegravir plasma concentrations in samples from patients with raltegravir-resistant virus were determined by a validated high-performance liquid chromatography (HPLC) assay with ultraviolet detection [7]. Baseline plasma HIV-1 RNA, baseline and nadir CD4 cell count, baseline genotypic sensitivity score (GSS) according to the 2008 ANRS resistance algorithm (defined as the number of fully active antiretroviral drugs used as part of the regimen excluding raltegravir and enfuvirtide), number of episodes of low-level viremia, maximum HIV-1 RNA during episodes of low-level viremia and treatment adherence (measured by self-administrated questionnaire [8]) were reviewed for each patient.
Results
Of 170 Easier patients, 49 (29%) had at least one episode of low-level viremia while receiving raltegravir, including 32 in the switch arm and 17 in the enfuvirtide arm who switched to raltegravir after week 24. Table 1 shows baseline characteristics of these patients. Patients had a median nadir CD4 cell count of 37 cells/µl; 59% were classified as CDC stage C; viruses harbored a median of 4.5 resistance associated mutations; and patients were receiving a median of three drugs with a median of GSS of 1.5. In addition, 19 of 84 patients (23%) experienced at least one episode of low-level viremia while receiving enfuvirtide, including 10 who also experienced low-level viremia after switching to raltegravir.
Table 1.
Characteristics of the patients with low-level viremia.
| IN genotype available (n = 39) | |||||
|---|---|---|---|---|---|
| No INSTI resistance mutation (n = 36) |
INSTI resistance mutation (n = 3) |
IN genotype not available (n = 10) |
Total (N = 49) | ||
| Baseline characteristics | |||||
| Median age (years) | 48 (43;58)a | 48 (45;60) | 46 (41;60) | 48 (42;59)] | |
| CDC classification | A | 8 | 33 | 10 | 10 |
| (% of patients) | B | 28 | 0 | 50 | 31 |
| C | 64 | 67 | 40 | 59 | |
| Median CD4 cell count (cells/µl) | |||||
| Baseline | 422 (252;545) | 358 (113;370) | 301 (263;552) | 370 (263;535) | |
| Nadir | 30 (17;86) | 60 (14;106) | 88 (30;126) | 37 (17;102) | |
| ART at study entry | |||||
| (%) | NRTI/PI | 92 | 67 | 100 | 92 |
| NRTI/NNRTI/PI | 6 | 0 | 0 | 4 | |
| PI | 3 | 33 | 0 | 4 | |
| Median number of antiretroviral drugs | 3 (3;3) | 3 (1;5) | 3 (2;4) | 3 (3;4) | |
| Median number of resistance mutations to: | |||||
| NRTI | 4.5 (4;5.5) | 3.5 (3;5.5) | 4.5 (4.5;4.5) | 4.5 (4;5.5) | |
| NNRTI | 2 (2;2) | 2 (2;2) | 2 (2;2) | 2 (2;2) | |
| PI | 6 (5;6) | 5 (3.5;6) | 5.5 (4.5;6) | 6 (5;6) | |
| Median baseline GSSb | 1.5 (1;2) | 1.5 (1;2.5) | 1.5 (1;1.5) | 1.5 (1;2) | |
| RAL-containing treatment study period | |||||
| Median time spent on RAL-containing treatment study period (days) | 335 (174;336) | 336 (335;336) | 336 (182;337) | 335 (179;336) | |
| Median virus load during low-level viremia | |||||
| (copies/ml) | 104 (79;173) | 108 (98;230) | 73 (63;98) | 100 (75;167) | |
| Adherence scoring | Undetermined | 10 | 16 | 8 | 10 |
| (% of patients) | Low (< 80%) | 2 | 0 | 1 | 2 |
| Moderate (80–99%) | 13 | 17 | 7 | 12 | |
| High (100%) | 75 | 67 | 84 | 76 | |
ART, antiretroviral therapy; GSS, genotypic susceptibility score; IN, integrase; INSTI, integrase strand-transfer inhibitor; NNRTI, non-nucleoside reverse transcriptase inhibitor; NRTI, nucleoside reverse transcriptase inhibitor; PI, protease inhibitor; RAL, raltegravir.
Values in parentheses denote interquartile range.
Genotype sensitivity score: raltegravir and enfuvirtide were not included in calculation of GSS.
Plasma samples from a total of 94 episodes of low-level viremia were analyzed. Nineteen patients (39%) had more than one episode of low-level viremia (range 1–8 episodes/patient). The overall median viral load during episodes of low-level viremia was 100 copies/ml [interquartile range (IQR) 75–167 copies/ml). The median time between starting raltegravir-containing ART and the first episode of low-level viremia was 165 days (range 24–341 days) and the median time spent on raltegravir-containing treatment was 335 days (IQR 179–336 days).
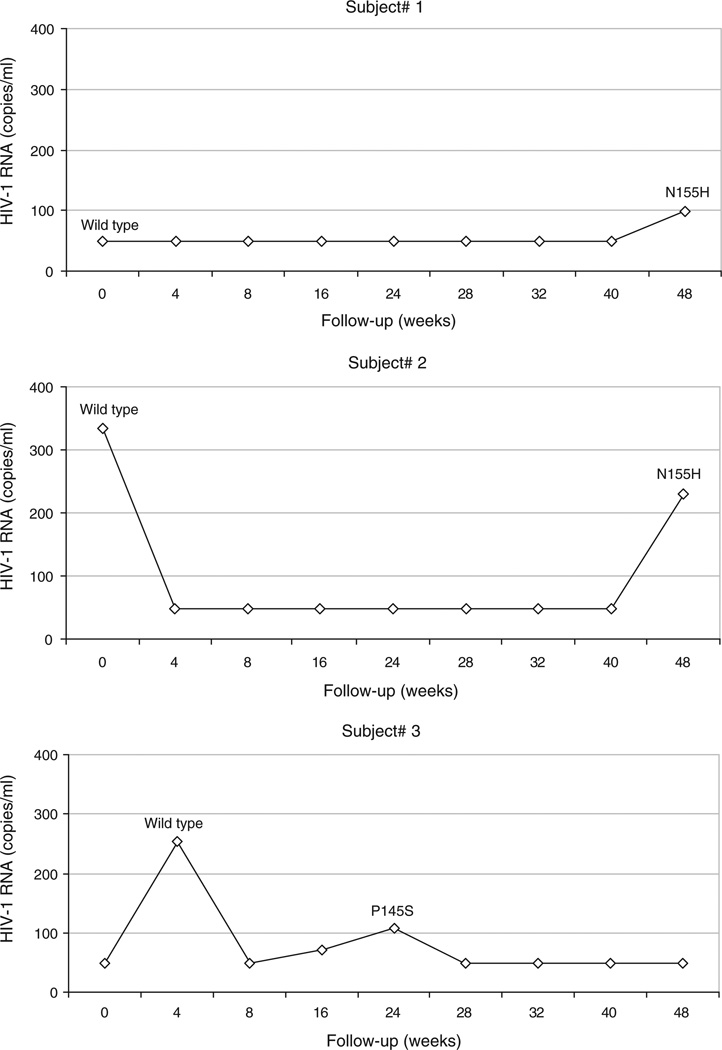
Integrase gene sequencing was successfully accomplished on 73 samples (78%), corresponding to 39 patients (80%). Characteristics of these patients were similar to those of patients in whom integrase sequencing was unsuccessful (Table 1). Among the patients with a genotype result, three [7.7%, 95% confidence interval (CI) 1.6–20.9%] had INSTI resistance associated mutations consisting of N155H in two and P145S in one; all three were enrolled in the switch arm (Fig. 1). The N155H mutation was detected in samples from two patients (#1 and #2) and the P145S mutation in one patient (#3). The N155H mutation was not detected in proviral DNA from PBMC obtained at entry from either patient. A baseline DNA sample was not available for patient 3, but the P145S mutations was absent from 25 integrase clones obtained from plasma virus at week 4.
Fig. 1.
Plasma HIV-1 RNA levels for the three patients for whom INSTI resistance-associated mutations were detected during treatment with raltegravir (W0: beginning of RAL).
Long-term follow-up data for the three patients with raltegravir-resistant virus were obtained from their medical providers. Plasma HIV-1 RNA levels for the two patients with N155H mutant virus remained below 200 copies/ml at months 34 and 17, respectively, on continued raltegravir treatment. After week 24 patient #3 had four consecutive samples with plasma HIV-1 RNA levels below 50 copies/ml through week 48.
Self-reported adherence to treatment was high in all patients while receiving raltegravir-containing antiretroviral therapy, including for those in whom resistance mutations were found (Table 1). Median raltegravir through plasma concentrations were 126 ng/ml (IQR 90–172 ng/ml), 155 ng/ml (IQR 87–183 ng/ml), 355 ng/ml (IQR 166–469 ng/ml) for patients 1, 2, and 3, respectively. No factors significantly associated with emergence of INSTI resistance mutations during episodes of low-level viremia were identified among baseline characteristics, number of low-level viremia episodes, level of viremia during these episodes or adherence score.
Discussion
We showed that episodes of low-level HIV-1 viremia in treatment-experienced HIV-1-infected patients receiving a raltegravir-containing regimen may be associated with the emergence of INSTI resistance mutations in some patients.
The N155H mutation, which is associated in vitro with a modest decrease in raltegravir sensitivity [9], emerged in two patients. It has been shown that the N155H mutation impairs viral replication capacity compared to wild type [9,10]. This finding could explain the absence of virological failure in these patients despite continued raltegravir therapy. We also found the unexpected emergence of the P145S mutation, which confers elvitegravir resistance but has not been associated with resistance to raltegravir [11,12]. Although detection of this mutation in a virus from a single patient must be interpreted with caution, this finding suggests that the P145S mutation can be selected in vivo by raltegravir.
In a previous study, 68% of treatment-experienced patients receiving a raltegravir-containing regimen with virological failure developed raltegravir resistance-associated mutations [2], but no samples from patients with plasma HIV-1 RNA below 1000 copies/ml were tested in that study. Two other studies did perform resistance testing on patients with low-level viremia, but did not detect emergence of raltegravir resistance [13,14].
Although we examined samples from 94 episodes of low-level viremia, the overall sample size was modest. The relatively small number of patients studied could explain our failure to identify factors associated with the selection of INSTI resistance in these patients. Previous studies evaluating the risks of developing antiretroviral drug resistance in patients with low-level viremia identified virus load and a history of prior antiretroviral therapy as predictors of virological failure but not of the emergence of resistance-associated mutations [15,16]. In addition, the reduced replication capacity of the multidrug-resistant viruses present in the heavily pretreated patients studied here could have contributed to the low level of viral replication, and hence to the low rate of emergence of INSTI resistance mutations.
In conclusion, low-level viremia in patients receiving a raltegravir-containing antiretroviral regimen was associated in some patients with the emergence of INSTI resistance mutations. Identifying those patients with low-level viremia in whom a regimen switch is indicated remains a significant clinical challenge.
Acknowledgements
We thank Merck Sharp & Dohme-Chibret Laboratories for graciously providing raltegravir for this trial. We also thank the participating Easier investigators, clinical trials sites and study participants for their contributions to this work.
The work was supported by a grant from the Agence Nationale de Recherches sur le SIDA et les Hépatites Virales (ANRS) and by NIH grants AI068636 (through a Virology Specialty Laboratory contract from the ACTG) and K24 RR016482 to D.R.K.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lennox JL, DeJesus E, Lazzarin A, Pollard RB, Madruga JV, Berger DS, et al. Safety and efficacy of raltegravir-based versus efavirenz-based combination therapy in treatment-naive patients with HIV-1 infection: a multicentre, double-blind randomised controlled trial. Lancet. 2009;374:796–806. doi: 10.1016/S0140-6736(09)60918-1. [DOI] [PubMed] [Google Scholar]
- 2.Steigbigel RT, Cooper DA, Kumar PN, Eron JE, Schechter M, Markowitz M, et al. Raltegravir with optimized background therapy for resistant HIV-1 infection. N Engl J Med. 2008;359:339–354. doi: 10.1056/NEJMoa0708975. [DOI] [PubMed] [Google Scholar]
- 3.Cooper DA, Steigbigel RT, Gatell JM, Rockstroh JK, Katlama C, Yeni P, et al. Subgroup and resistance analyses of raltegravir for resistant HIV-1 infection. N Engl J Med. 2008;359:355–365. doi: 10.1056/NEJMoa0708978. [DOI] [PubMed] [Google Scholar]
- 4.Eron JJ, Young B, Cooper DA, Youle M, Dejesus E, Andrade-Villanueva J, et al. Switch to a raltegravir-based regimen versus continuation of a lopinavir-ritonavir-based regimen in stable HIV-infected patients with suppressed viraemia (SWITCHMRK 1 and 2): two multicentre, double-blind, randomised controlled trials. Lancet. 2010;375:396–407. doi: 10.1016/S0140-6736(09)62041-9. [DOI] [PubMed] [Google Scholar]
- 5.De Castro N, Braun J, Charreau I, Pialoux G, Cotte L, Katlama C, et al. Switch from enfuvirtide to raltegravir in virologically suppressed multidrug-resistant HIV-1-infected patients: a randomized open-label trial. Clin Infect Dis. 2009;49:1259–1267. doi: 10.1086/605674. [DOI] [PubMed] [Google Scholar]
- 6.De Castro N, Braun J, Charreau I, De Truchis P, Jeanblanc F, Verdon R, et al. Switch from enfuvirtide to raltegravir in virologically suppressed multidrug-resistant HIV-1 infected patients: final results of the randomized ANRS 138 Trial (EASIER). 5th IAS Conference on HIV Pathogenesis, Treatment and Prevention; Cape Town, South Africa. 2009. [Google Scholar]
- 7.Goldwirt L, Barrail-Tran A, Da Cruz M, Taburet AM, Furlan V. Quantification of raltegravir (MK0518) in human plasma by high-performance liquid chromatography with photodiode array detection. J Chromatogr B Analyt Technol Biomed Life Sci. 2010;878:456–460. doi: 10.1016/j.jchromb.2009.10.029. [DOI] [PubMed] [Google Scholar]
- 8.Le Moing V, Chene G, Carrieri MP, Alioum A, Brun-Vezinet F, Piroth L, et al. Predictors of virological rebound in HIV-1-infected patients initiating a protease inhibitor-containing regimen. AIDS. 2002;16:21–29. doi: 10.1097/00002030-200201040-00004. [DOI] [PubMed] [Google Scholar]
- 9.Fransen S, Gupta S, Danovich R, Hazuda D, Miller M, Witmer M, et al. Loss of raltegravir susceptibility by human immunodeficiency virus type 1 is conferred via multiple nonoverlapping genetic pathways. J Virol. 2009;83:11440–11446. doi: 10.1128/JVI.01168-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu Z, Kuritzkes DR. Effect of raltegravir resistance mutations in HIV-1 integrase on viral fitness. J Acquir Immune Defic Syndr. 2010 doi: 10.1097/QAI.0b013e3181e9a87a. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garvey EP, Johns BA, Gartland MJ, Foster SA, Miller WH, Ferris RG, et al. The naphthyridinone GSK364735 is a novel, potent human immunodeficiency virus type 1 integrase inhibitor and antiretroviral. Antimicrob Agents Chemother. 2008;52:901–908. doi: 10.1128/AAC.01218-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kobayashi M, Nakahara K, Seki T, Miki S, Kawauchi S, Suyama A, et al. Selection of diverse and clinically relevant integrase inhibitor-resistant human immunodeficiency virus type 1 mutants. Antiviral Res. 2008;80:213–222. doi: 10.1016/j.antiviral.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 13.Caby F, Valin N, Marcelin AG, Schneider L, Andrade R, Guiguet M, et al. Raltegravir as functional monotherapy leads to virological failure and drug resistance in highly treatment-experienced HIV-infected patients. Scand J Infect Dis. 2010;42:527–532. doi: 10.3109/00365541003621502. [DOI] [PubMed] [Google Scholar]
- 14.Charpentier C, Roquebert B, Colin C, Taburet AM, Fagard C, Katlama C, et al. Resistance analyses in highly experienced patients failing raltegravir, etravirine and darunavir/ritonavir regimen. AIDS. 2010 doi: 10.1097/QAD.0b013e32833ed2a7. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 15.Garcia-Gasco P, Maida I, Blanco F, Barreiro P, Martin-Carbonero L, Vispo E, et al. Episodes of low-level viral rebound in HIV-infected patients on antiretroviral therapy: frequency, predictors and outcome. J Antimicrob Chemother. 2008;61:699–704. doi: 10.1093/jac/dkm516. [DOI] [PubMed] [Google Scholar]
- 16.Sungkanuparph S, Groger RK, Overton ET, Fraser VJ, Powderly WG. Persistent low-level viraemia and virological failure in HIV-1-infected patients treated with highly active antiretroviral therapy. HIV Med. 2006;7:437–441. doi: 10.1111/j.1468-1293.2006.00403.x. [DOI] [PubMed] [Google Scholar]