Abstract
Among developmental control genes, transcription factor-target gene “linkages” — the direct connections between target genes and the factors that control their patterns of expression — can show remarkable evolutionary stability. However, the specific binding sites that mediate and define these regulatory connections are themselves often subject to rapid turnover. Here we describe several instances in which particular transcription factor binding motif combinations have evidently been conserved upstream of orthologous target genes for extraordinarily long evolutionary periods. This occurs against a backdrop in which other binding sites for the same factors are coming and going rapidly. Our examples include a particular Dpp Silencer Element upstream of insect brinker genes, in combination with a novel motif we refer to as the Downstream Element; combinations of a Suppressor of Hairless Paired Site (SPS) and a specific proneural protein binding site associated with arthropod Notch pathway target genes; and a three-motif combination, also including an SPS, upstream of deuterostome Hes repressor genes, which are also Notch targets. We propose that these stable motif architectures have been conserved intact from a deep ancestor, in part because they mediate a special mode of regulation that cannot be supplied by the other, unstable motif instances.
Keywords: Cis-regulatory evolution, Conserved motifs, Notch pathway, Dpp signaling, Hairy/Enhancer of split repressor genes, Bearded family genes
Introduction
It is now well recognized that changes in transcriptional cis-regulatory elements, particularly those that direct the expression of developmental control genes, represent a fundamental mechanism underlying animal evolution (Davidson, 2006; Wray, 2007). Such cis-regulatory novelties have been shown to confer both loss (Chan et al., 2010; Jeong et al., 2008; Prud’homme et al., 2006) and gain (Gompel et al., 2005; Prud’homme et al., 2006; Rebeiz et al., 2011) on a gene’s repertoire of expression specificities. But cis-regulatory evolution is not restricted to the generation of major alterations in gene activity. Even orthologous enhancer modules that drive very similar patterns of expression in two species can differ enormously in their cis-regulatory architecture — the number, order, spacing, and orientation of their component transcription factor binding sites (Hare et al., 2008; Ludwig et al., 2000; Markstein et al., 2004; Romano and Wray, 2003; Swanson et al., 2011).
In this context, it is important to distinguish between a transcription factor-target gene “linkage” — the direct regulatory connection between factor and target — and the specific binding site instances that mediate and define this connection. A transcriptional regulatory linkage might be quite stable evolutionarily even as the relevant binding sites are turning over.
We have previously suggested that transcriptional linkages that confer abstract or generic developmental regulatory capabilities, of general utility to all metazoans, might be expected to be retained for especially long evolutionary periods (Rebeiz et al., 2005). We described one such example, the direct transcriptional repression of genes encoding proneural basic helix-loop-helix (bHLH) activator proteins by bHLH repressor factors of the Hairy/Enhancer of split (Hes) class. We found that bilaterian proneural genes belonging to both the achaete-scute and atonal classes (representing an ancient division that predates the cnidarian-bilaterian divergence) are consistently associated with a high-affinity binding site for a Hes repressor, suggesting that this linkage might be more than 500 million years (My) old. The generic ability to shape spatial patterns of proneural gene expression by direct repression would in principle be valuable regardless of the specific nature of a given species’ nervous system, and we suggested that this might be the basis for the long-term maintenance of this regulatory linkage. To our surprise, we also saw evidence in this phylogenetic study that not only was the Hes repressor-proneural gene linkage being retained in evolution, but that in some cases the specific binding site itself was also conserved over very long periods (Rebeiz et al., 2005).
Here we investigate the evolutionary history of two other transcriptional regulatory linkages involving developmental control genes. Insect genomes include a single gene encoding the transcriptional repressor protein Brinker, which plays an important role in regulating other genes that are targets of the Decapentaplegic (Dpp) signaling pathway (Affolter and Basler, 2007). Transcription of the brinker (brk) gene is itself subject to repression in response to Dpp signaling (Muller et al., 2003). This is mediated by cis-regulatory motifs upstream of brk known as Dpp Silencer Elements (SEs) (Pyrowolakis et al., 2004), which bind a tetrameric complex that includes the transcription factors Mothers against dpp (Mad), Medea (Med), and Schnurri (Shn) (Gao et al., 2005). Remarkably, the brk gene in some species is associated with multiple SEs; the fruit fly Drosophila melanogaster has 11, while the mosquito Anopheles gambiae has 12, leading to the suggestion that this architecture has been evolutionarily conserved (Yao et al., 2008). We show here that other species have only a single SE upstream of their brk ortholog. Moreover, we have identified in nine species representing five insect orders a unique SE upstream of brk that is not only unusually related between species but is also uniquely associated with a novel motif we refer to as the Downstream Element (DE). We propose that this SE+DE motif combination has been conserved from a common insect ancestor, even as the number of other SEs upstream of brk has been changing rapidly in evolution.
The second regulatory linkage we have analyzed involves target genes of the Notch cell-cell signaling pathway (Bray, 2006; Fiuza and Arias, 2007). Suppressor of Hairless [Su(H); CBF1 in vertebrates] functions as the transducing transcription factor for this pathway. In the absence of signaling through the Notch receptor, Su(H) acts to repress Notch target genes. Activation of the receptor leads to the cleavage of its intracellular domain (NICD), which enters the nucleus and forms a trimeric complex with Su(H) and the co-activator protein Mastermind (Mam); this complex now transcriptionally activates the formerly repressed targets.
The known repertoire of Notch pathway targets in both protostomes and deuterostomes includes genes encoding members of the Hes family of bHLH transcriptional repressor (bHLHR) proteins (Bailey and Posakony, 1995; Jarriault et al., 1995; Lecourtois and Schweisguth, 1995). These factors function to inhibit the expression of genes associated with cell fates that are antagonized by Notch signaling. Arthropods also have a second class of Notch targets, the Bearded (Brd) family genes (BFMs) (Bailey and Posakony, 1995; Fontana and Posakony, 2009; Lai et al., 2000; Nellesen et al., 1999). The Notch ligands Delta and Serrate require mono-ubiquitination of their ICDs by the E3 ligase Neuralized (Neur) in order to be active in signaling (Le Bras et al., 2011). In cells that receive and respond to Notch signals, Brd proteins act as competitive inhibitors of the Neur-ligand binding interaction, thereby preventing ligand activation (Bardin and Schweisguth, 2006; Fontana and Posakony, 2009). This helps keep Notch responder cells from themselves becoming effective signalers, thus ensuring the desired directionality of the signaling event.
Su(H) typically binds to its targets via one or more occurrences of an eight-nucleotide motif (Tun et al., 1994), but a small subset of target genes are associated with a special regulatory element known as the Su(H) Paired Site (SPS) (Bailey and Posakony, 1995; Nellesen et al., 1999). This consists of two high-affinity binding sites in opposite orientations, typically separated by 15-17 base pairs. By mediating the cooperative binding of two Su(H)/NICD/Mam trimers, the SPS drives an especially sensitized response to low levels of Notch signaling (Arnett et al., 2010; Nam et al., 2007; Ong et al., 2006). We show that an SPS motif is associated with certain orthologous Hes repressor and Brd family genes that last had a common ancestor hundreds of millions of years ago. In each instance, the SPS is consistently accompanied by specific binding sites for one or more other key regulatory factors. As with the insect brk SE+DE motif combination, we propose that these SPS-containing motif ensembles are ancestral and have been conserved for extraordinarily long evolutionary periods.
Why might a subset of the binding motifs that constitute the cis-regulatory architecture of a developmental control gene be conserved from a deep ancestor, while other motifs are changing freely in evolution? We extend our earlier proposal to include not only the principle of the linkage’s utility to a diverse range of organisms, but also the concept that these ancestral and conserved motifs mediate special modes of transcriptional regulation that are not conferred by other binding sites, even for the same factor or factors.
Materials and methods
Genome sequences
The following genome sequences were utilized in this study: Drosophila melanogaster (Adams et al., 2000); Anopheles gambiae (Holt et al., 2002); Aedes aegypti (Nene et al., 2007); Apis mellifera (Honeybee Genome Sequencing Consortium, 2006); Bombyx mori (International Silkworm Genome Consortium, 2008; Mita et al., 2004); Tribolium castaneum (Tribolium Genome Sequencing Consortium, 2008); Nasonia vitripennis (Werren et al., 2010); Rhodnius prolixus (http://genome.wustl.edu/genomes/view/rhodnius_prolixus/); Acyrthosiphon pisum (International Aphid Genomics Consortium, 2010); Mayetiola destructor (http://www.hgsc.bcm.tmc.edu/); Pediculus humanus corporis (http://www.vectorbase.org/); Daphnia pulex (Colbourne et al., 2011); Homo sapiens (Venter et al., 2001); Xenopus tropicalis (Hellsten et al., 2010); Danio rerio (http://www.sanger.ac.uk/); Gallus gallus (International Chicken Genome Sequencing Consortium, 2004); Strongylocentrotus purpuratus (Sea Urchin Genome Sequencing Consortium, 2006); Saccoglossus kowalevskii (http://www.hgsc.bcm.tmc.edu/); Branchiostoma floridae (Putnam et al., 2008); Amphimedon queenslandica (Srivastava et al., 2010); Trichoplax adhaerens (Srivastava et al., 2008); Nematostella vectensis (Putnam et al., 2007); Acropora digitifera (Shinzato et al., 2011).
Gene annotation and figure preparation
Gene structure annotation, detection of transcription factor binding motifs, and gene diagram figure preparation was carried out using the GenePalette software tool (Rebeiz and Posakony, 2004) (www.genepalette.org).
Hierarchical clustering analysis
A total of 32 Dpp Silencer Element (SE) motifs conforming to the original GRCGNCN5GTCTG definition (Pyrowolakis et al., 2004) were detected upstream of brk in the nine insect species shown in Fig. 1A. Sequence relationships between the motifs were investigated by multiple alignment using ClustalX version 2.1 (Larkin et al., 2007); the resulting phylogenetic tree was displayed using NJplot version 2.3 (Perriere and Gouy, 1996).
Fig. 1.
A Silencer Element (SE) + Downstream Element (DE) cis-regulatory motif combination is a shared feature of insect brk genes. A: Annotated scale diagrams of the brk gene and its upstream non-coding sequences in representatives of various insect orders. Blue boxes represent brk protein coding sequences; white boxes represent untranslated regions. Arrows denote direction of transcription and are positioned at either the transcription start site or the start codon of the gene. Except in the case of Ag and Aa, the entire intergenic region between brk and the next upstream gene is shown. Dpp Silencer Elements (Yao et al., 2008) are indicated by “S”; Downstream Elements are denoted by “D”. Shared S+D motif combinations are shown in red. Lower-case “s” upstream of Rp brk denotes single-base mismatch to the S motif definition; lower-case “d” upstream of Ag, Nv, and Ap brk denotes single-base mismatch to the D motif definition (see C for alignment). Other single-base mismatches to either motif are omitted. Sequence scale indicated in upper right corner. Dm, Drosophila melanogaster (Diptera); Ag, Anopheles gambiae (Diptera); Aa, Aedes aegypti (Diptera); Bm, Bombyx mori (Lepidoptera); Tc, Tribolium castaneum (Coleoptera); Am, Apis mellifera (Hymenoptera); Nv, Nasonia vitripennis (Hymenoptera); Ap, Acyrthosiphon pisum (Hemiptera); Rp, Rhodnius prolixus (Hemiptera). B: Logo plots of the “special” SE subset (upper, 10 occurrences) and all “other” SE motifs (lower, 32 occurrences) upstream of the insect brk genes shown in A. Note the additional sequence constraint in the “special” subset in both unconstrained (5, 8, 10, 12-14) and partially degenerate (7) positions within the motif, as well as in flanking positions (3, 20, 21), compared to that in the “other” set. The “special” subset has an uncorrected sequence information content of 30.35 bits, far higher than the mean of 21.01 bits for 1000 randomly chosen sets of 10 SE motif instances drawn from the non-coding portion of the fly genome (see Materials and methods). By contrast, the “other” subset has an uncorrected sequence information content of 20.43 bits, compared to a mean of 18.62 bits for 1000 randomly chosen sets of 32 SE motif instances. C: Sequence alignment of the SE+DE motif combinations shown in red in A. Except for the two occurrences in Ag, actual distances between SEs and the corresponding DEs are shown. D: Logo plot of the DE motifs associated with the 10 “special” SEs (see A, C).
Logo plots
Sequence logo plots were generated using WebLogo version 2.8.2 (Crooks et al., 2004).
Comparison of sequence information content
A total of 42 SE motifs conforming to the modified GNCKNCN5GTCTG definition suggested by Yao et al. (Yao et al., 2008) were detected upstream of brk in the nine species shown in Fig. 1A. Ten of these belong to the “special” subset. To compare the information content of the “special” subset to that of the remaining 32 motif instances (“other”), we inventoried the Drosophila melanogaster genome sequence (version R5/dm3) for all occurrences of the modified SE motif in noncoding regions; 1251 were found. The sequence information content (SIC, uncorrected for small sample size) of 1000 randomly chosen sets of 10 and 1000 randomly chosen sets of 32 (all drawn from the 1251 genomic instances) were computed, and the mean SIC values of these sets were then compared to those of the “special” (10) and “other” (32) subsets, respectively.
Identification of shared sequence motifs
To confirm that our identification of conserved sequence motifs upstream of Hes and BFM genes is substantially complete, we made use of the San Diego Supercomputer Center (SDSC) implementation of MEME (Bailey and Elkan, 1994) (http://meme.sdsc.edu/).
Transgene construction
A wild-type GFP reporter transgene bearing the "E+SPS+P" enhancer module from upstream of the Anopheles gambiae Brd family gene (Ag BFM-GFP) was constructed as follows. A 1.0-kb genomic DNA fragment covering −37 to −1037 upstream of Ag BFM was PCR-amplified using the following primers: Fwd: 5′-gaattcCTCTGAATAGCGCAAAACACAACACAATCGCAGGAC-3′ and Rev: 5′-ctcgagCCGACCCCGACCCCGACCCTTTCCACG-3′ (lowercase letters represent added EcoRI and XhoI restriction sites, respectively). The fragment was inserted into the multiple cloning site of the insulated P-element transformation vector pH-Stinger (Barolo et al., 2004). Mutant versions of this reporter transgene were prepared by changing the single proneural protein binding site (P) from GCAGGTG to GAAGCTT (Van Doren et al., 1992) (Ag BFM-Pm-GFP and Ag BFM-UmPm-GFP) and/or by changing the two Su(H) binding sites (U) in the SPS (of the form YGTGGGAA) to YGTGGCAA (Bailey and Posakony, 1995) (Ag BFM-Um-GFP and Ag BFM-UmPm-GFP).
The brk enhancer module “7/8/9” described by Yao et al. (Yao et al., 2008) was amplified by PCR from genomic DNA using the following primers: Fwd: 5′-gcatgcTATATACATGGGGTGGCATGAGCATGTGCG-3′ and Rev: 5′-ggcgcgccCCACAAGGCGCTAGAACGAGATGGCGCACA-3′ (lowercase letters represent added SphI and AscI restriction sites, respectively). Following sequence verification, the fragment was introduced into the multiple cloning site of the attB-H-Stinger GFP reporter vector (S.W. Miller, UC San Diego, unpublished; further information available upon request). The single Downstream Element (DE) in the 7/8/9 module was mutated by changing the wild-type sequence GCAACGTTGCCACTT to TCAACTTGGACCCTG.
Generation of transgenic fly lines
Wild-type and mutant versions of the Ag BFM-GFP enhancer-reporter construct were introduced into the w1118 recipient strain via P element-mediated germline transformation (Rubin and Spradling, 1982).
Wild-type and mutant versions of the brk 7/8/9 enhancer-reporter construct were introduced into the genome using the ϕC31-integrase system (Bischof et al., 2007). The recipient strain carries an attP docking site on the third chromosome (attP2 site at 68A4) and the nanos-ϕC31 integrase gene on the X chromosome (Stock #25710, Bloomington Drosophila Stock Center). Multiple independent insertion lines were obtained for both the wild-type and DEm versions. Integration events were confirmed by PCR according to Venken et al. (Venken et al., 2006).
Tissue preparation, antibody staining, and confocal microscopy
Wing imaginal discs from late third-instar larvae and nota from pupae at 14 hours after puparium formation (APF) were dissected in PBT (1X PBS, 0.1% Triton-X100) and fixed in 4% paraformaldehyde (in PBT) for 30 minutes at room temperature. Discs were washed in PBT, mounted, and imaged using a Leica TCS SP2 confocal microscope. Nota were first stained with anti-Hnt (monoclonal, Developmental Studies Hybridoma Bank) primary antibody and Alexa 555 (Molecular Probes) secondary antibody before mounting. Confocal average projections were generated with Z-axis sections at 2-μm (discs) or 1-μm (nota) intervals using Leica Confocal Software version 2.5 (Leica Microsystems). To compare fluorescence between wild-type and DEm brk 7/8/9 enhancer-reporter constructs, wing discs of the two genotypes were processed side-by-side and imaged using the same gain.
Results
Conservation of a unique Dpp Silencer Element upstream of insect brinker genes
The 16.6-kb intergenic region upstream of the Drosophila melanogaster brk gene (Fig. 1A) includes no fewer than nine instances of the original GRCGNCN5GTCTG definition of the SE motif (Pyrowolakis et al., 2004) and 11 occurrences of the modified motif GNCKNCN5GTCTG suggested by Yao et al. (Yao et al., 2008). Based on their thorough analyses using reporter transgenes in vivo, Yao et al. concluded that these 11 SE motifs probably correspond to nine or ten distinct cis-regulatory modules, each of which, by integrating activator and SE inputs, contributes independently to the brk expression pattern (Yao et al., 2008).
Their identification of 12, 11, and 11 SE sites upstream of brk in D. pseudoobscura (23 kb), D. virilis (24 kb), and the mosquito Anopheles gambiae (95 kb), respectively, prompted Yao et al. to suggest that this unusual cis-regulatory organization is evolutionarily conserved. However, a different picture emerges when the brk upstream regions of other insects are examined (Table 1; Fig. 1A). The 20 kb upstream of brk in Bombyx mori (silk moth) and the 8.5 kb of upstream sequence in Apis mellifera (honeybee) each include three original SE motifs (Bombyx has four matches to the Yao et al. site definition). In both Tribolium castaneum (red flour beetle) and Acyrthosiphon pisum (pea aphid), the brk upstream region (4.1 kb and 31 kb, respectively) contains only a single occurrence of either SE motif. And while another mosquito (Aedes aegypti) has 311 kb of intergenic sequence upstream of brk, this very large region includes only three and six instances of the original and the Yao et al. SE motifs, respectively, and only one and two instances, respectively, in the first 200 kb upstream. Thus, even if we assume that all of these motif occurrences represent functional SEs in vivo, the presence of large numbers of them is not a shared characteristic of insect brk genes.
Table 1.
| Speciesa | Upstream (kb)b | #oSEc | #mSEd | #DEe | SE+DE Motif Combination(s)f |
|---|---|---|---|---|---|
| Dm | 17 | 9 | 11 | 1 | 2×SEg+DE |
| Ag | 95 | 8 | 12 | 2 | SE+DE, 2×SEg+DEmmh |
| Aa | 311 | 3(1)i | 6(2)i | 1 | SE+DE |
| Bm | 20 | 3 | 4 | 1 | SE+DE |
| Tc | 4.1 | 1 | 1 | 1 | SE+DE |
| Am | 8.5 | 3 | 3 | 2 | SE+DE |
| Nv | 12 | 3 | 3 | 2mmh | SE+DEmmh |
| Ap | 31 | 1 | 1 | 1, 5mmh,j | SE+DEmmh,j |
| Rp | 17 | 1mmh | 1mmh | 1 | SEmmh+DE |
Species symbols are as listed in the legend to Fig. 1.
Indicates the size of the intergenic region upstream of brk, based on current genome annotations.
Number of Dpp Silencer Elements (SE) upstream of brk, using the original (o) GRCGNCN5GTCTG motif definition (Pyrowolakis et al., 2004).
Number of Dpp Silencer Elements (SE) upstream of brk, using the modified (m) GNCKNCN5GTCTG motif definition (Yao et al., 2008).
Number of Downstream Elements (DE; GCN3GTTGCCRY) upstream of brk.
Nature of SE+DE motif combination(s) found.
Dm and Ag both have two closely spaced SEs associated with a DE (Dm) or a DEmm (Ag); see Fig. 1A.
One-base mismatch (mm) to the motif definition.
Shown in parentheses is the number of SEs within the first 200 kb upstream of brk.
Ap has one exact match to the DE motif definition, not associated with its single SE; the SE is associated with a single-base-mismatch DE motif (see Fig. 1A,C).
To investigate further the question of evolutionary conservation, we asked if it was possible to discern any exceptional similarity between brk-associated SE motifs in the different species that might be suggestive of site orthology. Indeed, using ClustalX for hierarchical motif clustering, we identified ten SE motifs from nine species that comprise a distinct subset of the total ensemble of occurrences (see Fig. S1). First, these ten sites define an unusually constrained version of the SE motif (Fig. 1B). Significant information content is evident at three positions flanking the motif (3, 20, and 21 in Fig. 1B), while two partially degenerate and several fully unconstrained positions within the motif show strong (8, 10, 12, 13) and even complete (5, 7, 14) sequence bias. By contrast, the remaining 22 (32 by the Yao et al. definition) SE motif occurrences upstream of brk in the nine species contain little more information than that embodied in the motif definitions (Fig. 1B) (Pyrowolakis et al., 2004; Yao et al., 2008).
A second and unique characteristic of this distinctive SE motif class is the presence, at a variable but typically quite short distance downstream from the SE, of a novel sequence we refer to as the Downstream Element (DE) (Fig. 1A,C,D). The DE is always found in the same orientation with respect to the SE, regardless of the SE’s orientation with respect to the direction of brk transcription (see Fig. 1A). With the exception of Anopheles gambiae brk, no brk gene includes more than one SE+DE combination, even when the more relaxed Yao et al. (Yao et al., 2008) SE definition and single-base mismatches to the DE definition (GCN3GTTGCCRY) are both permitted. Thus, in each species (again, Anopheles being the sole exception), the single sequence-constrained SE motif described above is paired uniquely with a DE.
The nine species we have considered in our analysis represent five insect orders (Diptera, Lepidoptera, Coleoptera, Hymenoptera, and Hemiptera). That the brk upstream region in each species includes a single SE (Anopheles has two) that is both a member of the sequence-constrained set and uniquely associated with a DE is likely to be highly significant both evolutionarily and functionally. Particularly noteworthy is the observation that the single SE found upstream of brk in three species (Tribolium, Acyrthosiphon, and Rhodnius) is in each case a member of this special set. We suggest that this phenomenon represents the long-term evolutionary conservation of an ancestral SE+DE unit that, by comparison to other SE motif occurrences, confers a unique regulatory functionality.
The brk Downstream Element (DE) functions in activation
The long-term conservation of the DE motif upstream of insect brk genes strongly implies its functionality, and we sought to test this expectation directly. Yao et al. showed previously that a 784-bp region upstream of Drosophila brk that includes three SEs drives a pattern of lacZ reporter expression in the wing imaginal disc similar to that of endogenous brk (Yao et al., 2008) (Fig. 1A; Fig. 2). The DE of Drosophila brk lies adjacent to the middle SE in this fragment (see Fig. 1A,C; Fig. 2A). We compared the activities of reporter transgenes in which GFP expression is driven by either wild-type or DE-mutant versions of the fragment (Fig. 2). We find that, while their spatial patterns of GFP activity in imaginal discs appear very similar, the DE-mutant reporter (Fig. 2F-H) is expressed at a much lower level than the wild-type reporter (Fig. 2B-E). This result implies that the conserved DE does indeed have a functional role in the transcriptional activation of brk expression in this tissue.
Fig. 2.
The Downstream Element (DE) contributes to activation of a brk enhancer module in Drosophila. A: Diagram of the 0.8-kb “7/8/9” module from upstream of Dm brk (Yao et al., 2008). This fragment (bounded by red boxes) contains three SEs (“S”; the seventh, eighth, and ninth upstream of the transcription start site) plus the DE (“D”) that accompanies the middle SE (#8; see Fig. 1A,C). Wild-type (brk789wt) and DE-mutant (brk789DEm) versions of the fragment are shown. B-H: Expression in late third-instar imaginal disc tissue of GFP reporters driven by either the wild-type (B-E; four independent transgene insertions) or the DE-mutant (F-H; three independent transgene insertions) version of the 7/8/9 module (see A). All transgenes are present in one copy, inserted into the attP2 docking site (see Materials and methods). Mutation of the DE results in severe reduction of the GFP reporter signal.
Long-term evolutionary conservation of SPS-containing cis-regulatory architectures upstream of Notch pathway target genes
In previous reports, we have described the utilization of a “P+S” cis-regulatory code by genes that are activated via Notch signaling during lateral inhibition in proneural clusters (Bailey and Posakony, 1995; Castro et al., 2005; Nellesen et al., 1999; Singson et al., 1994). S sites mediate the activation and repression functions of the Notch-regulated transcription factor Su(H), while P sites mediate activation by proneural proteins. In Drosophila, Notch targets known to employ this code include bHLH repressor genes of the Hes class, as well as Brd family genes (Bailey and Posakony, 1995; Lai et al., 2000; Lecourtois and Schweisguth, 1995; Nellesen et al., 1999). The number and location of S and P motifs varies greatly from one target gene to another (Nellesen et al., 1999).
A subset of Notch target genes that use the “P+S” code are characterized by the presence of a special motif called the Su(H) Paired Site or SPS, which consists of two high-affinity Su(H) binding sites in opposite orientations, separated by 15-17 bp (Bailey and Posakony, 1995; Nellesen et al., 1999). This distinctive element is often accompanied by one or more single, or “lone”, Su(H) sites.
Examination of the upstream regulatory regions of orthologous Hes bHLH repressor and Brd family genes in various arthropods reveals in each case the apparent long-term conservation of a particular P+SPS motif combination (Fig. 3; highlighted in red). In the case of the Hes genes, the SPS, which occurs at various locations with respect to the transcription start site, is closely flanked on the upstream side by an “upper strand” P site (Fig. 3A). Brd family genes, by contrast, are associated with an SPS (again at various distances upstream) accompanied by a “lower strand” P motif located closer to the transcription start site (Fig. 3B). BFMs are also characterized [except in Drosophila and other Brachyceran flies, such as the tsetse fly Glossina morsitans (not shown)] by the presence, upstream of the SPS, of an extended “E box” motif (RRCAGATGGY) that we have found by in vitro assays to be a variant proneural protein binding site (S.W. Miller, unpublished). Note that, in both Hes and Brd family genes, additional “lone” Su(H) sites and/or P sites may also be present, but these are not widely conserved, if at all. At a minimum, though, the distinctive P+SPS or E+SPS+P combination is present (e.g., Ap bHLHR-1, Ag BFM).
Fig. 3.
Evolutionary conservation of “P+SPS” motif combinations upstream of orthologous arthropod Hes bHLH repressor and Brd family genes. A: Diagrams of orthologous Hes-class bHLH repressor genes located in the E(spl)-C of various insect species; these belong to the “E(spl)-C bHLH-1” clade described by Duncan and Dearden (Duncan and Dearden, 2010), and are labeled here as “bHLHR-1” for clarity. B: Diagrams of orthologous Brd family genes located in the E(spl)-C of various arthropod species. Blue boxes represent protein coding sequences; white boxes represent untranslated regions (UTRs). Arrows denote direction of transcription and are positioned at the transcription start site of the gene (or at the start codon in the case of Md and Rp genes). Genes are aligned on the start codon. High-affinity Su(H) binding sites (YGTGDGAA) are indicated by “U”; Achaete/Scute-type proneural protein binding sites (RCAGSTG) are denoted by “P”; an extended E box motif identified by MEME (RRCAGATGGY) upstream of Brd family genes (see B) is represented by “E”. Conserved P+SPS (A) and E+SPS+P (B) motif combinations are shown in red. In B, note the absence of the E motif in Dm m4 (and in the ortholog in other Brachyceran flies), and its presence on the “upper” instead of the “lower” strand in Am BFM (and in the ortholog in other Hymenopterans). Lower-case “u” in the SPS upstream of Phc bHLHR-1 (see A) denotes single-base mismatch to the U motif definition (CATGGGAA); Su(H) binds this site with somewhat reduced affinity (Nellesen et al., 1999). B, G, and K symbols in 3′ UTRs represent Brd box, GY box, and K box miRNA binding motifs (Lai and Posakony, 1997; Lai et al., 1998; Lai et al., 2005). Species symbols are listed in the legend to Fig. 1A, except Md, Mayetiola destructor (Diptera); Phc, Pediculus humanus corporis (Phthiraptera); Dp, Daphnia pulex (Crustacea). Sequence scale is shown in upper left corner of each panel.
We suggest that these observations reflect the evolutionary conservation, over more than 400 My, of a specific P+SPS or E+SPS+P cis-regulatory architecture that was present in the common ancestors of these genes. This interpretation is strengthened by the fact that in most species we can clearly establish orthology between the genes we are comparing. Both the bHLH repressor and Brd family genes shown in Fig. 3 typically occupy the same positions in the respective Enhancer of split gene complexes [E(spl)-Cs] of these species; moreover, Bayesian phylogenetic analysis fully supports the orthology of the Hes genes (Duncan and Dearden, 2010). We emphasize that, because of their low complexity, we cannot argue strongly for or against the conservation of the P and E sites that we have indicated as part of the shared architectures (see, however, Fig. S2), but we suggest that the highly constrained SPS is much more likely to have been conserved than repeatedly evolved anew.
Brd family genes have not been found in deuterostomes, but Hes repressor genes are widespread among metazoans, being present even in the placozoan Trichoplax adherens (see Discussion and Fig. 6). As in the arthropods, we find that an orthologous set of these Notch-regulated genes is associated in deuterostomes with the presence of three specific upstream cis-regulatory motifs (Fig. 4A; highlighted in red). First, an SPS element is found immediately upstream of the transcription start site. Next, at various distances upstream of the SPS, a predicted high-affinity binding site for bHLH repressors themselves (an "R" site) occurs. Finally, yet further upstream but often near the R site is a novel motif we refer to as the X element (XE) (Fig. 4B,C). As in the case of arthropod Hes repressor and Brd family genes, additional Su(H) lone sites may be present, but they do not show long-term conservation.
Fig. 6.
Direct regulation of Hey- and Hes-class bHLH repressor genes by Su(H) is apparently ancient. Shown are diagrams of Hey and Hes genes in three non-bilaterians. Blue boxes represent protein coding sequences; white boxes represent untranslated regions. Arrows denote direction of transcription and are positioned at either the transcription start site or the start codon of the gene. Genes are aligned on the start codon. High-affinity Su(H) binding sites (YGTGDGAA) are indicated by “U”; Achaete/Scute-type proneural protein binding sites (RCAGSTG) are denoted by “P”. Sequence scale is shown in upper left corner. The demosponge Amphimedon queenslandica (Amq) has a Hey gene, but no Hes genes (Simionato et al., 2007; Srivastava et al., 2010). The placozoan Trichoplax adhaerens (Ta) has one Hey ortholog, one Hey-related gene (not shown), and one Hes gene (Srivastava et al., 2008). The cnidarian Nematostella vectensis (Nv) has 11 Hes genes (Putnam et al., 2007); a representative example is shown [this corresponds to Nem52 (Simionato et al., 2007)]. Nv also has one Hey ortholog (shown) and one Hey-related gene (not shown). Note conservation of exon-intron structure between Hey and Hes genes, respectively. High-affinity Su(H) sites occur in the proximal upstream regions of all of these genes, but no SPSs are found. Significantly, the three-Su(H)-site configuration shown for Nv Hes1 is conserved in the orthologous gene of the distantly related anthozoan Acropora digitifera (stony coral) (see Fig. S3).
Fig. 4.
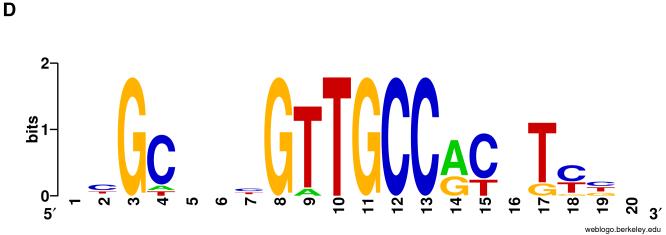
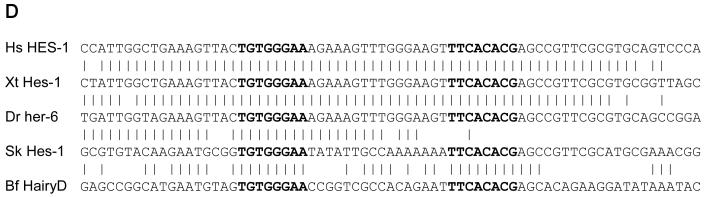
Evolutionary conservation of an “X+R+SPS” motif combination upstream of Hes1 bHLH repressor gene orthologs in various deuterostomes. A: Diagrams of Hes1 orthologs in representative deuterostomes. Blue boxes represent protein coding sequences; white boxes represent untranslated regions. Arrows denote direction of transcription and are positioned at the transcription start site of the gene. Genes are aligned on the start codon. High-affinity Su(H)/CBF1 binding sites (YGTGDGAA) are indicated by “U”; Achaete/Scute-type proneural protein binding sites (RCAGSTG) are denoted by “P”; high-affinity bHLH repressor binding sites (GGCACGYGHY) are designated “R”; the novel X Element motif (see B, C) is also indicated (X). Only a truncated version of the X Element is present in Sp, indicated by a lowercase “x”. Conserved X+R+SPS motif combinations are shown in red. Note also the conservation of a palindromic P site (brown) located between the X and R sites in mammals, amphibians, and fish. Hs, Homo sapiens (Chordata; Mammalia); Xt, Xenopus tropicalis (Chordata; Amphibia); Dr, Danio rerio (Chordata; Actinopteryggii); Gg, Gallus gallus (Chordata; Aves); Sp, Strongylocentrotus purpuratus (Echinodermata; Echinoidea); Sk, Saccoglossus kowalevskii (Hemichordata; Enteropneusta); Bf, Branchiostoma floridae (Cephalochordata). The Bf genome has undergone a large paralogous expansion of the Hes repressor gene family (Minguillon et al., 2003); the hairyD gene is shown here. Sequence scale is shown in upper left corner. B: Sequence alignment of the novel “X Element” motif upstream of deuterostome Hes1 genes; strictly conserved nucleotides are shown in bold. Note the 18/18 sequence identity between Hs and Bf (underlined). As shown, Sp has a strong partial match to this motif in the corresponding location (see A); note that the missing component GTTTTC occurs a short distance upstream. Species symbols as in A. C: Logo plot corresponding to the XE motif alignment shown in B (Sp omitted). D: Sequence alignment of the Hes1 SPS motif and flanking regions from Hs, Xt, Dr, Sk, and Bf, suggesting orthology between these elements in deuterostomes. Su(H)/CBF1 binding sites comprising the SPS are shown in bold; vertical bars indicate sequence identity. Note exceptionally strong sequence conservation between mammals, amphibians, and fish. Sequences of the two 8-bp Su(H)/CBF1 binding sites are strictly conserved in all seven species (not shown; see A). Species symbols as in A.
A number of lines of evidence support the interpretation that this shared cis-regulatory motif configuration upstream of deuterostome Hes repressor genes reflects long-term evolutionary conservation of an ancestral architecture. First, the genes themselves are generally unambiguous orthologs, so direct comparison of their putative regulatory motifs is valid and informative. Second, the nearly identical positioning of the SPS motif with respect to the transcription start sites of these genes is strongly suggestive of conservation. The frequent proximity of the X and R sites, as well as the common sequential order of the three motifs (X, R, SPS) is likewise consistent with this interpretation. Finally, strong conservation of the sequences of the various motifs, and even their flanking sequences, clearly suggests that the individual elements are orthologous. Thus, the various R sites denoted in Fig. 4A are identical in 10/10 positions (GGCACGTGCT), despite the fact that several variants of this motif are compatible with high-affinity binding by Hes repressor proteins (Jennings et al., 1999; Rebeiz et al., 2005; Van Doren et al., 1994). Even more strikingly, we observe strong sequence identity of the SPS elements and flanking sequences in various deuterostomes (Fig. 4D).
Conservation of “P+SPS” cis-regulatory function in Diptera
The foregoing analysis establishes the long-term evolutionary conservation of particular “P+SPS” motif combinations associated with Notch pathway target genes in arthropods. To investigate whether the functional properties of this cis-regulatory architecture are likewise conserved, we tested the behavior of a 1.0-kb non-coding DNA fragment from upstream of the Anopheles gambiae (Ag) BFM gene (Schlatter and Maier, 2005) in a reporter assay in transgenic Drosophila (Fig. 5). As shown and described above, this region contains only a single P site and an SPS (Fig. 5A). When placed upstream of a minimal Hsp70 promoter and an eGFP reporter gene in the pH-Stinger vector (Barolo et al., 2004), this fragment successfully recapitulates the specificity of previously studied Notch-regulated cis-regulatory modules from Drosophila BFM genes (Bailey and Posakony, 1995; Castro et al., 2005; Lai et al., 2000); i.e., it directs expression selectively in the non-SOP cells of proneural clusters (Fig. 5B-D). As in the case of the Drosophila enhancers, this expression is dependent on the integrity of the lone P site, since reporter gene expression is virtually abolished when this motif is mutated (Pm; Fig. 5E). Also mimicking the fly enhancers, two major effects are observed when the two Su(H) sites in the Ag BFM fragment’s SPS are mutated: Expression in non-SOP cells is greatly reduced, and ectopic expression in SOPs appears (Um; Fig. 5F-H). Finally, the ectopic SOP activity of the Um fragment is fully dependent on the P site, as this expression (along with the residual non-SOP expression) is lost in the UmPm triple site mutant (Fig. 5I).
Fig. 5.
Conservation of the regulatory activity of the SPS+P motif combination between Anopheles and Drosophila. A: Diagram of the lone member of the Brd gene family (BFM) in the mosquito Anopheles gambiae (Ag); shown also in Fig. 3B. The indicated upstream region (1.0 kb), which includes the conserved SPS+P motif combination, was tested for enhancer activity using a GFP reporter transgene in Drosophila (Ag BFM-GFP). The two Su(H) binding sites in the SPS are indicated by “U”; the lone proneural protein binding site is labeled “P”. B-I: Images of pupal thoraces at 14 hours APF, centered on the midline; anterior is toward the top. GFP expressed by reporter transgenes is shown in green in B, D, E, F, H, and I. B: The wild-type Ag BFM upstream fragment drives reporter gene expression in the two microchaete “proneural rows” flanking the thoracic midline. Note “holes” in the GFP pattern. C, G: Microchaete SOPs are labeled with anti-Hindsight (Hnt) antibody (magenta). D: Merge of B and C, showing exclusion of reporter activity from SOPs. E: Mutation of the lone P site in the enhancer (Ag BFM-Pm-GFP) extinguishes expression. F: Mutation of the two Su(H) binding sites in the SPS (Ag BFM-Um-GFP) reduces expression in the non-SOPs of the proneural rows. H: Merge of F and G reveals ectopic expression of Ag BFM-Um-GFP in SOPs (white arrows). I: Both residual non-SOP and ectopic SOP expression displayed by Ag BFM-Um-GFP (see F, H) is dependent on the lone P site, as shown by the lack of activity of the triple-mutant construct Ag BFM-UmPm-GFP.
We conclude that the Ag BFM non-coding DNA fragment, bearing a single P site and a single SPS, does indeed encompass an enhancer module that exhibits all of the key regulatory properties of similar modules from both BFM and Hes Notch pathway target genes in Drosophila (Castro et al., 2005): It directs expression specifically in non-SOP cells of proneural clusters; it requires activating inputs from both proneural proteins and Su(H); and it mediates “default repression” by Su(H) in SOP cells. Thus, not only has the core “P+SPS” architecture of the module been conserved for the ~235 My separating the fly and the mosquito, but so have its function and cis-regulatory logic.
Discussion
Deep origin and long-term evolutionary conservation of specific cis-regulatory motifs in developmental control genes
We have previously described the phylogenetically widespread occurrence of single, high-affinity bHLH repressor (R) binding sites upstream of bilaterian proneural genes (Rebeiz et al., 2005). We noted that we could not rule out the possibility that only the “linkage” (direct transcription factor-target gene relationship) has been maintained, and that the binding site itself has been replaced repeatedly in the course of animal evolution. However, we pointed to several lines of evidence suggesting that these R sites have been conserved from a deep common ancestor. These included the stability of the precise 10-bp sequence of the site over very long intervals, and the strong conservation of both the motif and flanking sequences in some instances, clearly suggesting that the sites are indeed orthologous.
The present report substantially expands the inventory of such apparently ancient and conserved cis-regulatory motifs in developmental control genes. We have described here five additional cases in which specific motif combinations have evidently been retained over hundreds of millions of years of evolution. With the exception of two novel elements (the insect brk DE and the deuterostome Hes XE), these motifs represent high-affinity binding sites for known transcription factors. The retention of these specific motif instances is especially striking when considered against the background of rapid appearance and disappearance of other binding sites for the same factors (Figs. 1A, 3A-B, 4A, S3B).
The conservation of the distinctive SE+DE motif combination upstream of insect brk genes extends over perhaps 270-300 My, reflecting the fact that the brk gene itself is found only in insects (Copley, 2008). A similar (minimum) age can be assigned to the P+SPS architecture found upstream of insect bHLH repressor genes, while the E+SPS+P combination associated with arthropod BFM genes is even older, in excess of 400 My, in view of its occurrence in the crustacean Daphnia pulex. Finally, it is likely that the X+R+SPS ensemble upstream of deuterostome Hes1 genes was present in the common ancestor, over 500 My ago. It is also possible that an SPS element was associated with an ancestral bilaterian Hes repressor gene, which would make this feature close to 600 My old.
Our analyses do not permit us to discern the population genetic/microevolutionary processes by which the distinctive cis-regulatory architectures we describe first arose and became fixed in an ancestral population (Lynch, 2007). However, we believe we can offer some useful insights into why these architectures have endured over such lengthy timescales.
Distinctive regulatory capabilities mediated by deeply conserved cis-regulatory motifs
What characteristics of ancient and conserved motifs drive their long-term preservation by selection, even as other binding sites for the same factors come and go rapidly in evolution? We first reiterate our earlier proposal that such deeply conserved motifs mediate abstract or generic regulatory functions of fundamental utility to all or most members of an ancient clade (Rebeiz et al., 2005). It is certainly plausible that, once established, the capacity to repress brk transcription in response to a Dpp signal remained of great utility to all the descendants of the common insect ancestor, as diverse as they became. Similarly, the abstract ability to activate a Hes repressor gene via Notch signaling would remain of exceptional utility to descendants of a bilaterian (or earlier) ancestor that had evolved it. Finally, a generic capability for autorepression of a Hes bHLH repressor gene (Brend and Holley, 2009; Hirata et al., 2002; Lewis, 2003) might very well be retained by descendants of a deuterostome ancestor.
But it is certainly sensible to argue that, to retain such abstract and valuable regulatory capabilities, it would suffice to preserve only the linkage between the appropriate transcription factors and their targets. In this view, individual factor-binding motifs need not be retained; they would be free to turn over during evolution. However, the examples we have described here suggest a second important reason for the long-term evolutionary retention of particular motifs or motif combinations. We propose that these conserved sequence elements mediate a distinctive regulatory capability not conferred by other instances of the same motif or motifs. In the case of the SPS element, we can be quite confident that this perspective is correct. The SPS has been shown to mediate cooperative binding of two Su(H)/Mam/NICD trimers, thus conferring on the associated target gene unusually high sensitivity to Notch signaling (Arnett et al., 2010; Nam et al., 2007). While two “lone” Su(H) sites are indeed able to contribute to a target gene’s response to activated Notch, they would not do so in a cooperative manner. In a similar vein, it seems plausible to suggest that while all SE motifs may be able to participate in signal-dependent repression of brk, the SE+DE combination offers a unique and valuable version of this capability (e.g., greater signal sensitivity), possibly conferring a fitness advantage. We hypothesize that in both cases, once the specialized motif architecture (SPS or SE+DE) had evolved to confer a distinctive capacity, it would be selectively retained in evolution. As we have seen, other instances of the SE or Su(H) binding motifs do arise and become fixed in individual clades, but these would not be expected to exhibit the same durability, since (according to the hypothesis) they confer no unique capability. The foregoing interpretation is particularly supported, we believe, by the frequent observation that if only one element mediating a particular response [either SE or Su(H) site] is present upstream of an orthologous gene in a given species, it is of the “special” type (SE+DE or SPS). Examples include the SE+DE combination in Tribolium castaneum brk and the SPS motifs in the Anopheles gambiae bHLHR1 gene, the Apis mellifera BFM gene, and Homo sapiens HES1.
Another factor that may contribute to the long-term evolutionary conservation of the specialized motif architectures we have considered is their very complexity. Both the SE+DE unit and the SPS represent unusually extended and constrained motif combinations. While in principle this does not prevent them from turning over by duplication/degeneration, they are unlikely to evolve de novo.
Finally, we note an intriguing feature of the conserved motif architectures described here that involve the SPS: the apparently conserved order and even orientation of the individual sequence elements. The arthropod BFM genes are associated with a “lower-strand“ E motif followed by an SPS followed by a “lower-strand” P site; insect Hes repressor genes bear an “upper-strand” P site followed by an SPS; and deuterostome Hes1 genes have an “upper-strand” X site followed by an “upper-strand” R site followed by an SPS, which also has fixed orientation. Inter-site distances are often not conserved; consider the varying separation of the SPS and the P site in the BFM genes, or the different distances between the X+R combination and the SPS in the deuterostome Hes1 genes. Evidently, the motif order and orientation of these architectures have functional significance, consistent with an “enhanceosome” model for the structure of these regions (Arnosti and Kulkarni, 2005). Alternatively, these features may suggest the existence of a “scanning” mechanism for optimal enhancer-promoter interaction. Such a property might be a particular characteristic of promoter-proximal cis-regulatory modules such as these, as contrasted with more distal enhancers. In the latter case, interaction with the promoter by “looping” may impose fewer architectural constraints.
Evolution and conservation of distinctive developmental regulatory capabilities
We have proposed here that the distinctive cis-regulatory architectures we describe are ancient ones that have been conserved from a deep ancestor. However, it also seems likely that, because of their very complexity, they may not represent the “original” version of their respective regulatory linkages. We believe that these two realizations can be reconciled via the following general evolutionary scenario.
The direct linkage of an ancestral Hes gene to Su(H) and the Notch pathway evidently originated in a deep metazoan ancestor, and was very likely mediated by a lone Su(H) binding site or sites. The genome of the demosponge Amphimedon queenslandica includes one member of the closely related Hey repressor family, but no Hes genes (Simionato et al., 2007; Srivastava et al., 2010); this Amphimedon Hey gene has one high-affinity Su(H) site 600 bp upstream of the transcription start site (Fig. 6). The placozoan Trichoplax adhaerens has one Hey ortholog, one Hey-related gene, and one Hes gene (Srivastava et al., 2008). The Hey ortholog has three high-affinity Su(H) sites in the first 800 bp upstream of the ATG start codon, while the Hes gene includes a single such site within 500 bp of its ATG (Fig. 6). The genome of the cnidarian Nematostella vectensis (sea anemone) is endowed with a large paralogous family of 11 Hes genes (Putnam et al., 2007; Simionato et al., 2007), many of them with multiple lone Su(H) sites immediately upstream (Fig. 6; see also Fig. S3). Likewise, the Nematostella Hey ortholog has two upstream Su(H) sites. The SPS evidently did not appear upstream of a Hey/Hes gene until after the cnidarian-bilaterian divergence, but as we have seen, this association is now widespread among both protostomes and deuterostomes.
We suggest, then, that what appeared first was the simple capacity to regulate a Hey/Hes gene directly by Su(H) (presumably linked to the Notch pathway), via one or more lone Su(H) binding sites. Then, in a bilaterian ancestor, an SPS came into being upstream of an individual Hes gene, making possible a cooperative and thus highly sensitive response to Notch-activated Su(H). Once this novel regulatory capacity was established, it bestowed a sufficient selective advantage to ensure its subsequent retention in a wide variety of bilaterian taxa. Such a scenario can account for the phylogenetic distribution of the SPS-containing cis-regulatory architectures we have described. We cannot, however, rule out more complex histories, including the possibility that the SPS arose independently more than once in association with Hes genes.
Duplication-divergence of developmental control genes and their cis-regulatory architectures
It is important to note our finding that, in the case of target genes that are part of paralogous families (Hes repressor and BFMs), only one particular paralog in a given species is typically associated with the conserved motif architectures we have described. This is true even if other paralogs make use of the same overall cis-regulatory “code” (combination of transcription factor binding sites) to direct a similar expression specificity. For example, of the seven unambiguous Hes repressor paralogs in Homo sapiens (Simionato et al., 2007), only HES1 bears the X+R+SPS motif combination, though four others have upstream S sites and two of these also have upstream R sites. Likewise, the Drosophila melanogaster genome includes nine BFM genes (Lai et al., 2000), most of which employ the S+P code, but only one, E(spl)m4, is associated with an SPS+P combination (Bailey and Posakony, 1995; Singson et al., 1994). It seems likely that, while the distinctive regulatory capability conferred by an ancient and conserved motif combination is of long-term selective value, it suffices for a single paralog in the genome to retain it.
This observation is consistent with a duplication-divergence model for the evolution of Hes and BFM paralogs. The special cis-regulatory architectures we have described, along with the associated protein coding sequences, comprise functional units that have been conserved from deep common ancestors because (we propose) of the unique regulatory capabilities they confer. Paralogous genes that arise by duplication within various taxa (this is a widespread phenomenon in the case of Hes genes) would not be subject to the same stringent constraints on their cis-regulatory architecture, since the ancestral gene would be present to provide the distinctive capabilities. The paralogs would thus be free to evolve their cis-regulatory motifs according to other selective pressures or genetic drift (Brown et al., 2007), yielding the many variations on a basic theme (e.g., S+P) that we observe within a single species today.
Supplementary Material
Highlights.
>Conservation of transcription factor binding sites for extraordinarily long periods.
>Includes motifs upstream of developmental regulatory genes in diverse species.
>Three novel conserved motifs are defined.
>Propose that conserved motifs confer distinctive regulatory capabilities.
Acknowledgments
We are grateful to Steve Miller for providing his attB-H-Stinger GFP reporter vector, for permitting us to describe unpublished results, and for valuable comments on the manuscript. We thank Scott Rifkin for his very useful suggestions for improving the Discussion. The Developmental Studies Hybridoma Bank provided antibodies; the Bloomington Drosophila Stock Center provided fly stocks. This work was supported by NIH grant R01 GM046993 to J.W.P.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, George RA, Lewis SE, Richards S, Ashburner M, Henderson SN, Sutton GG, Wortman JR, Yandell MD, Zhang Q, Chen LX, Brandon RC, Rogers YH, Blazej RG, Champe M, Pfeiffer BD, Wan KH, Doyle C, Baxter EG, Helt G, Nelson CR, Miklos G.L. Gabor, Abril JF, Agbayani A, An HJ, Andrews-Pfannkoch C, Baldwin D, Ballew RM, Basu A, Baxendale J, Bayraktaroglu L, Beasley EM, Beeson KY, Benos PV, Berman BP, Bhandari D, Bolshakov S, Borkova D, Botchan MR, Bouck J, Brokstein P, Brottier P, Burtis KC, Busam DA, Butler H, Cadieu E, Center A, Chandra I, Cherry JM, Cawley S, Dahlke C, Davenport LB, Davies P, de Pablos B, Delcher A, Deng Z, Mays AD, Dew I, Dietz SM, Dodson K, Doup LE, Downes M, Dugan-Rocha S, Dunkov BC, Dunn P, Durbin KJ, Evangelista CC, Ferraz C, Ferriera S, Fleischmann W, Fosler C, Gabrielian AE, Garg NS, Gelbart WM, Glasser K, Glodek A, Gong F, Gorrell JH, Gu Z, Guan P, Harris M, Harris NL, Harvey D, Heiman TJ, Hernandez JR, Houck J, Hostin D, Houston KA, Howland TJ, Wei MH, Ibegwam C, Jalali M, Kalush F, Karpen GH, Ke Z, Kennison JA, Ketchum KA, Kimmel BE, Kodira CD, Kraft C, Kravitz S, Kulp D, Lai Z, Lasko P, Lei Y, Levitsky AA, Li J, Li Z, Liang Y, Lin X, Liu X, Mattei B, McIntosh TC, McLeod MP, McPherson D, Merkulov G, Milshina NV, Mobarry C, Morris J, Moshrefi A, Mount SM, Moy M, Murphy B, Murphy L, Muzny DM, Nelson DL, Nelson DR, Nelson KA, Nixon K, Nusskern DR, Pacleb JM, Palazzolo M, Pittman GS, Pan S, Pollard J, Puri V, Reese MG, Reinert K, Remington K, Saunders RD, Scheeler F, Shen H, Shue BC, Siden-Kiamos I, Simpson M, Skupski MP, Smith T, Spier E, Spradling AC, Stapleton M, Strong R, Sun E, Svirskas R, Tector C, Turner R, Venter E, Wang AH, Wang X, Wang ZY, Wassarman DA, Weinstock GM, Weissenbach J, Williams SM, Woodage T, Worley KC, Wu D, Yang S, Yao QA, Ye J, Yeh RF, Zaveri JS, Zhan M, Zhang G, Zhao Q, Zheng L, Zheng XH, Zhong FN, Zhong W, Zhou X, Zhu S, Zhu X, Smith HO, Gibbs RA, Myers EW, Rubin GM, Venter JC. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2196. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Affolter M, Basler K. The Decapentaplegic morphogen gradient: from pattern formation to growth regulation. Nat. Rev. Genet. 2007;8:663–674. doi: 10.1038/nrg2166. [DOI] [PubMed] [Google Scholar]
- Arnett KL, Hass M, McArthur DG, Ilagan MX, Aster JC, Kopan R, Blacklow SC. Structural and mechanistic insights into cooperative assembly of dimeric Notch transcription complexes. Nat. Struct. Mol. Biol. 2010;17:1312–1317. doi: 10.1038/nsmb.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnosti DN, Kulkarni MM. Transcriptional enhancers: Intelligent enhanceosomes or flexible billboards? J. Cell. Biochem. 2005;94:890–898. doi: 10.1002/jcb.20352. [DOI] [PubMed] [Google Scholar]
- Bailey AM, Posakony JW. Suppressor of Hairless directly activates transcription of Enhancer of split Complex genes in response to Notch receptor activity. Genes Dev. 1995;9:2609–2622. doi: 10.1101/gad.9.21.2609. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Second International Conference on Intelligent Systems for Molecular Biology. 1994;28:36. [PubMed] [Google Scholar]
- Bardin AJ, Schweisguth F. Bearded family members inhibit Neuralized-mediated endocytosis and signaling activity of Delta in Drosophila. Dev. Cell. 2006;10:245–255. doi: 10.1016/j.devcel.2005.12.017. [DOI] [PubMed] [Google Scholar]
- Barolo S, Castro B, Posakony JW. New Drosophila transgenic reporters: insulated P-element vectors expressing fast-maturing RFP. Biotechniques. 2004;36:436–442. doi: 10.2144/04363ST03. [DOI] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray SJ. Notch signalling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- Brend T, Holley SA. Expression of the oscillating gene her1 is directly regulated by Hairy/Enhancer of Split, T-box, and Suppressor of Hairless proteins in the zebrafish segmentation clock. Dev. Dyn. 2009;238:2745–2759. doi: 10.1002/dvdy.22100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CD, Johnson DS, Sidow A. Functional architecture and evolution of transcriptional elements that drive gene coexpression. Science. 2007;317:1557–1560. doi: 10.1126/science.1145893. [DOI] [PubMed] [Google Scholar]
- Castro B, Barolo S, Bailey AM, Posakony JW. Lateral inhibition in proneural clusters: Cis-regulatory logic and default repression by Suppressor of Hairless. Development. 2005;132:3333–3344. doi: 10.1242/dev.01920. [DOI] [PubMed] [Google Scholar]
- Chan YF, Marks ME, Jones FC, Villarreal GJ, Shapiro MD, Brady SD, Southwick AM, Absher DM, Grimwood J, Schmutz J, Myers RM, Petrov D, Jonsson B, Schluter D, Bell MA, Kingsley DM. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science. 2010;327:302–305. doi: 10.1126/science.1182213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbourne JK, Pfrender ME, Gilbert D, Thomas WK, Tucker A, Oakley TH, Tokishita S, Aerts A, Arnold GJ, Basu MK, Bauer DJ, Caceres CE, Carmel L, Casola C, Choi JH, Detter JC, Dong Q, Dusheyko S, Eads BD, Frohlich T, Geiler-Samerotte KA, Gerlach D, Hatcher P, Jogdeo S, Krijgsveld J, Kriventseva EV, Kultz D, Laforsch C, Lindquist E, Lopez J, Manak JR, Muller J, Pangilinan J, Patwardhan RP, Pitluck S, Pritham EJ, Rechtsteiner A, Rho M, Rogozin IB, Sakarya O, Salamov A, Schaack S, Shapiro H, Shiga Y, Skalitzky C, Smith Z, Souvorov A, Sung W, Tang Z, Tsuchiya D, Tu H, Vos H, Wang M, Wolf YI, Yamagata H, Yamada T, Ye Y, Shaw JR, Andrews J, Crease TJ, Tang H, Lucas SM, Robertson HM, Bork P, Koonin EV, Zdobnov EM, Grigoriev IV, Lynch M, Boore JL. The ecoresponsive genome of Daphnia pulex. Science. 2011;331:555–561. doi: 10.1126/science.1197761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copley RR. The animal in the genome: comparative genomics and evolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2008;363:1453–1461. doi: 10.1098/rstb.2007.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH. Gene regulatory networks in development and evolution. Academic Press; San Diego: 2006. The regulatory genome. [Google Scholar]
- Dolfini D, Zambelli F, Pavesi G, Mantovani R. A perspective of promoter architecture from the CCAAT box. Cell Cycle. 2009;8:4127–4137. doi: 10.4161/cc.8.24.10240. [DOI] [PubMed] [Google Scholar]
- Duncan EJ, Dearden PK. Evolution of a genomic regulatory domain: The role of gene co-option and gene duplication in the Enhancer of split complex. Genome Res. 2010;20:917–928. doi: 10.1101/gr.104794.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiuza UM, Arias AM. Cell and molecular biology of Notch. J. Endocrinol. 2007;194:459–474. doi: 10.1677/JOE-07-0242. [DOI] [PubMed] [Google Scholar]
- Fontana JR, Posakony JW. Both inhibition and activation of Notch signaling rely on a conserved Neuralized-binding motif in Bearded proteins and the Notch ligand Delta. Dev. Biol. 2009;333:373–385. doi: 10.1016/j.ydbio.2009.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S, Steffen J, Laughon A. Dpp-responsive silencers are bound by a trimeric Mad-Medea complex. J. Biol. Chem. 2005;280:36158–36164. doi: 10.1074/jbc.M506882200. [DOI] [PubMed] [Google Scholar]
- Gompel N, Prud’homme B, Wittkopp PJ, Kassner VA, Carroll SB. Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature. 2005;433:481–487. doi: 10.1038/nature03235. [DOI] [PubMed] [Google Scholar]
- Hare EE, Peterson BK, Iyer VN, Meier R, Eisen MB. Sepsid even-skipped enhancers are functionally conserved in Drosophila despite lack of sequence conservation. PLoS Genet. 2008;4:e1000106. doi: 10.1371/journal.pgen.1000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellsten U, Harland RM, Gilchrist MJ, Hendrix D, Jurka J, Kapitonov V, Ovcharenko I, Putnam NH, Shu S, Taher L, Blitz IL, Blumberg B, Dichmann DS, Dubchak I, Amaya E, Detter JC, Fletcher R, Gerhard DS, Goodstein D, Graves T, Grigoriev IV, Grimwood J, Kawashima T, Lindquist E, Lucas SM, Mead PE, Mitros T, Ogino H, Ohta Y, Poliakov AV, Pollet N, Robert J, Salamov A, Sater AK, Schmutz J, Terry A, Vize PD, Warren WC, Wells D, Wills A, Wilson RK, Zimmerman LB, Zorn AM, Grainger R, Grammer T, Khokha MK, Richardson PM, Rokhsar DS. The genome of the Western clawed frog Xenopus tropicalis. Science. 2010;328:633–636. doi: 10.1126/science.1183670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata H, Yoshiura S, Ohtsuka T, Bessho Y, Harada T, Yoshikawa K, Kageyama R. Oscillatory expression of the bHLH factor Hes1 regulated by a negative feedback loop. Science. 2002;298:840–843. doi: 10.1126/science.1074560. [DOI] [PubMed] [Google Scholar]
- Holt RA, Subramanian GM, Halpern A, Sutton GG, Charlab R, Nusskern DR, Wincker P, Clark AG, Ribeiro JM, Wides R, Salzberg SL, Loftus B, Yandell M, Majoros WH, Rusch DB, Lai Z, Kraft CL, Abril JF, Anthouard V, Arensburger P, Atkinson PW, Baden H, de Berardinis V, Baldwin D, Benes V, Biedler J, Blass C, Bolanos R, Boscus D, Barnstead M, Cai S, Center A, Chaturverdi K, Christophides GK, Chrystal MA, Clamp M, Cravchik A, Curwen V, Dana A, Delcher A, Dew I, Evans CA, Flanigan M, Grundschober-Freimoser A, Friedli L, Gu Z, Guan P, Guigo R, Hillenmeyer ME, Hladun SL, Hogan JR, Hong YS, Hoover J, Jaillon O, Ke Z, Kodira C, Kokoza E, Koutsos A, Letunic I, Levitsky A, Liang Y, Lin JJ, Lobo NF, Lopez JR, Malek JA, McIntosh TC, Meister S, Miller J, Mobarry C, Mongin E, Murphy SD, O’Brochta DA, Pfannkoch C, Qi R, Regier MA, Remington K, Shao H, Sharakhova MV, Sitter CD, Shetty J, Smith TJ, Strong R, Sun J, Thomasova D, Ton LQ, Topalis P, Tu Z, Unger MF, Walenz B, Wang A, Wang J, Wang M, Wang X, Woodford KJ, Wortman JR, Wu M, Yao A, Zdobnov EM, Zhang H, Zhao Q, Zhao S, Zhu SC, Zhimulev I, Coluzzi M, della Torre A, Roth CW, Louis C, Kalush F, Mural RJ, Myers EW, Adams MD, Smith HO, Broder S, Gardner MJ, Fraser CM, Birney E, Bork P, Brey PT, Venter JC, Weissenbach J, Kafatos FC, Collins FH, Hoffman SL. The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002;298:129–149. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- Honeybee Genome Sequencing Consortium Insights into social insects from the genome of the honeybee Apis mellifera. Nature. 2006;443:931–949. doi: 10.1038/nature05260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Aphid Genomics Consortium Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol. 2010;8:e1000313. doi: 10.1371/journal.pbio.1000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Chicken Genome Sequencing Consortium Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- International Silkworm Genome Consortium The genome of a lepidopteran model insect, the silkworm Bombyx mori. Insect Biochem. Mol. Biol. 2008;38:1036–1045. doi: 10.1016/j.ibmb.2008.11.004. [DOI] [PubMed] [Google Scholar]
- Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- Jennings BH, Tyler DM, Bray SJ. Target specificities of Drosophila Enhancer of split basic helix-loop-helix proteins. Mol. Cell. Biol. 1999;19:4600–4610. doi: 10.1128/mcb.19.7.4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong S, Rebeiz M, Andolfatto P, Werner T, True J, Carroll SB. The evolution of gene regulation underlies a morphological difference between two Drosophila sister species. Cell. 2008;132:783–793. doi: 10.1016/j.cell.2008.01.014. [DOI] [PubMed] [Google Scholar]
- Lai EC, Bodner R, Posakony JW. The Enhancer of split Complex of Drosophila includes four Notch-regulated members of the Bearded gene family. Development. 2000;127:3441–3455. doi: 10.1242/dev.127.16.3441. [DOI] [PubMed] [Google Scholar]
- Lai EC, Burks C, Posakony JW. The K box, a conserved 3′ UTR sequence motif, negatively regulates accumulation of Enhancer of split Complex transcripts. Development. 1998;125:4077–4088. doi: 10.1242/dev.125.20.4077. [DOI] [PubMed] [Google Scholar]
- Lai EC, Posakony JW. The Bearded box, a novel 3′ UTR sequence motif, mediates negative post-transcriptional regulation of Bearded and Enhancer of split Complex gene expression. Development. 1997;124:4847–4856. doi: 10.1242/dev.124.23.4847. [DOI] [PubMed] [Google Scholar]
- Lai EC, Tam B, Rubin GM. Pervasive regulation of Drosophila Notch target genes by GY-box-, Brd-box-, and K-box-class microRNAs. Genes Dev. 2005;19:1067–1080. doi: 10.1101/gad.1291905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Le Bras S, Loyer N, Le Borgne R. The multiple facets of ubiquitination in the regulation of Notch signaling pathway. Traffic. 2011;12:149–161. doi: 10.1111/j.1600-0854.2010.01126.x. [DOI] [PubMed] [Google Scholar]
- Lecourtois M, Schweisguth F. The neurogenic Suppressor of Hairless DNA-binding protein mediates the transcriptional activation of the Enhancer of split Complex genes triggered by Notch signaling. Genes Dev. 1995;9:2598–2608. doi: 10.1101/gad.9.21.2598. [DOI] [PubMed] [Google Scholar]
- Lewis J. Autoinhibition with transcriptional delay: a simple mechanism for the zebrafish somitogenesis oscillator. Curr. Biol. 2003;13:1398–1408. doi: 10.1016/s0960-9822(03)00534-7. [DOI] [PubMed] [Google Scholar]
- Ludwig MZ, Bergman C, Patel NH, Kreitman M. Evidence for stabilizing selection in a eukaryotic enhancer element. Nature. 2000;403:564–567. doi: 10.1038/35000615. [DOI] [PubMed] [Google Scholar]
- Lynch M. The frailty of adaptive hypotheses for the origins of organismal complexity. Proc. Natl. Acad. Sci. USA. 2007;104(Suppl. 1):8597–8604. doi: 10.1073/pnas.0702207104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein M, Zinzen R, Markstein P, Yee KP, Erives A, Stathopoulos A, Levine M. A regulatory code for neurogenic gene expression in the Drosophila embryo. Development. 2004;131:2387–2394. doi: 10.1242/dev.01124. [DOI] [PubMed] [Google Scholar]
- Minguillon C, Jimenez-Delgado S, Panopoulou G, Garcia-Fernandez J. The amphioxus Hairy family: differential fate after duplication. Development. 2003;130:5903–5914. doi: 10.1242/dev.00811. [DOI] [PubMed] [Google Scholar]
- Mita K, Kasahara M, Sasaki S, Nagayasu Y, Yamada T, Kanamori H, Namiki N, Kitagawa M, Yamashita H, Yasukochi Y, Kadono-Okuda K, Yamamoto K, Ajimura M, Ravikumar G, Shimomura M, Nagamura Y, Shin-I T, Abe H, Shimada T, Morishita S, Sasaki T. The genome sequence of silkworm, Bombyx mori. DNA Res. 2004;11:27–35. doi: 10.1093/dnares/11.1.27. [DOI] [PubMed] [Google Scholar]
- Muller B, Hartmann B, Pyrowolakis G, Affolter M, Basler K. Conversion of an extracellular Dpp/BMP morphogen gradient into an inverse transcriptional gradient. Cell. 2003;113:221–233. doi: 10.1016/s0092-8674(03)00241-1. [DOI] [PubMed] [Google Scholar]
- Nam Y, Sliz P, Pear WS, Aster JC, Blacklow SC. Cooperative assembly of higher-order Notch complexes functions as a switch to induce transcription. Proc. Natl. Acad. Sci. USA. 2007;104:2103–2108. doi: 10.1073/pnas.0611092104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nellesen DT, Lai EC, Posakony JW. Discrete enhancer elements mediate selective responsiveness of Enhancer of split Complex genes to common transcriptional activators. Dev. Biol. 1999;213:33–53. doi: 10.1006/dbio.1999.9324. [DOI] [PubMed] [Google Scholar]
- Nene V, Wortman JR, Lawson D, Haas B, Kodira C, Tu ZJ, Loftus B, Xi Z, Megy K, Grabherr M, Ren Q, Zdobnov EM, Lobo NF, Campbell KS, Brown SE, Bonaldo MF, Zhu J, Sinkins SP, Hogenkamp DG, Amedeo P, Arensburger P, Atkinson PW, Bidwell S, Biedler J, Birney E, Bruggner RV, Costas J, Coy MR, Crabtree J, Crawford M, Debruyn B, Decaprio D, Eiglmeier K, Eisenstadt E, El-Dorry H, Gelbart WM, Gomes SL, Hammond M, Hannick LI, Hogan JR, Holmes MH, Jaffe D, Johnston JS, Kennedy RC, Koo H, Kravitz S, Kriventseva EV, Kulp D, Labutti K, Lee E, Li S, Lovin DD, Mao C, Mauceli E, Menck CF, Miller JR, Montgomery P, Mori A, Nascimento AL, Naveira HF, Nusbaum C, O’leary S, Orvis J, Pertea M, Quesneville H, Reidenbach KR, Rogers YH, Roth CW, Schneider JR, Schatz M, Shumway M, Stanke M, Stinson EO, Tubio JM, Vanzee JP, Verjovski-Almeida S, Werner D, White O, Wyder S, Zeng Q, Zhao Q, Zhao Y, Hill CA, Raikhel AS, Soares MB, Knudson DL, Lee NH, Galagan J, Salzberg SL, Paulsen IT, Dimopoulos G, Collins FH, Birren B, Fraser-Liggett CM, Severson DW. Genome sequence of Aedes aegypti, a major arbovirus vector. Science. 2007;316:1718–1723. doi: 10.1126/science.1138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong CT, Cheng HT, Chang LW, Ohtsuka T, Kageyama R, Stormo GD, Kopan R. Target selectivity of vertebrate Notch proteins. Collaboration between discrete domains and CSL-binding site architecture determines activation probability. J. Biol. Chem. 2006;281:5106–5119. doi: 10.1074/jbc.M506108200. [DOI] [PubMed] [Google Scholar]
- Perriere G, Gouy M. WWW-query: an on-line retrieval system for biological sequence banks. Biochimie. 1996;78:364–369. doi: 10.1016/0300-9084(96)84768-7. [DOI] [PubMed] [Google Scholar]
- Prud’homme B, Gompel N, Rokas A, Kassner VA, Williams TM, Yeh SD, True JR, Carroll SB. Repeated morphological evolution through cis-regulatory changes in a pleiotropic gene. Nature. 2006;440:1050–1053. doi: 10.1038/nature04597. [DOI] [PubMed] [Google Scholar]
- Putnam NH, Butts T, Ferrier DE, Furlong RF, Hellsten U, Kawashima T, Robinson-Rechavi M, Shoguchi E, Terry A, Yu JK, Benito-Gutierrez EL, Dubchak I, Garcia-Fernandez J, Gibson-Brown JJ, Grigoriev IV, Horton AC, de Jong PJ, Jurka J, Kapitonov VV, Kohara Y, Kuroki Y, Lindquist E, Lucas S, Osoegawa K, Pennacchio LA, Salamov AA, Satou Y, Sauka-Spengler T, Schmutz J, Shin-I T, Toyoda A, Bronner-Fraser M, Fujiyama A, Holland LZ, Holland PW, Satoh N, Rokhsar DS. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453:1064–1071. doi: 10.1038/nature06967. [DOI] [PubMed] [Google Scholar]
- Putnam NH, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A, Terry A, Shapiro H, Lindquist E, Kapitonov VV, Jurka J, Genikhovich G, Grigoriev IV, Lucas SM, Steele RE, Finnerty JR, Technau U, Martindale MQ, Rokhsar DS. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- Pyrowolakis G, Hartmann B, Muller B, Basler K, Affolter M. A simple molecular complex mediates widespread BMP-induced repression during Drosophila development. Dev. Cell. 2004;7:229–240. doi: 10.1016/j.devcel.2004.07.008. [DOI] [PubMed] [Google Scholar]
- Rebeiz M, Jikomes N, Kassner VA, Carroll SB. Evolutionary origin of a novel gene expression pattern through co-option of the latent activities of existing regulatory sequences. Proc. Natl. Acad. Sci. USA. 2011;108:10036–10043. doi: 10.1073/pnas.1105937108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeiz M, Posakony JW. GenePalette: A universal software tool for genome sequence visualization and analysis. Dev. Biol. 2004;271:431–438. doi: 10.1016/j.ydbio.2004.04.011. [DOI] [PubMed] [Google Scholar]
- Rebeiz M, Reeves NL, Posakony JW. SCORE: A computational approach to the identification of cis-regulatory modules and target genes in whole-genome sequence data. Proc. Natl. Acad. Sci. USA. 2002;99:9888–9893. doi: 10.1073/pnas.152320899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeiz M, Stone T, Posakony JW. An ancient transcriptional regulatory linkage. Dev. Biol. 2005;281:299–308. doi: 10.1016/j.ydbio.2005.03.004. [DOI] [PubMed] [Google Scholar]
- Romano LA, Wray GA. Conservation of Endo16 expression in sea urchins despite evolutionary divergence in both cis and trans-acting components of transcriptional regulation. Development. 2003;130:4187–4199. doi: 10.1242/dev.00611. [DOI] [PubMed] [Google Scholar]
- Rubin GM, Spradling AC. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- Schlatter R, Maier D. The Enhancer of split and Achaete-Scute complexes of Drosophilids derived from simple ur-complexes preserved in mosquito and honeybee. BMC Evol. Biol. 2005;5:67. doi: 10.1186/1471-2148-5-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sea Urchin Genome Sequencing Consortium The genome of the sea urchin Strongylocentrotus purpuratus. Science. 2006;314:941–952. doi: 10.1126/science.1133609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinzato C, Shoguchi E, Kawashima T, Hamada M, Hisata K, Tanaka M, Fujie M, Fujiwara M, Koyanagi R, Ikuta T, Fujiyama A, Miller DJ, Satoh N. Using the Acropora digitifera genome to understand coral responses to environmental change. Nature. 2011;476:320–323. doi: 10.1038/nature10249. [DOI] [PubMed] [Google Scholar]
- Simionato E, Ledent V, Richards G, Thomas-Chollier M, Kerner P, Coornaert D, Degnan BM, Vervoort M. Origin and diversification of the basic helix-loop-helix gene family in metazoans: insights from comparative genomics. BMC Evol. Biol. 2007;7:33. doi: 10.1186/1471-2148-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singson A, Leviten MW, Bang AG, Hua XH, Posakony JW. Direct downstream targets of proneural activators in the imaginal disc include genes involved in lateral inhibitory signaling. Genes Dev. 1994;8:2058–2071. doi: 10.1101/gad.8.17.2058. [DOI] [PubMed] [Google Scholar]
- Srivastava M, Begovic E, Chapman J, Putnam NH, Hellsten U, Kawashima T, Kuo A, Mitros T, Salamov A, Carpenter ML, Signorovitch AY, Moreno MA, Kamm K, Grimwood J, Schmutz J, Shapiro H, Grigoriev IV, Buss LW, Schierwater B, Dellaporta SL, Rokhsar DS. The Trichoplax genome and the nature of placozoans. Nature. 2008;454:955–960. doi: 10.1038/nature07191. [DOI] [PubMed] [Google Scholar]
- Srivastava M, Simakov O, Chapman J, Fahey B, Gauthier ME, Mitros T, Richards GS, Conaco C, Dacre M, Hellsten U, Larroux C, Putnam NH, Stanke M, Adamska M, Darling A, Degnan SM, Oakley TH, Plachetzki DC, Zhai Y, Adamski M, Calcino A, Cummins SF, Goodstein DM, Harris C, Jackson DJ, Leys SP, Shu S, Woodcroft BJ, Vervoort M, Kosik KS, Manning G, Degnan BM, Rokhsar DS. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature. 2010;466:720–726. doi: 10.1038/nature09201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson CI, Schwimmer DB, Barolo S. Rapid evolutionary rewiring of a structurally constrained eye enhancer. Curr. Biol. 2011;21:1186–1196. doi: 10.1016/j.cub.2011.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tribolium Genome Sequencing Consortium The genome of the model beetle and pest Tribolium castaneum. Nature. 2008;452:949–955. doi: 10.1038/nature06784. [DOI] [PubMed] [Google Scholar]
- Tun T, Hamaguchi Y, Matsunami N, Furukawa T, Honjo T, Kawaichi M. Recognition sequence of a highly conserved DNA binding protein RBP-Jk. Nucleic Acids Res. 1994;22:965–971. doi: 10.1093/nar/22.6.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Doren M, Bailey AM, Esnayra J, Ede K, Posakony JW. Negative regulation of proneural gene activity: hairy is a direct transcriptional repressor of achaete. Genes Dev. 1994;8:2729–2742. doi: 10.1101/gad.8.22.2729. [DOI] [PubMed] [Google Scholar]
- Van Doren M, Powell PA, Pasternak D, Singson A, Posakony JW. Spatial regulation of proneural gene activity: auto- and cross-activation of achaete is antagonized by extramacrochaetae. Genes Dev. 1992;6:2592–2605. doi: 10.1101/gad.6.12b.2592. [DOI] [PubMed] [Google Scholar]
- Venken KJ, He Y, Hoskins RA, Bellen HJ. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science. 2006;314:1747–1751. doi: 10.1126/science.1134426. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Miklos G.L. Gabor, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N, Levine AJ, Roberts RJ, Simon M, Slayman C, Hunkapiller M, Bolanos R, Delcher A, Dew I, Fasulo D, Flanigan M, Florea L, Halpern A, Hannenhalli S, Kravitz S, Levy S, Mobarry C, Reinert K, Remington K, Abu-Threideh J, Beasley E, Biddick K, Bonazzi V, Brandon R, Cargill M, Chandramouliswaran I, Charlab R, Chaturvedi K, Deng Z, Di Francesco V, Dunn P, Eilbeck K, Evangelista C, Gabrielian AE, Gan W, Ge W, Gong F, Gu Z, Guan P, Heiman TJ, Higgins ME, Ji RR, Ke Z, Ketchum KA, Lai Z, Lei Y, Li Z, Li J, Liang Y, Lin X, Lu F, Merkulov GV, Milshina N, Moore HM, Naik AK, Narayan VA, Neelam B, Nusskern D, Rusch DB, Salzberg S, Shao W, Shue B, Sun J, Wang Z, Wang A, Wang X, Wang J, Wei M, Wides R, Xiao C, Yan C, Yao A, Ye J, Zhan M, Zhang W, Zhang H, Zhao Q, Zheng L, Zhong F, Zhong W, Zhu S, Zhao S, Gilbert D, Baumhueter S, Spier G, Carter C, Cravchik A, Woodage T, Ali F, An H, Awe A, Baldwin D, Baden H, Barnstead M, Barrow I, Beeson K, Busam D, Carver A, Center A, Cheng ML, Curry L, Danaher S, Davenport L, Desilets R, Dietz S, Dodson K, Doup L, Ferriera S, Garg N, Gluecksmann A, Hart B, Haynes J, Haynes C, Heiner C, Hladun S, Hostin D, Houck J, Howland T, Ibegwam C, Johnson J, Kalush F, Kline L, Koduru S, Love A, Mann F, May D, McCawley S, McIntosh T, McMullen I, Moy M, Moy L, Murphy B, Nelson K, Pfannkoch C, Pratts E, Puri V, Qureshi H, Reardon M, Rodriguez R, Rogers YH, Romblad D, Ruhfel B, Scott R, Sitter C, Smallwood M, Stewart E, Strong R, Suh E, Thomas R, Tint NN, Tse S, Vech C, Wang G, Wetter J, Williams S, Williams M, Windsor S, Winn-Deen E, Wolfe K, Zaveri J, Zaveri K, Abril JF, Guigo R, Campbell MJ, Sjolander KV, Karlak B, Kejariwal A, Mi H, Lazareva B, Hatton T, Narechania A, Diemer K, Muruganujan A, Guo N, Sato S, Bafna V, Istrail S, Lippert R, Schwartz R, Walenz B, Yooseph S, Allen D, Basu A, Baxendale J, Blick L, Caminha M, Carnes-Stine J, Caulk P, Chiang YH, Coyne M, Dahlke C, Mays A, Dombroski M, Donnelly M, Ely D, Esparham S, Fosler C, Gire H, Glanowski S, Glasser K, Glodek A, Gorokhov M, Graham K, Gropman B, Harris M, Heil J, Henderson S, Hoover J, Jennings D, Jordan C, Jordan J, Kasha J, Kagan L, Kraft C, Levitsky A, Lewis M, Liu X, Lopez J, Ma D, Majoros W, McDaniel J, Murphy S, Newman M, Nguyen T, Nguyen N, Nodell M, Pan S, Peck J, Peterson M, Rowe W, Sanders R, Scott J, Simpson M, Smith T, Sprague A, Stockwell T, Turner R, Venter E, Wang M, Wen M, Wu D, Wu M, Xia A, Zandieh A, Zhu X. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Werren JH, Richards S, Desjardins CA, Niehuis O, Gadau J, Colbourne JK, Werren JH, Richards S, Desjardins CA, Niehuis O, Gadau J, Colbourne JK, Beukeboom LW, Desplan C, Elsik CG, Grimmelikhuijzen CJ, Kitts P, Lynch JA, Murphy T, Oliveira DC, Smith CD, van de Zande L, Worley KC, Zdobnov EM, Aerts M, Albert S, Anaya VH, Anzola JM, Barchuk AR, Behura SK, Bera AN, Berenbaum MR, Bertossa RC, Bitondi MM, Bordenstein SR, Bork P, Bornberg-Bauer E, Brunain M, Cazzamali G, Chaboub L, Chacko J, Chavez D, Childers CP, Choi JH, Clark ME, Claudianos C, Clinton RA, Cree AG, Cristino AS, Dang PM, Darby AC, de Graaf DC, Devreese B, Dinh HH, Edwards R, Elango N, Elhaik E, Ermolaeva O, Evans JD, Foret S, Fowler GR, Gerlach D, Gibson JD, Gilbert DG, Graur D, Grunder S, Hagen DE, Han Y, Hauser F, Hultmark D, Hunter H.C.t., Hurst GD, Jhangian SN, Jiang H, Johnson RM, Jones AK, Junier T, Kadowaki T, Kamping A, Kapustin Y, Kechavarzi B, Kim J, Kim J, Kiryutin B, Koevoets T, Kovar CL, Kriventseva EV, Kucharski R, Lee H, Lee SL, Lees K, Lewis LR, Loehlin DW, Logsdon JMJ, Lopez JA, Lozado RJ, Maglott D, Maleszka R, Mayampurath A, Mazur DJ, McClure MA, Moore AD, Morgan MB, Muller J, Munoz-Torres MC, Muzny DM, Nazareth LV, Neupert S, Nguyen NB, Nunes FM, Oakeshott JG, Okwuonu GO, Pannebakker BA, Pejaver VR, Peng Z, Pratt SC, Predel R, Pu LL, Ranson H, Raychoudhury R, Rechtsteiner A, Reese JT, Reid JG, Riddle M, Robertson HM, Romero-Severson J, Rosenberg M, Sackton TB, Sattelle DB, Schluns H, Schmitt T, Schneider M, Schuler A, Schurko AM, Shuker DM, Simoes ZL, Sinha S, Smith Z, Solovyev V, Souvorov A, Springauf A, Stafflinger E, Stage DE, Stanke M, Tanaka Y, Telschow A, Trent C, Vattathil S, Verhulst EC, Viljakainen L, Wanner KW, Waterhouse RM, Whitfield JB, Wilkes TE, Williamson M, Willis JH, Wolschin F, Wyder S, Yamada T, Yi SV, Zecher CN, Zhang L, Gibbs RA. Functional and evolutionary insights from the genomes of three parasitoid Nasonia species. Science. 2010;327:343–348. doi: 10.1126/science.1178028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray GA. The evolutionary significance of cis-regulatory mutations. Nat. Rev. Genet. 2007;8:206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- Yao LC, Phin S, Cho J, Rushlow C, Arora K, Warrior R. Multiple modular promoter elements drive graded brinker expression in response to the Dpp morphogen gradient. Development. 2008;135:2183–2192. doi: 10.1242/dev.015826. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.