Abstract
In order to investigate regulation and redundancy within the sloppy paired (slp) locus, we analyzed 30 kilobases of DNA encompassing the tandem, coordinately regulated slp1 and slp2 transcription units. We found a remarkable array of stripe enhancers with overlapping activities surrounding the slp1 transcription unit, and, unexpectedly, glial cell enhancers surrounding slp2. The slp stripe regulatory region generates 7 stripes at blastoderm, and later 14 stripes that persist throughout embryogenesis. Phylogenetic analysis among drosophilids suggests that the multiplicity of stripe enhancers did not evolve through recent duplication. Most of the direct integration among cis-regulatory modules appears to be simply additive, with one notable exception. Despite the apparent redundancy among stripe enhancers, transgenic rescue suggests that most are required for full function, to maintain wingless expression and parasegment boundaries throughout embryogenesis. Transgenic rescue also reveals indirect positive autoregulation by the 7 early stripes, without which alternate stripes within the 14-stripe pattern are lost, leading to embryos with a pair-rule phenotype.
Keywords: functional redundancy, segmentation, cis-regulatory module, gene regulation, wingless, even skipped
Introduction
The sloppy paired (slp) locus contains two tandem transcription units, slp1 and slp2, that both encode transcription factors with a forkhead domain (Grossniklaus et al., 1992). During segmentation of the germ band, they act downstream of primary pair-rule genes such as even skipped (eve) (Fujioka et al., 1995), and have been classified as secondary pair-rule genes (Akam, 1987; Cadigan et al., 1994b; Ingham, 1988).
Like several other pair-rule genes, including eve, slp1 and slp2 are expressed in both 7- and 14-stripe patterns (Grossniklaus et al., 1992; Macdonald et al., 1986). The 7-stripe pattern is established, in part, through repression by the 7-stripe pattern of eve: in eve mutants, each slp stripe expands posteriorly into the eve domain (Fujioka et al., 1995). In turn, slp helps to restrict the late eve stripe pattern and to maintain the engrailed (en) pattern of 14 stripes (Jaynes and Fujioka, 2004). More specifically, slp expression in 14 stripes helps to maintain the parasegment (PS) boundary by preventing the expansion of en stripes anteriorly into the slp domain, and by maintaining wingless (wg) expression in the slp-expressing cells (Cadigan et al., 1994a, b; Jaynes and Fujioka, 2004). In turn, En represses slp in an interaction that is likely to be direct (Kobayashi et al., 2003). Specific sites of En action in the slp locus have not yet been localized. This function in establishing and maintaining PS boundaries is conserved in insects that do not have a clear pair-rule stage of segmentation (Choe and Brown, 2007).
Early transgenic studies suggested that regulatory DNA upstream of slp1 is required for segmentation function (Grossniklaus et al., 1992). Despite the fact that both slp1 and slp2 are expressed in the same striped pattern (with slp2 appearing to start a bit later), a rescue construct containing this region along with only the slp1 transcription unit can rescue most of the segmentation defects caused by a deficiency of the entire slp locus (Cadigan et al., 1994a), suggesting that slp2 may be dispensable for segmentation. In further transgenic studies, several cis-regulatory modules (CRMs) were identified. The 6 kb just upstream of slp1 was shown to contain at least 3 CRMs, producing a head stripe at blastoderm, germband stripes at stages 10-11, and stripes in the ventral ectoderm at stage 11 and later (Lee and Frasch, 2000). A genome-wide search for Bicoid binding site clusters helped to identify 3 slp CRMs, of which two (located about 1 kb 5′ and 3 kb 3′ of slp1) were shown to drive head stripes at blastoderm (Ochoa-Espinosa et al., 2005). Using consensus binding site information for segmentation gene products, another head stripe CRM was identified about 2 kb 5′ of slp2 (Schroeder et al., 2004).
Most recently, an interaction between two CRMs further upstream of slp1 was studied (Prazak et al., 2010). One region drives 14 stripes beginning at blastoderm, and shows ectopic activation in some cells within odd-numbered parasegments that normally do not express detectable levels of slp RNA. However, when combined with another CRM, which drives properly restricted expression within even-numbered parasegments, ectopic expression is repressed, suggesting that an interaction between distant CRMs plays an important role in slp regulation.
Motivated by a desire to more fully understand the regulation and function of the slp locus, we conducted a systematic transgenic analysis of a 30 kilobase (kb) region surrounding the slp transcription units. This analysis revealed a surprising degree of overlap in both space and time in the striped expression driven by CRMs surrounding slp1, as well as unexpected neuronal regulatory CRMs surrounding slp2. Phylogenetic analysis suggests that the multiplicity of stripe CRMs did not evolve through recent duplication. Extensive dissection of the regulatory region showed that integration of this CRM information is mostly additive, with the exception noted above. We rescued slp mutants with transgenes carrying various CRMs. These experiments confirm the conclusions of Prazak et al. (2010), and show that the improper pattern driven by the upstream region produces significant embryonic defects. These experiments also reveal that autoregulation, through repression of a repressor, is a primary function of the early 7-stripe pattern. They further suggest that the extensive apparent redundancy among stripe elements actually provides for fully functional levels of expression across the many stages of slp expression.
Materials and Methods
Plasmids construction and production of transgenic flies
To analyze CRM activities, conventional P-element transgenesis was used (Fujioka et al., 1998; Spradling and Rubin, 1982). To generate DNA fragments, PCR was performed using BAC clone 06H02 as template (obtained from the Berkeley Drosophila Genome Project (Hoskins et al., 2000)). PCR fragments were cloned into a modified P-element vector (Fujioka et al., 1999) upstream of a lacZ reporter gene. For slp1-promoter-lacZ, the region from –261 (SfiI) to +121 bp relative to the slp1 transcription start site (TSS), or for slp2-promoter-lacZ, the region from –314 to +373 bp (relative to the slp2 TSS), was fused to the lacZ coding region followed by the eve 3′ UTR from +1306 to +1521 bp (KpnI). The mini-white gene is positioned so that the two genes are divergently transcribed. Several independent insertion sites were analyzed for each construct, and the expression patterns shown were seen consistently.
To analyze the rescue ability of u8100, a region from -8.1 to +1.5 kb relative to the slp1 TSS, which includes 78 bp 3′ of the slp1 mRNA polyA signal, was cloned into a conventional P-element vector. Five independent insertion sites were analyzed, and showed similar rescue ability. To compare the rescue ability of different CRMs, ΦC31 recombinase-mediated cassette exchange (ΦC31-RMCE) was used (Bateman et al., 2006). Various CRM regions were cloned into attBΔ2 (Fujioka et al., 2008). The regions used for the rescue constructs are described in the figure legends. ΦC31-RMCE was performed as previously described (Bateman et al., 2006), except that chromosomally integrated ΦC31 recombinase (Bischof et al., 2007) was used, instead of co-injection of ΦC31 mRNA. Successful RMCE events were first identified by loss of mini-white-dependent eye color. The presence and direction of the exchanged region were confirmed by PCR. The attP-docking site at cytological location 95E5 (Fujioka et al., 2008) was used.
Embryo analysis
Embryos were subjected to in situ hybridization using anti-sense RNA probes against lacZ, slp1 (which may cross-react with slp2), and wg mRNA, or to antibody staining with anti-pgalactosidase (β-gal, ICN) as previously described (Fujioka et al., 1999). For glial cell expression, anti-β-gal, and anti-Reversed polarity (Repo) (Alfonso and Jones, 2002) obtained from the Developmental Studies Hybridoma Bank, were visualized with DyLight549-conjugated anti-mouse IgG and DyLight488-conjugated anti-rabbit IgG (Jackson Immuno Research). Rescue constructs were analyzed in a CyO,Δ34 mutant (Grossniklaus et al., 1992) background. Cuticle preparation was performed as previously described (Fujioka et al., 1995).
Sequence comparison and analysis
To identify conserved sequence blocks (CSBs) within each slp CRM, we used the phylogenetic analysis application EvoPrinter (Odenwald et al., 2005) on the cis-Decoder web site (Brody et al., 2007, 2008), with default settings. These CSBs were then used in cis-Decoder, with default settings, to identify conserved sequence clusters (CSCs), and to ask whether slp CRMs with overlapping expression patterns share CSCs.
We performed BLAST searches from FlyBase (Tweedie et al., 2009) with individual CRM sequences against other drosophilid genome sequences (Clark et al., 2007) using default settings. The most conserved subsequences were then BLAST searched against both the D. melanogaster and A. gambiae genomes, using an expect value of 1000. Matching sequences were placed on a map of the region to determine their relative positions and orientations. This methodology provided evidence for specific homologous sequences for most of the slp CRMs in a common ancestor of the drosophilids, but not between the drosophilids and A. gambiae.
To identify possible transcription factor binding to a 12 bp element shared between two mesodermally expressed CRMs (see Results), we searched Drosophila transcription factor binding site matrices in the JASPAR database (Bryne et al., 2008; Portales-Casamar et al., 2010) using each 6 bp subsequence with a relative profile score threshold of either 90% (described as “high stringency” in “Results”) or 80% (“low stringency”).
Results
Regulatory anatomy of the sloppy paired locus
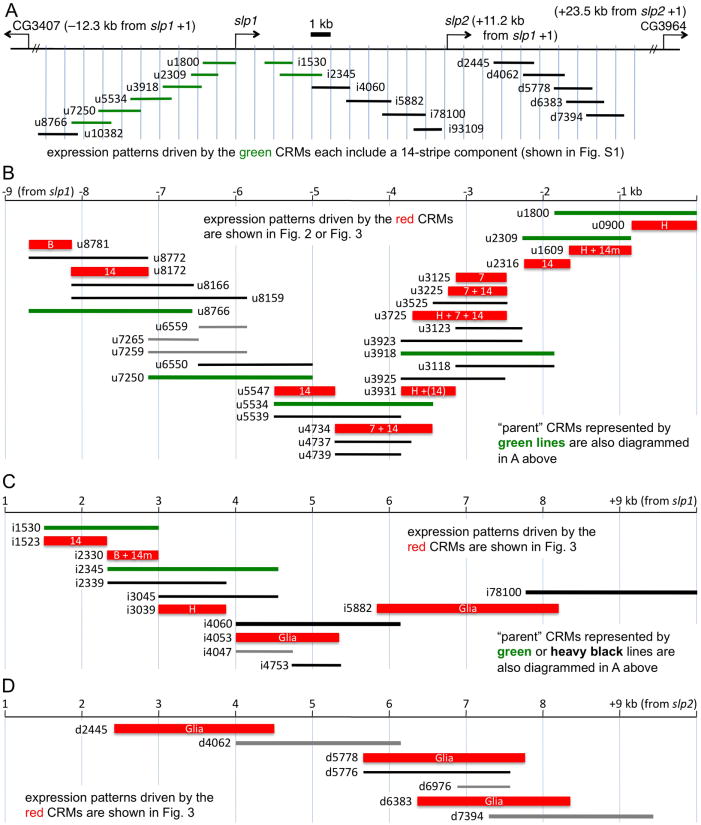
Motivated by a desire to better understand the regulation of the tandem slp1 and slp2 transcription units by pair-rule and segment polarity genes, we performed a detailed mapping of enhancer activity throughout the locus. Although several slp CRMs have been localized and studied (Lee and Frasch, 2000; Ochoa-Espinosa et al., 2005; Prazak et al., 2010; Schroeder et al., 2004), a locus-wide analysis had not been done. We surveyed the 30.9 kb genomic region from 10.3 kb upstream of slp1 to 9.4 kb downstream of slp2 for enhancer activity using reporter transgenes. We initially tested approximately 2 kb DNA fragments overlapping by about 500 bp, producing 18 transgenic constructs (see Fig. 1A for map, Fig. S1 for expression data).
Fig. 1. Mapping of CRMs in the slp locus.
The 30.9 kb genomic region from 10.3 kb upstream of slp1 to 9.4 kb downstream of slp2 was analyzed for enhancer activity in transgenic reporters. A: Large-scale mapping, using approximately 2 kb fragments overlapping by about 500bp. Bars indicate locations of the fragments. Locations are based on Flybase coordinates (Tweedie et al., 2009) (see Fig. S2). B-D: Diagram of fragments used for finer scale dissection of the region upstream of slp1 (B), between slp1 and slp2 (C), and downstream of slp2 (D). The smallest fragments found to drive consistent reporter gene expression in transgenic lines are diagrammed as red boxes, with internal lettering indicating the pattern: “B”, larval brain; “7”, 7 stripes (at stage 7); “14”, 14 stripes (at stage 7 or later); “14m”, 14 stripes restricted to the mesoderm; “(14)”, 14 weak stripes; “H”, strong embryonic head stripe (stage 6); “Glia”, glial cells (in late embryonic CNS). These expression patterns are shown in other figures. Those regions found to drive no consistent expression are indicated as gray lines. Thin black lines are regions that drive expression like the sum of the smaller elements that they contain, except where noted in the text.
In our CRM names, the initial letter indicates the location relative to transcription units: (u) upstream of slp1, (i) internal, between slp1 and slp2, and (d) downstream of slp2. The numbers following these letters indicate the end points in hundreds of bp, where the first 2 digits (or 3 for u10382) are the 5′ end point, and the remaining digits are the 3′ end point. For example, “u8172” extends from about –8.1 to –7.2 kb relative to the slp1 transcription start site (TSS), “i1523” from +1.5 to +2.3 kb relative to the slp1 TSS, and d2445 from +2.4 to +4.5 kb relative to the slp2 TSS. Regions upstream of slp1 were tested for CRM activity in the context of slp1-promoter-lacZ, while regions between slp1 and slp2, and those downstream of slp2, were analyzed in the context of slp2-promoter-lacZ (see Materials and Methods).
Apparent redundancy among stripe elements
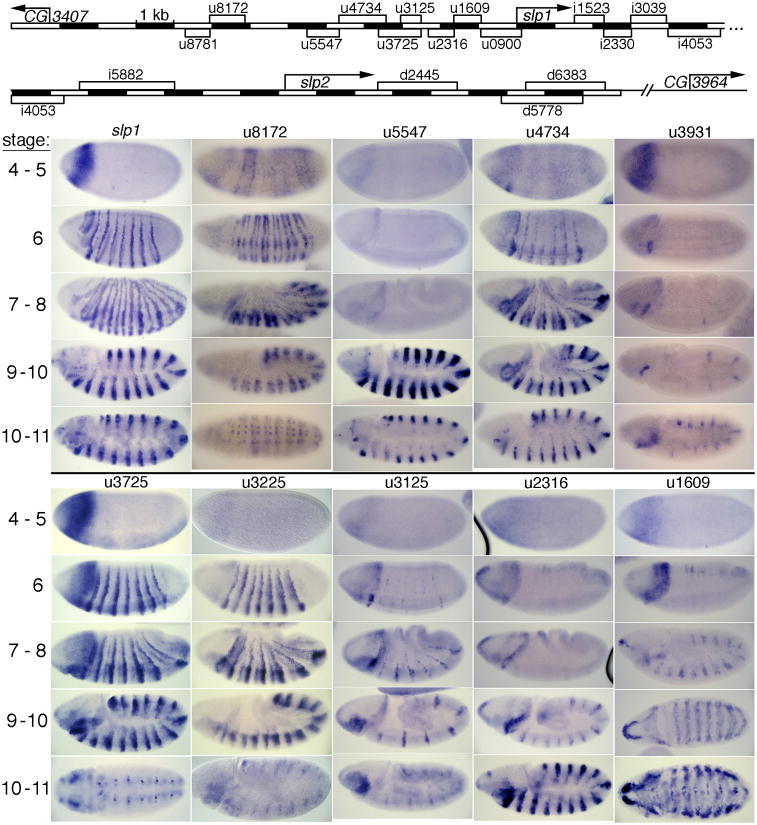
Of our 18 constructs carrying about 2-kb each of regulatory DNA (Fig. 1), 8 showed a 14-stripe pattern (Fig. S1), suggesting a surprising level of redundancy in producing this aspect of slp expression. The region represented by these 8 constructs span the slp1 TSS. Further dissection of these 8 constructs identified 8 non-overlapping CRMs that each give a 14-stripe pattern (Figs. 2, 3; Fig. 1B,C for maps and summary). Two of these (u1609, and i2330) are restricted to the mesoderm. Unlike early eve stripes, which are produced individually or in pairs by distinct CRMs (Fujioka et al., 1999; Goto et al., 1989; Harding et al., 1989; Sackerson, 1995), multiple, non-overlapping slp CRMs drive expression of 7 stripes (both u4734 and u3225 drive first 7 then 14 stripes, while u3125 drives only 7 stripes) or 14 stripes (u8172, u5547, u2316, and i1523) in the ectoderm. This is consistent with its role as a secondary pair-rule gene, as it is regulated by other 7- and 14-stripe patterns of primary and secondary pair-rule genes, as well as by segment polarity genes. Although we did identify the 7-stripe-specific u3125 within the 7-plus-14 stripe u3225, we did not identify a 7-stripe-specific subregion of u4734. In recent studies, CRMs u3125 and u8172 were analyzed in greater detail (Prazak et al., 2010), and a binding site for an activator of slp, Odd-paired, was found in u8172 (Sen et al., 2010).
Fig. 2. CRMs upstream of slp1 drive germband stripe and head expression.
Map at the top shows the locations of the shortest identified sequence blocks that drive aspects of the slp pattern. The 1st column shows endogenous slp expression. The other columns show lacZ expression from transgenes carrying each indicated CRM upstream of slp1-promoter-lacZ (see Materials and Methods) at the 5 embryonic stages shown at the left. Note that u3225 (which has the same 3′ end point as u3125) is not shown on the map.
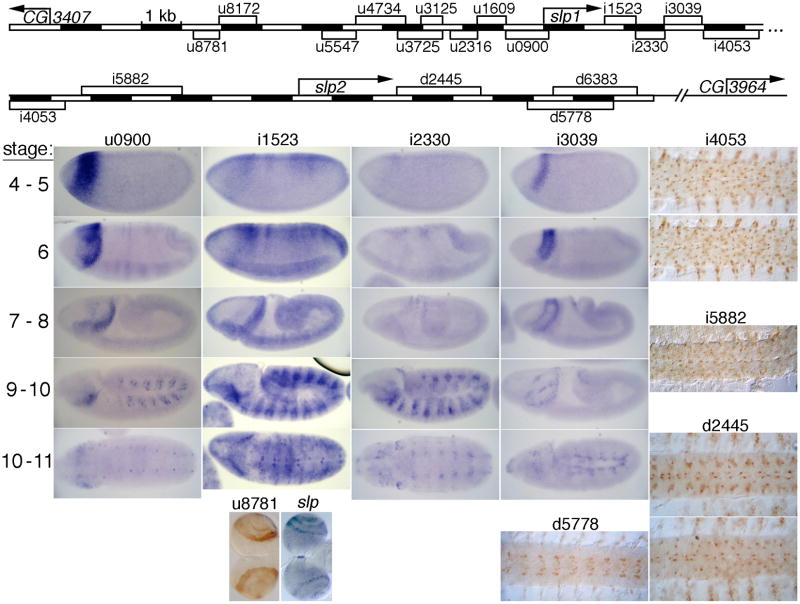
Fig. 3. CRMs downstream of but nearby slp1 drive germband stripe and head expression, while those closer to slp2 drive CNS expression.
Map at the top shows the locations of the shortest identified sequence blocks that drive aspects of the slp pattern. Both “i” and “d” CRMs were analyzed upstream of slp2-promoter-lacZ, while u8781 was upstream of slp1-promoter-lacZ. In the first 3 columns, expression patterns of lacZ RNA from transgenes carrying each CRM are shown at 5 (or 6) embryonic stages (listed at the left), while the last 2 columns show expression of p-galactosidase (p-gal) in the embryonic CNS, or, for u8781, in 3rd instar larval optic lobes. The panel next to u8781 shows endogenous slp expression in the optic lobes.
The non-overlapping CRMs u1609 (Figs. 1B and 2) and i2330 (Figs. 1C and 3) each drive a 14-stripe pattern in the mesoderm at stage 10. A similar pattern is seen with u8766 (Fig. S1), showing that there is apparent redundancy in mesodermal, as well as ectodermal, stripe expression. Shortening u8766 to create u8772 causes loss of this mesodermal stripe expression (data not shown; see Fig. S2 for detailed locations of slp CRMs and a summary of expression data). We note that our CRM u1609 is a more compact version of the previously described slp5′-1 (see Fig. S2), while u3931 corresponds roughly to slp5′-3 (Lee and Frasch, 2000).
After the stripe activities of u8172, u3725, u1609, i1523 (Figs. 2, 3), and u5547 (not shown, its activity is somewhat weaker than the others) fade, expression in some cells in the CNS, probably neuroblasts, becomes apparent. These CRMs are good candidates for providing the known function of slp in developing neuroblasts (Bhat et al., 2000). The CRMs u0900 and i2330 (Figs. 2, 3) also drive expression like that previously described for slp as ventrolateral cell clusters (Grossniklaus et al., 1992). We also saw this activity in 3 out of 7 independent transgenic lines with u3931 (data not shown). We were not able to separate these activities from the stripe activities, suggesting that they are regulated by overlapping sets of transcription factors.
There are 4 non-overlapping CRMs (u3931, u1609, u0900, and i3039) that drive a head stripe at the blastoderm stage (Figs. 2, 3). In addition, several other CRMs drive weak head expression (Fig. S2, data not shown). Both a region spanning the junction between u1609 and u0900 and a region within i3039 were identified by searching for Bicoid binding site clusters genome-wide (Ochoa-Espinosa et al., 2005). These clusters are presumably involved in activating the head stripes. The early head-stripe generating u3931 was not identified at high stringency by this method (Ochoa-Espinosa et al., 2005; Schroeder et al., 2004). However, at lower stringency, clustered Bicoid binding sites can be found there (Hongtao Chen and Stephen Small, personal communication). As u3525 also drives this head expression (data not shown, see Fig. S2), the region common to these constructs (–3455 to –3056 bp) is a good candidate for functional Bicoid binding sites. Bicoid binding in the vicinity of these CRMs has been confirmed in a genome-wide study using chromatin immunoprecipitation (Li et al., 2008), where one binding region encompasses u3931, another spans u1609 and u0900, and a third spans i3039.
Some stripe CRMs described above also drive expression in 3rd instar larvae. CRM u8766 drives reporter gene expression in dorsal and ventral sections of the eye disc (Fig. S3 A-E), and also affects mini-white expression within the transgene, causing patterned eye color (Fig. S3 F-J). In other lines with the same CRM, reporter gene expression behind the morphogenetic furrow was stronger, and was not associated with patterned eye color (Fig. S3 K-N). Perhaps strong, uniform late expression of mini-white masks the effect on eye color of earlier patterned mini-white expression.
Intriguingly, u8781 drives a ring of expression in the brain of 3rd instar larvae (Fig. 3). Although the slp locus is not known to have a function in this part of the nervous system, slp RNA is also seen there in a pattern similar to that of u8781 (Fig. 3). CRMs i1530, i2330, and i2339 each drive a stripe of expression closer to the ventral midline in the larval CNS and brain (Fig. S3 O-Q). However, we were unable to clearly detect endogenous slp expression there. Nonetheless, such a similar activity of multiple CRMs suggests functional significance.
Stripe element rescue of the slp mutant phenotype
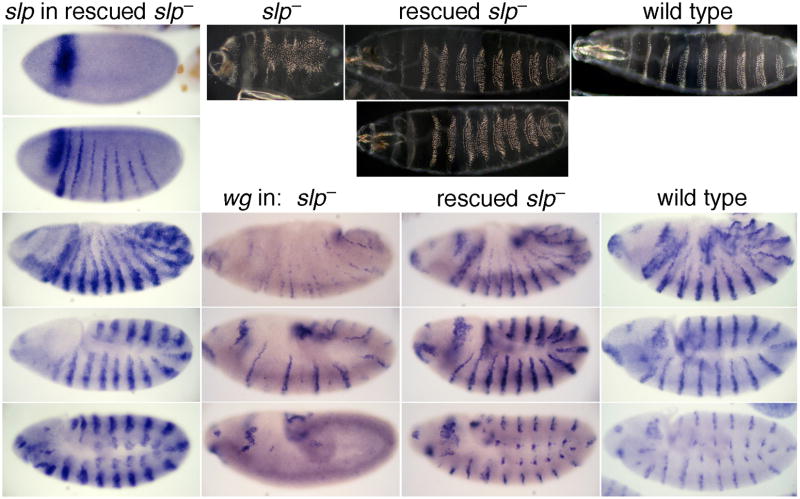
None of the slp stripe CRMs drive an expression pattern that continues until stage 13 (data not shown), when endogenous slp RNA can still be seen (Grossniklaus et al., 1992). A 9.6 kb construct spanning from –8.1 to +1.5 kb, including the slp1 transcription unit, was tested for the ability to rescue the slpΔ34 mutant chromosome, which is a modified CyO balancer chromosome with a deletion that removes the slp1 coding region, the intergenic region, and the 5′ half of the slp2 coding region, and thus is null for both transcripts (Grossniklaus et al., 1992). Consistent with a loss of expression at stage 13 driven by the individual lacZ reporter constructs, this rescue construct did not maintain slp expression to stage 13 (not shown). Nonetheless, it did rescue the slpΔ34 segmentation defects almost completely (Fig. 4) at several different P-element insertion sites, although mild abnormalities within the ventral denticle bands could still be seen in many embryos. Because CyO/CyO embryos do not hatch (but don't have segmentation defects), it is unclear whether this rescue construct would rescue hatching of a slp null mutation in an otherwise wildtype background (which does not exist).
Fig. 4. Transgenic rescue of slp expression and function.
1stcolumn: embryonic expression of slp RNA in a slp RNA null mutant (slpΔ34) with a transgene carrying the slp locus from –8.1 to +1.5 kb relative to the slp1 TSS, which includes 78 bp 3′ of the slp1 mRNA polyA signal. 2nd column: cuticle pattern (top) and wg RNA expression (lower) in the same slp RNA null mutant as in column 1, with no rescue construct. 3rd column: two cuticles representing the range of phenotypes seen (top), and the wg RNA pattern (lower), in the rescued slp mutant of column 1. Note the near-complete rescue. 4thcolumn: Cuticle pattern and wg RNA expression in wild type.
We tested several combinations of apparently redundant stripe CRMs for their ability to rescue the slp null mutant phenotype. Comparisons were made at the same chromosomal docking site, using the ΦC31 recombinase system (Bateman et al., 2006; Groth et al., 2004). We first tested the same region used in Fig. 4, spanning from –8.1 to +1.5 kb (u8100, Fig. 5). The rescue ability at this attP-docking site (at cytological location 95E5) was indistinguishable from that seen at several random chromosomal insertion sites using P-element transgenesis (Fig. 4). Therefore, this docking site was used for all subsequent rescue analysis. The rescue ability of this construct was very similar to that seen previously for a longer construct that included the two downstream stripe CRMs i1523 and i2330 (Cadigan et al., 1994a). This suggests that these two CRMs are functionally redundant with the upstream stripe CRMs.
Fig. 5. Transgenic rescue of slp expression and function by subsets of CRMs.
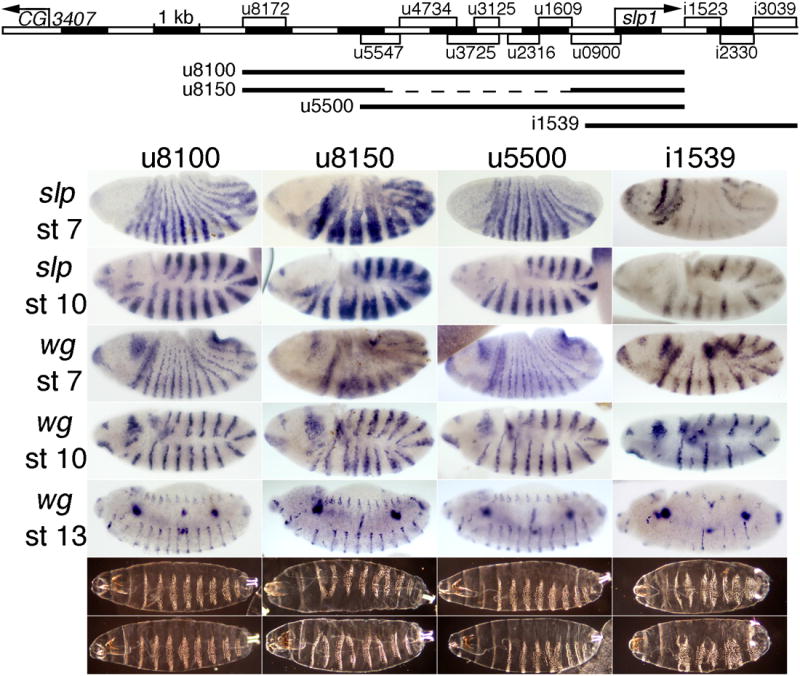
The map at the top indicates the extent of each rescue construct as a line below the map of all the stripe CRMs of the slp locus. The panels below show embryonic expression of slp RNA at stages 7 (top row) and 9-10 (2nd row), wg expression at stages 7, 10, and 13-14 (indicated on the left), and cuticles at the end of embryogenesis (bottom rows show the range of patterns seen) in a slp RNA null mutant (slpΔ34) with a transgene carrying the slp locus from: 1st column: –8076 to +1539 bp relative to the slp1 TSS, which includes 78 bp 3′ of the slp1 mRNA polyA signal. Note the near-complete rescue (see Fig. 4 for wild type). 2nd column: -8076 to -5000, fused with –940 to +1539 bp. Note the ectopic expression of both slp and wg at stages 7 and 10, and the partial loss of wg and naked cuticle at later stages. 3rd column: –5510 to +1539 bp. Note the near-complete rescue, with more severe defects in the denticle pattern in some embryos, relative to column 1. 4th column: –665 to +3934 bp. Note the loss of expression of both slp and wg, more severely in alternate parasegments, at all stages (see text), and the pair-rule loss of naked cuticle. All constructs were analyzed at the same chromosomal location (see Material and Methods).
To further test for redundancies within the stripe elements of the slp locus, we first tested the upstream-most 3 kb of the 9.6 kb rescue construct u8100, in combination with an extended promoter and slp1 coding region from –904 bp to +1536 bp (u8150, Fig. 5). This includes CRMs that give 14 stripes beginning at stage 7 (u8172, Fig. 2, which is contained within u8766, Fig. S1) and persisting until stage 12 (u7250, Fig. S1), as well as strong head expression (u900, Fig. 2). Consistent with the mild ectopic expression seen with u8172 (Fig. 2 and (Prazak et al., 2010)), this construct drove clear ectopic expression within the odd-numbered parasegments (confirmed by co-staining for Eve, data not shown). This results in an aberrant wg expression pattern at embryonic stage 7 (Fig. 5, 2nd column) that is largely, but not completed, corrected at later stages. Most rescued embryos end up with a pair-rule deletion of naked cuticle between ventral denticle bands at the end of embryogenesis (Fig. 5, bottom rows, 2nd column). Such a pair-rule phenotype was not seen with the u8100 rescue construct (described above). These results are consistent with those described previously (Prazak et al., 2010), and suggest that the activity of the upstream-most stripe CRMs is restricted through Eve-dependent repression, acting through regulatory regions closer to the TSS. Nonetheless, this construct rescues wg expression quite well, and the pair-rule defects that remain are relatively mild (compare to slp- in Fig. 4).
We also tested two other combinations of stripe CRMs for their rescue ability. One extends from –5.5 kb through the slp1 coding region (to +1536 bp). It also includes both early and later stripe CRMs, as well as a CRM that drives strong head expression. It drives approximately normal slp1 expression similar to that of the u8100 rescue construct, and rescues the wg and cuticle patterns quite well (u5500, Fig. 5). However, many more embryos show mild cuticle defects than with the u8100 rescue construct, indicating that the level of slp expression is not sufficient for full rescue. The fact that both of these rescue constructs, which share two CRMs, rescue the mutant phenotype well shows that there is some redundancy between them, as expected from the reporter analysis. On the other hand, the fact that neither one rescues as completely as the combination of the two (u8100) shows that this redundancy is only partial, when examined at the level of functional rescue.
Finally, we discovered an autoregulatory requirement for the 7 early slp stripes in activating 7 of the 14 late stripes. This was revealed when we tested a combination of the stripe CRMs downstream of slp1 along with an extended slp1 promoter and coding region (-665 through +1539 bp) for the ability to rescue the slpΔ34 mutant phenotype. As with the above rescue transgenes, the combination of these elements drives both 14-stripe lacZ expression and head expression (Figs. 3 and S1). However, the stripe expression does not begin until slightly later than with the other rescue constructs. In contrast to the other rescue transgenes, this one (i1539, Fig. 5) gives strong expression in only 7 stripes in the slp mutant background, and only weak expression in the other 7. This reveals a functional requirement for the early 7-stripe pattern, which is very weak in these embryos. Without these 7 early stripes of slp expression, half of the 14 later stripes do not form properly. These are the ones located just anterior to the 7 early stripes of eve expression, and in a slp mutant, odd-skipped (odd) stripes have been shown to expand into these cells, preventing activation of half of the wg stripes (Jaynes and Fujioka, 2004). This expanded odd expression can also prevent activation of 7 of the slp stripes within the 14-stripe pattern, accounting for our results. This loss of every other slp stripe, and the accompanying loss of wg, results in a pair-rule loss of naked cuticle between ventral denticle bands in the odd-numbered parasegments (Fig. 5, 4th column). Clearly, these downstream CRMs are not sufficient for rescue, apparently because they do not drive the 7 early stripes strongly enough. All in all, these results suggest that despite the seeming redundancy when stripe CRMs are tested individually, all of them contribute to full slp function in the native context.
Glial cell regulatory elements
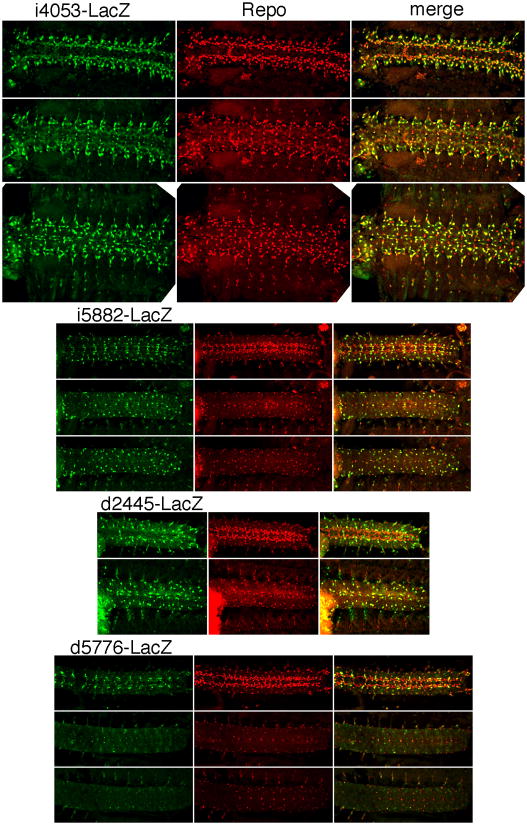
Strikingly, several CRMs that do not drive striped expression do drive patterns in the nervous system. CRMs i4053, i5882, d2445, and d5778 (as well as the partially overlapping d6383) drive expression in spindle-shaped cells in both the central and peripheral nervous systems (Fig. 3). Based on the cell shape, we suspected that these were glial cells. To test this, we double stained for expression of our reporter and a glial cell marker, Repo, product of the gene reversed polarity, or repo (Campbell et al., 1994; Halter et al., 1995; Xiong et al., 1994). As shown in Fig. 6, i4053 drives expression in most, but not all, Repo-expressing cells at embryonic stage 13. Few, if any, strongly reporter-expressing cells are Repo-negative, suggesting that reporter expression is limited to glial cells. Since slp CRMs i5882, d2445, and d5776 (a shorter version of d5778) also drive expression in glial cells (Fig. 6), including some that do not express i4053-lacZ (data not shown), it is likely that most, if not all, glial cells express one or more slp-lacZ reporter. Furthermore, transgenes carrying i4060, which contains i4053, gave reporter gene expression in eye disc cells in the position of glia (Fig. S3R, S) (Campbell et al., 1994; Xiong et al., 1994). Although we have not been able to see a consistent signal for slp RNA or Slp protein in these cells, a transgenic line that carried a BAC clone in which the slp2 coding region was fused to GFP showed expression (Venken et al., 2009) similar to our constructs (data not shown), suggesting that endogenous slp2 is expressed there, but either transiently or at a relatively low level. The lack of good Slp antibodies, however, leaves this an open question. The independent activities of several CRMs in glial cells suggest that they have been the subject of considerable evolutionary selection, consistent with the regulation of glial cell fates by slp. However, this appears to be a separate function from the previously described negative regulation of glial cell fates by slp (Mondal et al., 2007).
Fig. 6. Central nervous system CRMs both upstream and downstream of slp2 drive expression in glial cells.
1st column: β-gal expression from the indicated CRM-carrying transgene. 2nd column: expression of the glial cell-specific protein Repo. 3rdcolumn: merged view of p-gal (green) and Repo (red). Either 2 or 3 focal planes within a dissected embryonic CNS are shown for each: i4053 is at stage 13, the others are at stage 15 (when the CNS has become condensed). Note that each of these slp CRM activities overlap extensively with Repo expression.
Evolutionary origins of the slp stripe CRMs
We explored the evolution of the slp CRMs using two methodologies. First, we used BLAST searches to identify related regions among the sequenced drosophilid genomes (Clark et al., 2007). Each of the CRMs tested showed strong conservation among the more closely related drosophilids, all of which contain both a slp1- and a slp2-related coding region in a similar tandem arrangement to that in D. melanogaster. Sufficient similarity was found within each tested CRM to identify a related region in most of the drosophilid species. The locations of these sequence similarities are shown on maps of the slp locus in Figs. S4A (for the more closely related species) and S4B (for those more distantly related to D. melanogaster). Overall, this analysis suggests that separate elements related to each of these CRMs existed in the common ancestor of the drosophilids, about 40 million years ago (Russo et al., 1995).
We also performed BLAST searches with the most conserved elements of each stripe CRM against both the D. melanogaster and Anopheles gambiae (mosquito) genomes, as mosquitoes are the next most closely related lineage for which sequenced genomes are available. Within D. melanogaster, the best match found among co-expressed CRMs was between the two mesodermal stripe CRMs u1609 and i2330. This 12 bp sequence, GACGTCTTCATT, is highly conserved among drosophilids within u1609, but not within i2330. We used this sequence to search the JASPAR transcription factor database (Bryne et al., 2008; Portales-Casamar et al., 2010). The only high stringency predicted binding site was for the homeodomain-containing ventral veins lacking gene product, which has not been found to be expressed in a pattern (Tomancak et al., 2002; Tomancak et al., 2007) that overlaps with those driven by these CRMs. At lower stringency, possible binding by very many transcription factors is predicted. Likewise, many relatively low stringency matches could be found between conserved elements of our D. melanogaster CRMs and sequences surrounding the A. gambiae transcription unit that is most closely related to D. melanogaster slp. However, we did not find a pattern to these matches that suggested the existence of common ancestral regulatory regions. We also did not find another slp-related coding sequence on the same chromosome as that of the most closely related sequence. This closest sequence is more similar to slp2 than to slp1 (data not shown). This suggests that the twin slp coding regions in drosophilids arose from a duplication event occurring after the split from their last common ancestor with mosquitoes.
Lastly, because shared conserved sequence clusters have previously been found in some CRMs with similar expression patterns (Brody 2007), and a genome-wide database of conserved sequence clusters (CSCs) has been generated, we used the interactive resources Evoprinter and cis-Decoder to identified CSCs using each of our 15 minimal slp CRMs (listed in Fig. S2 in boldface) as a starting point. Although CSCs were found within all except 3 of them (u3931, i4053, and i5882), none of these CSCs were shared among co-expressed CRMs. The single case of a shared CSC was between the glial cell CRM d5778 and a CSC that spans the junction between u1609 and u2316. However, these three CRMs have little or no overlap in their expression patterns (Figs. 2, 3), so the functional significance of these results is unclear.
Discussion
Individual stripe enhancers
We did not identify CRMs that drive individual slp stripes in the germ band at any stage, consistent with the slp locus acting strictly downstream of the primary pair-rule genes, which are responsible for converting non-periodic patterns of the maternal and gap gene products into periodic 7- or 8-stripe patterns (Ingham, 1988). However, a head stripe is driven by several separable CRMs (u3931, u1609, u0900, and i3039; Figs. 1-3; see also Fig. S2). The early slp head stripe is regulated by the maternal gradient-generating gene bicoid, among other genes, and some of these CRMs contain previously identified clusters of Bicoid binding sites (Li et al., 2008; Ochoa-Espinosa et al., 2005; Schroeder et al., 2004).
Two distinct regulatory domains
The slp1, but not the slp2, transcription unit is surrounded by stripe CRMs. This situation may have arisen following a chromosomal duplication that gave rise to these twin transcription units. However, a simple duplication within an array of unique CRMs cannot explain the current regulatory landscape of this locus. In stark contrast to slp1, slp2 is surrounded by nervous system CRMs. These drive expression mostly, if not exclusively, in glial cells (Fig. 6). Consistent with this expression being dispensable for viability, an earlier study indicated that slp2 and the flanking region are not required for viability (Cadigan et al., 1994a; Grossniklaus et al., 1992). However, flies can survive in the laboratory with clear CNS defects (Fujioka et al., 2003). A previous study (Mondal et al., 2007) suggested negative regulation of glial cell specification by slp. Our data suggest that slp is expressed in glial cells alongside Repo. Because 4 non-overlapping CRMs drive expression in glial cells, we suggest that slp has a separate, positive function in glial cells following their specification. Previous analysis of slp-related protein-coding sequences in non-drosophilid insects and basally branching arthropods suggested that the common ancestral coding sequence of slp1 and slp2 was more similar to slp2 (Choe and Brown, 2007; Damen et al., 2005). We found that this is also true in the mosquito A. gambiae. A conserved nervous system function for slp2 might help to explain why the slp2 coding region has diverged more slowly than slp1 from their common ancestral sequence.
Some of the stripe CRMs surrounding slp1 also drive embryonic CNS expression (Figs. 1-3, S2), possibly in neuroblasts. Previous studies showed that slp is involved in specifying neuroblast identity (Bhat et al., 2000). These CRMs are good candidates for providing this function. In addition, multiple CRMs drive expression in the larval brain and in eye discs (Figs. 1, S2, S3).
Standard P-element transgenesis revealed that many of the CRMs surrounding slp1 can cause pairing-sensitive silencing of mini-white in some transgenic lines (Fig. S2), a rare phenomenon that is usually associated with Polycomb-response elements (PREs). This may indicate a set of dispersed PREs in this region that facilitate the association of Polycomb with the locus, and maintain a chromatin domain enriched in histone H3 tri-methylated at lysine 27, which was found to be present throughout the slp locus in embryos (Negre et al., 2011; Schuettengruber et al., 2009).
Transgenic rescue suggests minimal redundancy among stripe CRMs
Our rescue construct is shorter at both ends than one previously tested (Cadigan et al., 1994a), yet generates a similar degree of rescue. In addition to containing upstream sequences extending into neighboring genes, the previous construct included our i1523 and i2330 stripe CRMs. Both constructs included the slp1, and not the slp2, coding region. There are several possible explanations for the inability of both rescue constructs to completely rescue the denticle defects of the slpΔ34 null mutant chromosome. One possibility is that the slp2 transcript may be more stable than that of slp1, which would be consistent with the fact that slp2 RNA normally appears to both begin expression and reach its maximum levels later (Grossniklaus et al., 1992). This could explain not only our inability to completely rescue function, but also the premature disappearance of slp RNA from our construct. The sufficiency of this explanation is argued against, however, by the fact that a slp2-specific mutation is probably viable (Grossniklaus et al., 1992), although the mild denticle defects resulting from rescue by our transgene might not cause lethality. Finally, maintenance of stripe expression may require sequences in the slp locus that do not themselves have enhancer activity, such as the maintenance elements within the bithorax complex (Maeda and Karch, 2009) or the eve gene (Fujioka et al., 2008). Testing of this possibility will require further study.
Recent studies of genes with apparently redundant enhancers (Frankel et al., 2010; Perry et al., 2010) suggest that true redundancy may be rare, and that distinct enhancers with overlapping activities contribute to phenotypic robustness that is likely to be maintained by natural selection. Our results are consistent with this, although they suggest that there is some redundancy among the stripe CRMs, as those downstream of slp1 do not noticeably contribute to patterning the cuticle when all the upstream ones are present. However, the sequences within these elements appear to be conserved, suggesting they contribute to function, at least in the wild. Our results further suggest that even following a genomic duplication that generates partially redundant coding regions, redundant CRMs may be rapidly lost during subsequent evolution.
Interactions among CRMs
In almost all cases, our larger elements drive expression in all the places where expression is driven by smaller CRMs that they contain. As an example, the 2.1 kb u8766 drives expression both in the larval brain and in 14 stripes, consistent with the fact that it spans the 600 bp u8781 and the 900 bp u8172, which drive expression in the brain and in 14 stripes, respectively. Furthermore, most of the differences among partially redundant CRMs are consistent with their activities combining additively to generate endogenous slp expression. For example, while the regions u8172, u4734, and u3225 each drive a 14-stripe pattern in the ectoderm beginning at embryonic stage 7 or 8 (Figs. 3, 4), the regions u5547, u2316, and i1523 are expressed later, at stages 9–11. Thus, while there is considerable overlap among the striped patterns driven by these elements, they are not all redundant, and each may be important to produce the robust slp striped pattern in the endogenous context.
In contrast, some negative positional cues depend on more complex CRM interactions. A recent study (Prazak et al., 2010) described a detailed analysis of the u8172 region (whose 14-stripe pattern includes some cells outside the normal slp expression domain). That study showed that u3125 (which drives a 7-stripe pattern with no ectopic expression), and derivatives of it, can suppress ectopic expression from u8172 when combined in the same construct. Our rescue data show that ectopic expression driven by the upstream CRM disrupts normal function (Fig. 5, u8150), and so must be suppressed within the endogenous locus. Thus, non-additive interactions among individual CRMs have important roles in regulating slp expression, even though the general trend is for the activities of slp CRMs to combine additively.
Another kind of interaction among CRMs is revealed in slp mutants that are rescued using the stripe CRMs located downstream of slp1, which do not drive an early 7-stripe pattern. Although i1530 drives a regular 14-stripe pattern in wild-type embryos (Fig. S1), in a slp mutant the longer i1539 drives expression strongly in only 7 stripes, and weakly in the other 7 (Fig. 5). This difference is explained by positive autoregulation, in that the early slp stripes are required for functional levels of later slp expression in the same cells. This is reminiscent of the positive autoregulation of eve stripes, which is indirect (Fujioka et al., 1995). Here, the late loss of slp expression in the absence of early slp stripes can be explained by expanded odd expression (Jaynes and Fujioka, 2004), which apparently represses later slp expression in every other stripe of the 14 stripe pattern. We have not localized the site of action of this odd-dependent repression, which could be either in the stripe CRM region downstream of slp1, or within the slp1 promoter region, both of which are contained within this rescue construct. In either case, it is interesting to note that the 14-stripe pattern driven by these CRMs is regulated, at least initially, in a pair-rule fashion, with independent inputs to two interdigitated sets of 7 stripes.
The fact that there is such an indirect autoregulatory requirement for only half of the slp stripes highlights the pair-rule character of slp function in its intimate relationship with eve and odd (Jaynes and Fujioka, 2004), even though it is clearly also required in 14 stripes at later stages, where it has a similar mutual repressive relationship with engrailed (Cadigan et al., 1994b; Kobayashi et al., 2003). This example illustrates that the pair-rule genes are difficult to neatly classify into early and late classes because of the complexity of their interactions both with gap genes and with each other. A recent study (Schroeder et al., 2011) placed odd, which had traditionally been classified as a secondary pair-rule gene, into the “early” class, while slp was assigned to the “late” class. Despite the fact that odd participates directly in translating non-periodic pattern information into periodic pattern, while slp does not, slp nonetheless regulates odd after periodic pair-rule patterns have been established. This secondary cross-regulation, which formally goes “backwards” in the hierarchy, is essential for the correct transition to segment polarity gene control. Specifically, without early 7-stripe slp expression, half of the wg stripes are not established (those that coincide with the “missing” slp stripes), and the adjacent parasegment borders decay, resulting in pair-rule defects (Fig. 5, i1539 rescue). Thus, complex regulatory interactions occur at both the early pair-rule stage and the late pair-rule stage, and may be the norm for developmental processes.
Stage-dependent regulation of slp CRMs
The 7- and 14-stripe slp patterns occur at different stages, and are driven in part by separable elements. Among the 14-stripe CRMs, some drive earlier expression, which overlaps in time with expression driven by the later-acting CRMs. This suggests that different combinations of activators, and possibly different repressors, may be responsible for activating, and restricting the activity of, these elements at different stages. This, in turn, provides a rationale for the existence of multiple regulatory elements with temporally overlapping patterns. As the expression of activators change during development, maintenance of expression within a given cell is subject to changing constraints on the relevant CRMs. In particular, the need to maintain both the on state and the off state in the appropriate cells may limit the ability of a single CRM to respond properly at all stages, making it advantageous to utilize different CRMs as the milieu of trans-acting factors changes within the nucleus.
Evolution of slp CRMs
We used BLAST searches to map sequence similarities for each stripe CRM among the sequenced drosophilid genomes, all of which contain both slp1 and slp2 coding regions, in a similar arrangement to that in D. melanogaster. The highest-stringency similarity was found between two CRMs expressed in stripes in the presumptive mesoderm, u1609 and i2330. Analysis of likely transcription factor binding to this 12 bp sequence based on known specificities did not reveal any specific factors with a pattern of expression suggesting regulation of these CRMs. However, the arrangements of best-match sequences to each stripe CRM in the most distantly related drosophilids suggest that ancestral sequences for each stripe CRM existed separately in their common ancestor (see Fig. S4 for a map of the relative locations of these cross-species similarities). However, whether these apparently conserved sequences represent distinct, ancestral CRMs with functions similar to those in D. melanogaster remains an open question.
We also tried, without success, to find clear evidence of homologies to stripe CRMs in the next-most closely related sequenced genome, that of A. gambiae, which might indicate an ancestral element from which more than one drosophilid CRM evolved. Although numerous short sequence similarities were found, their arrangements did not suggest any specific relationship to a drosophilid CRM. Presumably, future analysis will reveal how the locus evolved, when sequenced genomes become available for species that diverged from the drosophilids more recently than mosquitoes.
Shared conserved sequence clusters have previously been found in some CRMs with similar expression patterns (Brody et al., 2007, 2008). Therefore, we used Evoprinter and cis-Decoder to look for CSCs both within slp CRMs and between different CRMs. CSCs were found within many of them, but none of these CSCs were shared among co-expressed CRMs. We suggest that this may be different from the situation among enhancers active in neuroblasts, for example, because the slp CRMs may have evolved by convergent evolution under conditions where the available pool of DNA binding activators was large enough to preclude convergence to a similar set of sequence clusters.
Supplementary Material
Each column shows the lacZ RNA expression pattern driven by the CRM listed at the top, in the context of a reporter transgene, at various embryonic stages, depending on when the CRM is active. The “u” fragments were placed upstream of the slp1-promoter-lacZ reporter, while the “i” fragments were upstream of slp2-promoter-lacZ (see Material and Methods for details). See the main text for our naming scheme and Fig. 1A for a map of the CRMs used.
Columns A and O: names of constructs. slp1 -lacZ and slp2-lacZ indicate which promoter region was used for in vivo analysis of the construct. Columns B – C: coordinates relative to either the slp1 or slp2 (bold and underlined) TSS. Columns D – E: restriction enzymes used to create end points (if applicable). Columns F – G: Flybase sequence coordinates. Columns H – I: 5′ and 3′ end point sequences. Columns J – N: Synopses of expression patterns in various tissues.
A-E, K-N: pgal expression from u8766-lacZ in eye discs from different independent insertion lines. F-J: patterned eye color in each of the lines shown in A-E, respectively. Note the expression in dorsal and ventral sections of eye discs in A-E, which correlates roughly with the eye color patterns in F-J. These transgenes contain mini-white, which produces the eye color. Thus, u8766 may contain an eye disc enhancer that activates both lacZ and mini-white expression. In lines K-N, lacZ expression posterior to the morphogenetic furrow is relatively strong and uniform, and these lines do not have a patterned eye color (they have uniform eye colors ranging from dark orange to red, not shown). We note also that our transgenic vector contains Glass activator binding sites just upstream of the mini-white gene, to allow us to identify transformants with regulatory elements that tend to repress mini-white, such as Polycomb response elements, and the results might be influenced by these sites. O-Q: βgal expression in the 3rd instar larval CNS from i1530-lacZ, i2330-lacZ, and i2339-lacZ, respectively. Comparison with slp staining in Fig. 3 suggests that these CRMs may drive expression in cells that normally express slp, although we did not clearly detect slp expression in precisely these patterns. R,S: pgal expression of i4060-lacZ. The pattern suggests that these may be eye disc glial cells (Campbell et al., 1994; Xiong et al., 1994).
BLAST searches (see Materials and Methods) were used to identify sequences similar to each D. melanogaster slp CRM shown on the left, in a representative subset of the 11 other available drosophilid genomes. Colored bars represent the extent of homologous sequences identified, relative to the beginning of the slp1 coding region in each species. Black bars represent the slp1 and slp2 coding regions in the same species, in the order in which they are listed in the inset key. The length of each bar does not indicate the degree of homology, but only the distance over which homologous sequences were identified. That is, the ends of a long bar are defined by short similarities at those positions, rather than by extensive similarity between the ends. Sequence similarities identified in reverse orientation are indicated by arrows (or arrowheads for short sequences). A: Map of the locations of sequence similarities among the 5 most closely related species tested. Note that both the order and orientations of these similarities are the same in all these species, except for an inversion of u4734. The fact that this region is inverted in the melanogaster group of species, which includes D. ananassae and D. erecta, relative to all the other drosophilids (see also B) suggests that the inverted orientation was present in their common ancestor. B: Map of the locations of sequence similarities between D. melanogaster and 4 drosophilid species more diverged from it. Note that the relative orientations of similarities suggests multiple inversion events involving either individual CRMs (or parts of them), or more than one of them (where both the orientation and relative position of adjacent elements are inverted) during drosophilid divergence. The details of these sequence similarities are available on request.
Acknowledgments
We thank Guizhi Sun and Jian Zhou for excellent technical assistance. We thank Stephen Small for helpful comments on the manuscript, Kenneth M. Cadigan for slp1 and slp2 plasm ids, and the CyOΔ34B line, and the Developmental Studies Hybridoma Bank (supported by NICHD and maintained by the Univ. of Iowa Dept. of Biology) for anti-Repo. Confocal imaging and DNA sequencing were carried out in Kimmel Cancer Center facilities, which are supported in part by NCI Cancer Center Grant P30CA56036. This work was supported by NSF-MCB-0818118 and NIH-2R01GM050231 awards to J.B.J. and M.F.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akam M. The molecular basis for metameric pattern in the Drosophila embryo. Development. 1987;101:1–22. [PubMed] [Google Scholar]
- Alfonso TB, Jones BW. gcm2 promotes glial cell differentiation and is required with glial cells missing for macrophage development in Drosophila. Dev Biol. 2002;248:369–383. doi: 10.1006/dbio.2002.0740. [DOI] [PubMed] [Google Scholar]
- Bateman JR, Lee AM, Wu CT. Site-specific transformation of Drosophila via phiC31 integrase-mediated cassette exchange. Genetics. 2006;173:769–777. doi: 10.1534/genetics.106.056945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat KM, van Beers EH, Bhat P. Sloppy paired acts as the downstream target of wingless in the Drosophila CNS and interaction between sloppy paired and gooseberry inhibits sloppy paired during neurogenesis. Development. 2000;127:655–665. doi: 10.1242/dev.127.3.655. [DOI] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody T, Rasband W, Baler K, Kuzin A, Kundu M, Odenwald WF. cis-Decoder discovers constellations of conserved DNA sequences shared among tissue-specific enhancers. Genome Biology. 2007;8:R75. doi: 10.1186/gb-2007-8-5-r75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody T, Rasband W, Baler K, Kuzin A, Kundu M, Odenwald WF. Sequence conservation and combinatorial complexity of Drosophila neural precursor cell enhancers. BMC Genomics. 2008;9:371. doi: 10.1186/1471-2164-9-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryne JC, Valen E, Tang MHE, Marstrand T, Winther O, da Piedade I, Krogh A, Lenhard B, Sandelin A. JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update. Nucl Acids Res. 2008;36:D102–106. doi: 10.1093/nar/gkm955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadigan KM, Grossniklaus U, Gehring WJ. Functional redundancy: the respective roles of the two sloppy paired genes in Drosophila segmentation. Proc Natl Acad Sci. 1994a;91:6324–6328. doi: 10.1073/pnas.91.14.6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadigan KM, Grossniklaus U, Gehring WJ. Localized expression of sloppy paired protein maintains the polarity of Drosophila parasegments. Genes Dev. 1994b;8:899–913. doi: 10.1101/gad.8.8.899. [DOI] [PubMed] [Google Scholar]
- Campbell G, Goring H, Lin T, Spana E, Andersson S, Doe CQ, Tomlinson A. RK2, a glial-specific homeodomain protein required for embryonic nerve cord condensation and viability in Drosophila. Development. 1994;120:2957–2966. doi: 10.1242/dev.120.10.2957. [DOI] [PubMed] [Google Scholar]
- Choe CP, Brown SJ. Evolutionary flexibility of pair-rule patterning revealed by functional analysis of secondary pair-rule genes, paired and sloppy-paired in the short-germ insect, Tribolium castaneum. Dev Biol. 2007;302:281–294. doi: 10.1016/j.ydbio.2006.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, Markow TA, Kaufman TC, Kellis M, Gelbart W, Iyer VN, et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- Damen WGM, Janssen R, Prpic NM. Pair rule gene orthologs in spider segmentation. Evolution & Development. 2005;7:618–628. doi: 10.1111/j.1525-142X.2005.05065.x. [DOI] [PubMed] [Google Scholar]
- Frankel N, Davis GK, Vargas D, Wang S, Payre F, Stern DL. Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature. 2010;466:490–493. doi: 10.1038/nature09158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka M, Emi-Sarker Y, Yusibova GL, Goto T, Jaynes JB. Analysis of an even-skipped rescue transgene reveals both composite and discrete neuronal and early blastoderm enhancers, and multi-stripe positioning by gap gene repressor gradients. Development. 1999;126:2527–2538. doi: 10.1242/dev.126.11.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka M, Jaynes JB, Bejsovec A, Weir M. Production of Transgenic Drosophila. In: Tuan RS, Lo CW, editors. Developmental Biology Protocols. Humana Press; 1998. [Google Scholar]
- Fujioka M, Jaynes JB, Goto T. Early even-skipped stripes act as morphogenetic gradients at the single cell level to establish engrailed expression. Development. 1995;121:4371–4382. doi: 10.1242/dev.121.12.4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka M, Lear BC, Landgraf M, Yusibova GL, Zhou J, Riley KM, Patel NH, Jaynes JB. Even-skipped, acting as a repressor, regulates axonal projections in Drosophila. Development. 2003;130:5385–5400. doi: 10.1242/dev.00770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka M, Yusibova GL, Zhou J, Jaynes JB. The DNA-binding Polycomb-group protein Pleiohomeotic maintains both active and repressed transcriptional states through a single site. Development. 2008;135:4131–4139. doi: 10.1242/dev.024554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto T, Macdonald P, Maniatis T. Early and late periodic patterns of even skipped expression are controlled by distinct regulatory elements that respond to different spatial cues. Cell. 1989;57:413–422. doi: 10.1016/0092-8674(89)90916-1. [DOI] [PubMed] [Google Scholar]
- Grossniklaus U, Pearson RK, Gehring WJ. The Drosophila sloppy paired locus encodes two proteins involved in segmentation that show homology to mammalian transcription factors. Genes Dev. 1992;6:1030–1051. doi: 10.1101/gad.6.6.1030. [DOI] [PubMed] [Google Scholar]
- Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halter DA, Urban J, Rickert C, Ner SS, Ito K, Travers AA, Technau GM. The homeobox gene repo is required for the differentiation and maintenance of glia function in the embryonic nervous system of Drosophila melanogaster. Development. 1995;121:317–332. doi: 10.1242/dev.121.2.317. [DOI] [PubMed] [Google Scholar]
- Harding K, Hoey T, Warrior R, Levine M. Autoregulatory and gap gene response elements of the even-skipped promoter of Drosophila. EMBO J. 1989;8:1205–1212. doi: 10.1002/j.1460-2075.1989.tb03493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins RA, Nelson CR, Berman BP, Laverty TR, George RA, Ciesiolka L, Naeemuddin M, Arenson AD, Durbin J, David RG, Tabor PE, Bailey MR, DeShazo DR, Catanese J, Mammoser A, Osoegawa K, de Jong PJ, Celniker SE, Gibbs RA, Rubin GM, Scherer SE. A BAC-based physical map of the major autosomes of Drosophila melanogaster. Science. 2000;287:2271–2274. doi: 10.1126/science.287.5461.2271. Erratum appears in Science 2000 Jun 9;288(5472):1751. [DOI] [PubMed] [Google Scholar]
- Ingham PW. The molecular genetics of embryonic pattern formation in Drosophila. Nature. 1988;335:25–34. doi: 10.1038/335025a0. [DOI] [PubMed] [Google Scholar]
- Jaynes JB, Fujioka M. Drawing lines in the sand: even skipped et al. and parasegment boundaries. Dev Biol. 2004;269:609–622. doi: 10.1016/j.ydbio.2004.03.001. erratum appears in Dev Biol 2004 Aug 1;272(1):277-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Fujioka M, Tolkunova EN, Deka D, Abu-Shaar M, Mann RS, Jaynes JB. Engrailed cooperates with extradenticle and homothorax to repress target genes in Drosophila. Development. 2003;130:741–751. doi: 10.1242/dev.00289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HH, Frasch M. Wingless effects mesoderm patterning and ectoderm segmentation events via induction of its downstream target sloppy paired. Development. 2000;127:5497–5508. doi: 10.1242/dev.127.24.5497. [DOI] [PubMed] [Google Scholar]
- Li Xy, MacArthur S, Bourgon R, Nix D, Pollard DA, Iyer VN, Hechmer A, Simirenko L, Stapleton M, Luengo Hendriks CL, Chu HC, Ogawa N, Inwood W, Sementchenko V, Beaton A, Weiszmann R, Celniker SE, Knowles DW, Gingeras T, Speed TP, Eisen MB, Biggin MD. Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol. 2008;6:e27. doi: 10.1371/journal.pbio.0060027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald PM, Ingham P, Struhl G. Isolation, structure, and expression of even-skipped: a second pair-rule gene of Drosophila containing a homeo box. Cell. 1986;47:721–734. doi: 10.1016/0092-8674(86)90515-5. [DOI] [PubMed] [Google Scholar]
- Maeda RK, Karch F. The bithorax complex of Drosophila, an exceptional Hox cluster. Curr Top Dev Biol. 2009;88:1–33. doi: 10.1016/S0070-2153(09)88001-0. [DOI] [PubMed] [Google Scholar]
- Mondal S, Ivanchuk SM, Rutka JT, Boulianne GL. Sloppy paired 1/2 regulate glial cell fates by inhibiting Gcm function. Glia. 2007;55:282–293. doi: 10.1002/glia.20456. [DOI] [PubMed] [Google Scholar]
- Negre N, Brown CD, Ma L, Bristow CA, Miller SW, Wagner U, Kheradpour P, Eaton ML, Loriaux P, Sealfon R, Li Z, Ishii H, Spokony RF, Chen J, Hwang L, Cheng C, Auburn RP, Davis MB, Domanus M, Shah PK, Morrison CA, Zieba J, Suchy S, Senderowicz L, Victorsen A, Bild NA, Grundstad AJ, Hanley D, MacAlpine DM, Mannervik M, Venken K, Bellen H, White R, Gerstein M, Russell S, Grossman RL, Ren B, Posakony JW, Kellis M, White KP. A cis-regulatory map of the Drosophila genome. Nature. 2011;471:527–531. doi: 10.1038/nature09990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochoa-Espinosa A, Yucel G, Kaplan L, Pare A, Pura N, Oberstein A, Papatsenko D, Small S. The role of binding site cluster strength in Bicoid-dependent patterning in Drosophila. Proc Natl Acad Sci. 2005;102:4960–4965. doi: 10.1073/pnas.0500373102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odenwald WF, Rasband W, Kuzin A, Brody T. EVOPRINTER, a multigenomic comparative tool for rapid identification of functionally important DNA. Proc Natl Acad Sci. 2005;102:14700–14705. doi: 10.1073/pnas.0506915102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry MW, Boettiger AN, Bothma JP, Levine M. Shadow enhancers foster robustness of Drosophila gastrulation. Curr Biol. 2010;20:1562–1567. doi: 10.1016/j.cub.2010.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portales-Casamar E, Thongjuea S, Kwon AT, Arenillas D, Zhao X, Valen E, Yusuf D, Lenhard B, Wasserman WW, Sandelin A. JASPAR 2010: the greatly expanded open-access database of transcription factor binding profiles. Nucl Acids Res. 2010;38:D105–110. doi: 10.1093/nar/gkp950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prazak L, Fujioka M, Gergen JP. Non-additive interactions involving two distinct elements mediate sloppy-paired regulation by pair-rule transcription factors. Dev Biol. 2010;344:1048–1059. doi: 10.1016/j.ydbio.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo CA, Takezaki N, Nei M. Molecular phylogeny and divergence times of drosophilid species. Molecular Biology & Evolution. 1995;12:391–404. doi: 10.1093/oxfordjournals.molbev.a040214. [DOI] [PubMed] [Google Scholar]
- Sackerson C. Patterns of conservation and divergence at the even-skipped locus of Drosophila. Mech Dev. 1995;51:199–215. doi: 10.1016/0925-4773(95)00365-7. [DOI] [PubMed] [Google Scholar]
- Schroeder MD, Pearce M, Fak J, Fan H, Unnerstall U, Emberly E, Rajewsky N, Siggia ED, Gaul U. Transcriptional control in the segmentation gene network of Drosophila. PLoS Biol. 2004;2:E271. doi: 10.1371/journal.pbio.0020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B, Ganapathi M, Leblanc B, Portoso M, Jaschek R, Tolhuis B, van Lohuizen M, Tanay A, Cavalli G. Functional anatomy of polycomb and trithorax chromatin landscapes in Drosophila embryos. PLoS Biol. 2009;7:e13. doi: 10.1371/journal.pbio.1000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen A, Stultz BG, Lee H, Hursh DA. Odd paired transcriptional activation of decapentaplegic in the Drosophila eye/antennal disc is cell autonomous but indirect. Dev Biol. 2010;343:167–177. doi: 10.1016/j.ydbio.2010.04.003. [DOI] [PubMed] [Google Scholar]
- Spradling AC, Rubin GM. Transcription of the cloned P elements into Drosophila germ line chromosomes. Science. 1982;218:341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
- Tomancak P, Beaton A, Weiszmann R, Kwan E, Shu S, Lewis SE, Richards S, Ashburner M, Hartenstein V, Celniker SE, Rubin GM. Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome Biology. 2002;3 doi: 10.1186/gb-2002-3-12-research0088. RESEARCH0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomancak P, Berman BP, Beaton A, Weiszmann R, Kwan E, Hartenstein V, Celniker SE, Rubin GM. Global analysis of patterns of gene expression during Drosophila embryogenesis. Genome Biology. 2007;8:R145. doi: 10.1186/gb-2007-8-7-r145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tweedie S, Ashburner M, Falls K, Leyland P, McQuilton P, Marygold S, Millburn G, Osumi-Sutherland D, Schroeder A, Seal R, Zhang H, Consortium TF. FlyBase: enhancing Drosophila Gene Ontology annotations. Nucl Acids Res. 2009;37:D567–D570. doi: 10.1093/nar/gkn788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken KJT, Carlson JW, Schulze KL, Pan H, He Y, Spokony R, Wan KH, Koriabine M, de Jong PJ, White KP, Bellen HJ, Hoskins RA. Versatile P[acman] BAC libraries for transgenesis studies in Drosophila melanogaster. Nat Methods. 2009;6:431–434. doi: 10.1038/nmeth.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong WC, Okano H, Patel NH, Blendy JA, Montell C. repo encodes a glial-specific homeo domain protein required in the Drosophila nervous system. Genes Dev. 1994;8:981–994. doi: 10.1101/gad.8.8.981. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Each column shows the lacZ RNA expression pattern driven by the CRM listed at the top, in the context of a reporter transgene, at various embryonic stages, depending on when the CRM is active. The “u” fragments were placed upstream of the slp1-promoter-lacZ reporter, while the “i” fragments were upstream of slp2-promoter-lacZ (see Material and Methods for details). See the main text for our naming scheme and Fig. 1A for a map of the CRMs used.
Columns A and O: names of constructs. slp1 -lacZ and slp2-lacZ indicate which promoter region was used for in vivo analysis of the construct. Columns B – C: coordinates relative to either the slp1 or slp2 (bold and underlined) TSS. Columns D – E: restriction enzymes used to create end points (if applicable). Columns F – G: Flybase sequence coordinates. Columns H – I: 5′ and 3′ end point sequences. Columns J – N: Synopses of expression patterns in various tissues.
A-E, K-N: pgal expression from u8766-lacZ in eye discs from different independent insertion lines. F-J: patterned eye color in each of the lines shown in A-E, respectively. Note the expression in dorsal and ventral sections of eye discs in A-E, which correlates roughly with the eye color patterns in F-J. These transgenes contain mini-white, which produces the eye color. Thus, u8766 may contain an eye disc enhancer that activates both lacZ and mini-white expression. In lines K-N, lacZ expression posterior to the morphogenetic furrow is relatively strong and uniform, and these lines do not have a patterned eye color (they have uniform eye colors ranging from dark orange to red, not shown). We note also that our transgenic vector contains Glass activator binding sites just upstream of the mini-white gene, to allow us to identify transformants with regulatory elements that tend to repress mini-white, such as Polycomb response elements, and the results might be influenced by these sites. O-Q: βgal expression in the 3rd instar larval CNS from i1530-lacZ, i2330-lacZ, and i2339-lacZ, respectively. Comparison with slp staining in Fig. 3 suggests that these CRMs may drive expression in cells that normally express slp, although we did not clearly detect slp expression in precisely these patterns. R,S: pgal expression of i4060-lacZ. The pattern suggests that these may be eye disc glial cells (Campbell et al., 1994; Xiong et al., 1994).
BLAST searches (see Materials and Methods) were used to identify sequences similar to each D. melanogaster slp CRM shown on the left, in a representative subset of the 11 other available drosophilid genomes. Colored bars represent the extent of homologous sequences identified, relative to the beginning of the slp1 coding region in each species. Black bars represent the slp1 and slp2 coding regions in the same species, in the order in which they are listed in the inset key. The length of each bar does not indicate the degree of homology, but only the distance over which homologous sequences were identified. That is, the ends of a long bar are defined by short similarities at those positions, rather than by extensive similarity between the ends. Sequence similarities identified in reverse orientation are indicated by arrows (or arrowheads for short sequences). A: Map of the locations of sequence similarities among the 5 most closely related species tested. Note that both the order and orientations of these similarities are the same in all these species, except for an inversion of u4734. The fact that this region is inverted in the melanogaster group of species, which includes D. ananassae and D. erecta, relative to all the other drosophilids (see also B) suggests that the inverted orientation was present in their common ancestor. B: Map of the locations of sequence similarities between D. melanogaster and 4 drosophilid species more diverged from it. Note that the relative orientations of similarities suggests multiple inversion events involving either individual CRMs (or parts of them), or more than one of them (where both the orientation and relative position of adjacent elements are inverted) during drosophilid divergence. The details of these sequence similarities are available on request.