Abstract
In this review, the role of S-nitrosylation (SNO) in cardioprotection will be discussed. This review will cover the methodology used to measure SNO levels, and the mechanisms by which SNO serves to modulate cell function and mediate protection. We will also consider whether SNO acts through many targets or whether there are a few key SNO proteins that mediate protection. Issues regarding the percentage of the total protein which is SNO and how this plays a role in the modulation of cell function will also be discussed. The role of nitric oxide synthase uncoupling in cardioprotection will also be addressed.
1. Introduction
Prolonged ischemia, with or without reperfusion, leads to cell death. Treatment with drugs or intermittent ischemia (ischemic preconditioning) has been shown to reduce cell death following ischemia and reperfusion. This protection is referred to as cardioprotection. Nitric oxide (NO) has been suggested to be an important signaling component of cardioprotection [1]. NO is well known to modulate cell function by activation of guanylyl cyclase. However, in the past decade it has become apparent that NO can also signal by post-translational modification of protein thiol groups [2–4]. This modification, referred to as S-nitrosylation or S-nitrosation (SNO), involves the covalent attachment of an NO group to protein thiols. S-nitrosylation and S-nitrosation often used interchangeably to refer to the same modification. However, some investigators have defined ” S-nitrosylation as the addition of NO without a change in the formal charge of the substrate, while S-nitrosation is defined as the formation of a covalent bond between a nitrosonium equivalent (NO+) and a nucleophilic amine”[5]. Because there are many cysteine containing proteins, NO signaling via SNO provides a large number of signaling targets. In fact, much of NO-mediated signaling appears to result from SNO. SNO has been shown to play an important role in the regulation of many cellular functions such as excitation-contraction coupling, cardioprotection, G-protein coupled receptor signaling, and apoptosis.
2. Measurement of S-nitrosylation
Given the large number of processes regulated by SNO signaling, it is important to have reliable methods for the measurement of protein SNO. The current methods for measuring SNO are reviewed. The advantages and disadvantages of the different methods are summarized in Table 1.
Table 1.
| Method | Advantages | Disadvantages |
|---|---|---|
| Biotin switch |
|
|
| Biotin/fluor label- 1D gel MS identification |
|
|
| Fluor label –2D gel MS identification |
|
|
| SNO-RAC – LC-MS/MS |
|
|
| His-tag - NI column enriched 1G gel - MS |
|
|
| Phenylmercury |
|
|
| Direct Mass spec. |
|
|
| Antibody |
|
|
Biotin Switch Based Methods
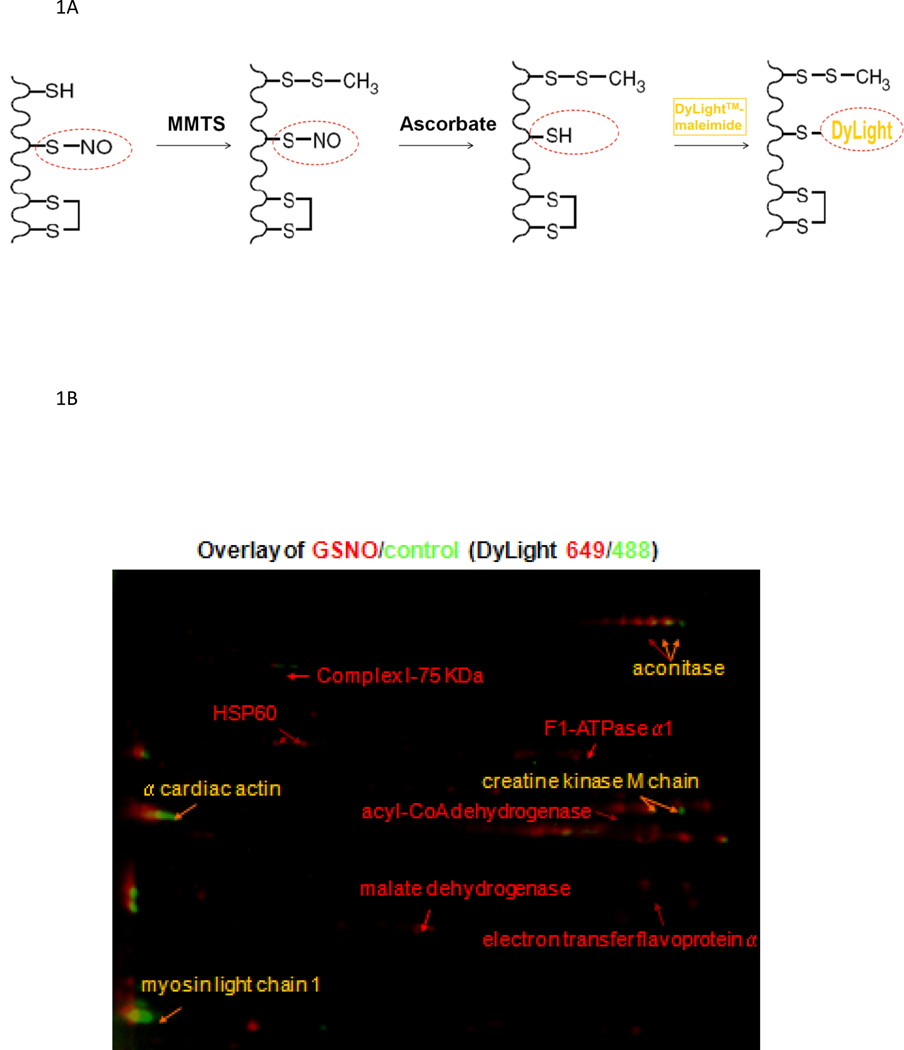
Most measurements of SNO are based on the biotin switch method that was published about a decade ago [6]. In this method, free SH groups are blocked, SNO groups are reduced with ascorbic acid and copper to SH groups, which are then reacted with a sulfhydryl-sensitive tag such as biotin-HPDP (N-[6-(biotinamido)-hexyl]-3’-(2’-pyridyldithio)-propionamide). As indicated in Table 1, methods based on the biotin switch protocol rely on complete blocking of free SH groups and the selective reduction of SNO with ascorbic acid. Appropriate controls need to be done to assure that these conditions are met. The biotin can then be detected using an antibody or enriched using a streptavidin column followed by mass spectrometry, which allows for the identification of proteins/peptides. Using the biotin switch method and mass spectrometry, it was shown that treating heart mitochondrial extracts with S-nitrosoglutathione (GSNO) resulted in SNO of 60 mitochondrial proteins[7]. There have been a number of variations on the biotin switch method. These include methods such as the DyLight switch (see figure 1A) [1] and SNO-DIGE (differential gel electrophoresis) [8], which utilizes a fluorescent maleimide tag to label SNO rather than biotin-HPDP. The DyLight switch and SNO-DIGE methods are compatible with 2D gel methods because the maleimide linkage is stable under reducing conditions used in 2D gels. The DyLight reagent is also available with multiple fluorescent labels, which allows differential labeling of samples in a 2D gel. The protocol and a representative Dylight switch gel are shown in Figure 1, which shows a control sample labeled with one fluorescent tag (e.g.green) and the GSNO treated sample can be labeled with a different fluorescent label (e.g.red) [1]. The samples were then run together in the 2D gel and scanned at the respective wavelengths. Proteins with equal concentration and therefore equal DyLight-labeling in both control and GSNO treated samples are pseudo-colored in yellow, whereas an increase in the control is pseudo-colored green and an increase in a treated sample is red.
Figure 1.
Panel 1A illustrates the protocol for the DyLight switch. Panel 1B shows a representative 2D-gel where control samples are pseudo-colored in green and GSNO treated samples are in red.
Other methods based on the biotin switch have also been used to measure SNO. Isotope coded affinity tags (ICAT), which are isobaric labels, can be added instead of biotin [9]. The ICAT label is then identified by mass spectrometry. Hao et al [10] described a method for SNO site identification that they termed SNOSID. The method uses the biotin switch to biotinylate proteins, followed by protein digestion with trypsin and affinity purification of the biotinylated peptide followed by LC-MS/MS. Torta et al [11] described a modification of the biotin switch called the His-tag switch method, in which a His-tag is added instead of biotin. Yang and Loscalzo [12] employed a modified biotin switch in which SNO proteins were labeled with a Texas Red derivative of methanethiosulfonate. Labeled samples are then examined via confocal microscopy to determine the localization of SNO proteins.
Recently, a resin assisted capture (RAC) method has been described to measure SNO [13]. With this biotin switch-based approach, SNO groups are reduced with ascorbic acid and the proteins are passed over a resin that binds free SH groups. The resin is washed to remove non-specific binding and bound peptides can be subjected to on-resin digestion, eluted and identified by mass spectrometry. This method has the advantage of identifying sites of SNO. Kohr et al [14] used this method to identify sites of SNO in the heart. In untreated mouse heart homogenates 116 unique endogenously SNO proteins were identified. When these mouse heart homogenates were treated with GSNO 951 potential SNO sites were identified. Kohr et al [15] also used this method to identify SNO proteins occurring with preconditioning.
Comparison of SNO-RAC and DyLight switch
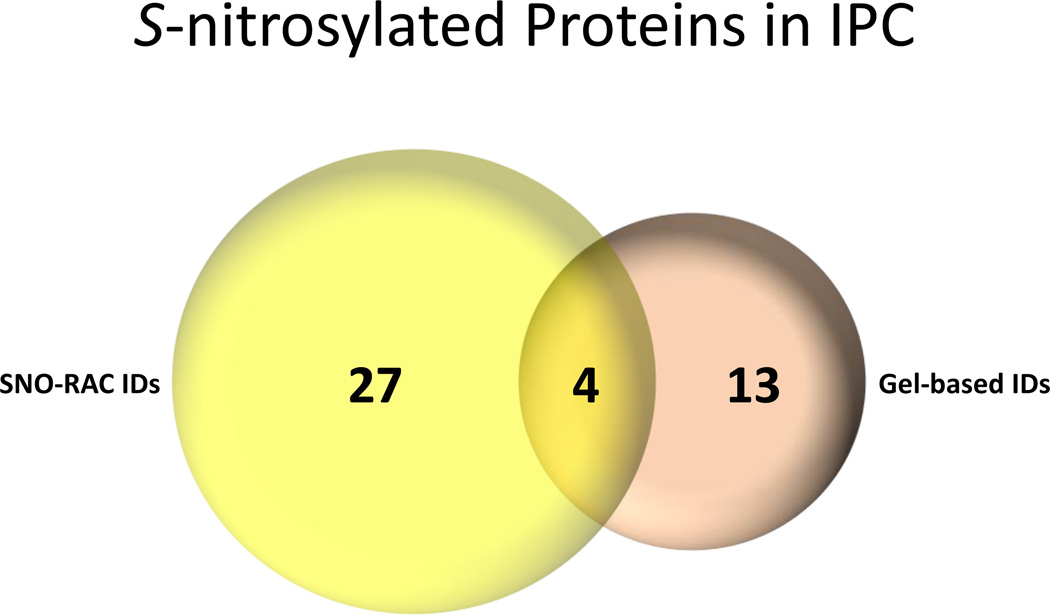
A comparison of SNO proteins identified with 2D-DyLight switch versus those identified with SNO-RAC following ischemic preconditioning is shown in Figure 2. Interestingly, there is little overlap between the proteins identified by the two methods. These data are similar to what was found in a comparison between ICAT and 2D-DIGE measurements of protein oxidation[9]. SNO-RAC identified a larger number of proteins than the 2D gel method. This is expected because high molecular weight proteins and hydrophobic membrane proteins are typically difficult to observe in 2D gel based methods. It was somewhat surprising that 13 proteins were only identified by the DyLight switch gel based method. Mass spectrometry identification is limited by dynamic range; mass spectrometry will identify only the most abundant proteins in the fraction. The gel based method separates on the protein level and the SNO-RAC separates on the peptide level and this might explain the different sets of proteins identified by the two methods. With separation on the protein level, abundant proteins such as actin and myosins will swamp out proteins at the same molecular weight. In contrast if these abundant proteins are digested, they can now overwhelm peptides from other proteins that fractionate similarly. Thus the separation of low abundance proteins from more abundant proteins will differ in these two methods. It is also worth noting that multiple proteins can be present in a spot identified in the 2D gel by fluorescence. The fluorescence, and hence the SNO, could be due to a protein in low abundance, but the more abundant protein will be identified by mass spectrometry. With the SNO-RAC method, proteins are often identified based on a single peptide, because that is all that binds to the resin. If this peptide does not ionize well in the mass spectrometer the protein will not be identified. In summary, the gel based methods and SNO-RAC method identify different sets of proteins. It is therefore important to use multiple approaches to identify a more complete set of SNO proteins.
Figure 2.
A comparison of proteins identified with 2D-DyLight switch versus those identified with SNO-RAC. There were 4 common proteins identified with both SNO-RAC and the 2D gel. There wer proteins that were only identified with SNO-RAC and 13 proteins that were only identified with the 2D-gel method.
Phenylmercury Based Methods
A recent paper by Doulias et al [16] describes a method to measure protein SNO using an organo-mercury reagent. Phenylmercury reacts with SNO resulting in a thiol-mercury bond. An organomercury resin was synthesized by conjugation of p-amino-phenylmercuric- acetate to N-hydroxysuccinimide-activated Affi-Gel 10 agarose beads and a phenylmercury-polyethleneglycol-biotin compound. Similar to the biotin switch, this method relies on complete blocking of free SH groups. However, instead of reducing SNO groups with ascorbic acid, the SNO groups are bound to the resin; thus this method also relies on the specificity of the reaction of the organo-mercury reagent with SNO groups and therefore appropriate controls are needed. The resin is then washed to remove unbound proteins followed by beta mercaptoethanol mediated release of proteins. Alternatively, resin capture can be followed by on-resin digestion, and performic acid treatment to release bound peptides and oxidize cysteine thiols to sulfonic acid allowing for site specific identification by LC-MS/MS. Using this method Doulias et al [16] identified 328 endogenously SNO peptides from 192 proteins in mouse liver extract. They reported that the SNO sites have an average pKa of 10.01. They also reported that 70% of the SNO cysteines were located near negatively or positively charged amino acids.
Direct Mass Spectrometry based Methods
Detection of cysteine-bound NO by direct mass spectrometry analysis is difficult due to the lability of the SNO modification. The NO group is typically lost under conditions associated with MALDI-MS, but the NO group can be preserved with electrospray mass spectrometry [17]. Neutral loss of 29 Da has been used to identify SNO proteins and peptides; however, this has mostly been done with purified proteins (see [11, 18]).
Antibody Based Methods
Antibodies recognizing S-nitrosylated proteins have also been reported. The use of antibodies to identify SNO can be limited because of the lability of the SNO modification. The modification is often lost during protein extraction and/or the procedures used for western blotting or immunoprecipitation [19, 20]. There have been also some questions with regard to antibody specificity, and therefore, it is important to compare the antibody results to those obtained with another method.
3. How Does SNO Alter Cell Signaling or Cell Function?
Analogous to phosphorylation and other post-translational modifications, SNO can alter cell function by altering activity of proteins, by altering cellular localization, or by altering the stability of the protein. In addition, SNO can protect the modified cysteine residues from irreversible oxidation. SNO fulfills the requirements for a signaling post-translational modification. SNO is stimulus mediated; it is initiated by activation of NOS. NO generated by NOS can directly lead to SNO of proteins. In addition, these SNO proteins can specifically bind to other proteins and by transnitrosylation can lead to SNO of additional proteins. Similar to phosphorylation and other post-translational modifications SNO appears to be targeted to specific cysteines. For example RyR1 has 100 cysteine residues, but only a few are targeted for SNO. SNO is reversible; a number of enzyme systems have been shown to function as denitrosylases [21–23]. GSH can remove SNO from proteins to generate GSNO which is then converted back to GSH by S-nitrosoglutathione reductase (GSNOR). Similarly thioredoxin can denitrosylate proteins and thioredoxin is then regenerated by thioredoxin reductase.
SNO can alter the activity of proteins and enzymes
There is a large and growing list of proteins for which SNO has been shown to alter activity and/or function (see Table 2 for a partial list). Analogous to other post-translational modifications, SNO can alter protein structure and thereby change protein activity. SNO has been shown to alter the activity of proteins in virtually all cellular pathways. As illustrated in Table 2, the activity of a large number of ion channels can be regulated by SNO [1, 24–29]; this might be due in part to SNO targeting to membranes.
Table 2.
Effects of SNO on protein activity/function of some proteins of interest in the cardiovascular system.
| Protein | Effect of SNO on Protein | Ref. |
|---|---|---|
| Ion channel and transporter | ||
| Cx43 | Increases open probability | [74] |
| KCNQ1 | Activates channel | [26] |
| LTCC | Inhibits and/or activates Ca2+ entry | [24, 50] |
| NCX1 | Stimulates activity | [75] |
| RyR1 | Increases open channel probability | [25, 76] |
| RyR2 | Increase open channel probability | [61, 77] |
| SERCA | Increases activity | [1] |
| TRP | Activates channel | [28] |
| Signaling and Cardioprotection | ||
| Aconitase | Inhibits activity | [8] |
| Akt | Inhibits activity | [31] |
| ALDH2 | Inhibits activity | [8, 78] |
| Beta-arrestin | Promotes binding to clathrin and receptor internalization | [79] |
| Caspase 3 | Inhibits activity | [80] |
| Complex IV | Inhibits activity | [81] |
| Complex I | Inhibits activity | [49] |
| COX-2 | Increases activity | [82] |
| Creatine kinase | Inhibits activity | [83] |
| DJ1 | Alters protein dimerization | [84] |
| DRP1 | Activates GTPase activity and activates fission | [85] |
| eNOS | Inhibits activity | [86, 87] |
| ER | Inhibits binding to DNA | [88] |
| F1-ATPase | Inhibits activity | [1] |
| GAPDH | Inhibits activity; increases binding to Siah & increases nuclear localization | [36, 38] |
| GRK2 | Decreased phosphorylation of β receptor | [30] |
| HIF-1 | Stabilizes protein | [23] |
| HSP90 | Inhibits ATPase activity | [89] |
| myosin | Inhibits activity | [32] |
| NF-kappaB/IKK | Inhibits activity | [90] |
| Parkin | Initially increased then represses E3 ubiquitin ligase activity | [91] |
| PTEN | Inhibits activity and increases degradation | [34] |
| PTP-1B | Inhibits activity | [42, 92] |
| Thioredoxin | Increases reductase activity/increase transnitrosylase activity | [22, 93] |
| Transglutaminase | Inhibits activity | [94] |
SNO modifies the activity of many other proteins, only a few of which are mentioned in Table 2. Whalen et al [30] examined the mechanism by which NO/SNO reduces β-adrenergic receptor desensitization. They found that SNO of G protein-coupled receptor kinase 2 (GRK2) at C340 decreases GRK2 phosphorylation of β-adrenergic receptors and its resultant desensitization. Lu et al [31] found evidence for SNO of C296 of Akt. The authors suggested that SNO of C296 would prevent the formation of a disulfide bond between C296 and C310 and that blocking disulfide bond formation attenuates the activity of Akt. Nogueira et al [32] reported that myosin is reversibly inhibited by SNO. GSNO treatment of isolated myosin inhibits myosin activity without affecting the affinity for actin. Kapadia et al [33]reported that SNO reversibly inhibits the 26S proteasome catalytic activity. PTEN (phosphatase and tensin homologue deleted on chromosome 10) is a phosphatase that dephosphorylates AKT, a kinase that has been shown to be important in signaling cardioprotection. SNO of PTEN results in its ubiquitination and degradation[34]. SNO can modify function directly, but SNO can also promote other redox modifications, such as disulfide bond formation or glutathiolation that can also regulate function. For example, a disulfide bond between the R1 subunits of protein kinase A (PKA) is formed after treatment with the NO donor, nitrocysteine. This disulfide bond results in activation of PKA [35].
SNO can also alter the localization of protein
One of the best described examples of SNO altering protein localization is illustrated by the glyceraldehyde phosphate dehydrogenase (GAPDH) GOSPEL (GAPDH's competitor of Siah Protein Enhances Life) pathway [36–38]. Hara et al [36] reported that high levels of NO lead to SNO of GAPDH resulting in an increased association with an E3 ubiquitin ligase, Siah. GAPDH stabilizes Siah and this complex enters the nucleus (Siah has a nuclear localization signal) where it leads to degradation of nuclear proteins. Degradation of nuclear receptor corepressor (NCoR) is increased by SNO of GAPDH and NCoR is involved in the binding of histone deacetylases (HDAC). Therefore, degradation of NCoR by Siah promotes acetylation which alters gene expression and ultimately leads to an increase in cell death [39]. Interestingly, low levels of NO lead to SNO of a protein known as GOSPEL, which subsequently binds GAPDH and sequesters GADPH away from Siah [37], thereby blocking the stabilization of nuclear Siah and its activation of cell death pathways. This example illustrates the dose dependent effects of NO/SNO, and shows that low levels of SNO reduce cell death whereas high levels of SNO promote cell death. These data suggest that GOSPEL is S-nitrosylated at lower levels of NO than GAPDH; the mechanism by which this occurs is not known. These data also show that SNO can be selectively targeted to proteins.
SNO has also been proposed to protect by shielding thiol groups from oxidation
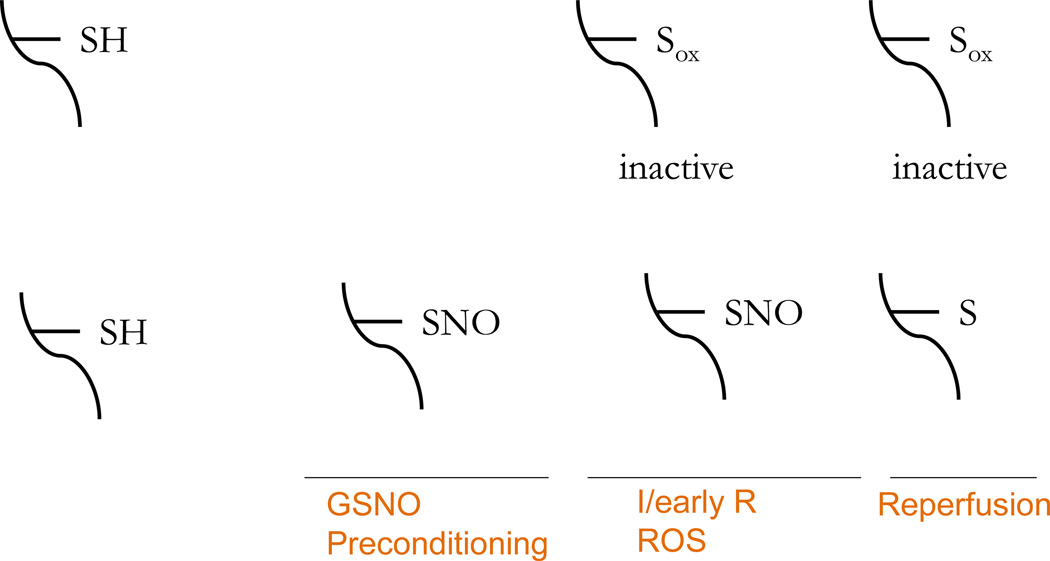
There are data suggesting that S-nitrosylated thiols are protected from irreversible oxidation [15, 40–42]. Chen et al [42] showed that SNO of protein-tyrosine phosphatase 1B (PTP-1B) at C215 protected PTP-1B from H2O2-induced irreversible oxidation. Because SNO is a transient modification, the SNO can be readily reversed, but during time of oxidative stress, SNO can shield modified cysteines from more irreversible states of oxidation. This could be important in conditions such as ischemia and reperfusion where there is a large burst of reactive oxygen species (ROS) during the first few seconds of reperfusion. This increase in ROS is well documented to result in the irreversible oxidation of a number of important proteins. These proteins are irreversibly modified and need to be degraded and re-synthesized to regain normal function. As discussed in detail in the section on cardioprotection, Kohr et al [15] recently combined the SNO-RAC method with an OX-RAC method to measure protein oxidation and showed that preconditioning leads to SNO of many proteins and that a large majority of these proteins are protected from oxidation (see Figure 3).
Figure 3.
Preconditioning increases SNO which can shield thiol groups from irreversible oxidation.
4. Role of SNO in Cardioprotection
An increase in NO is well established to play a role in cardioprotection [43, 44]. Initially much of the cardioprotection afforded by NO was attributed to activation of guanylyl cyclase [45]. Recent data has suggested a role for SNO in cardioprotection, and a number of studies have shown an association between an increase in SNO and cardioprotection [1, 4, 7, 14, 15, 23, 24, 46–49]. SNO modulates protein function/activity and like other post-translational modifications, depending on the context and the proteins that are modified, this can positively or negatively modulate cell death. As previously noted, low levels of NO result in SNO of a protein called GOSPEL which results in reduced cell death; however with higher levels of NO, GAPDH is SNO which leads to increased death [37]. Thus it is likely that the beneficial effects of SNO are dependent on the level of SNO and also dependent on the redox state of the cell.
Cardioprotection alters protein SNO
Sun et al. showed that an increase in SNO occurred with cardioprotection in females [24]. This study showed that females exhibited increased SNO of L-type Ca2+ channel (LTCC) and further showed that SNO of LTCC was associated with decreased Ca2+ entry following β-adrenergic stimulation. There are also reports that SNO can activate LTCC [50]; the difference may depend on levels of NO and redox state. During ischemia and reperfusion, inhibition of LTCC would lead to reduced Ca2+ entry via the LTCC and therefore reduce Ca2+ overload of the cell. It is worth noting that SNO is a very labile, transient post-translational modification; thus the alteration in protein activity is likely to be transient. This inhibition of Ca2+ entry via LTCC would be quickly reversed during reperfusion. Thus, the reduced Ca2+ entry would only occur during ischemia and early reperfusion; this is a time when it would be advantageous to limit Ca2+ entry. Taken together these data suggested that increased SNO of LTCC and corresponding reduction in its activity in females will reduce Ca2+ overload during ischemia and reperfusion thus contributing to cardioprotection [24]. Lin et al [48] using 2D-DyLight methods, followed up on these studies and identified a number of proteins in females which showed increased SNO. Consistent with these studies showing a correlation between cardioprotection and increased SNO, Sun et al [1] using 2D-DyLight methods found an increase in SNO in hearts extracted immediately after the preconditioning protocol. Preconditioning involves brief intermittent periods of ischemia followed by reperfusion and has been shown to significantly reduce ischemia-reperfusion injury [51]. Interestingly many of the same proteins that showed an increase in SNO in females also were increased following preconditioning [1, 48]. Sun et al found an increase in SNO of the F1-ATPase and showed that addition of GSNO, a transnitrosylating agent, to submitochondrial particles resulted in a decrease in F1-ATPase activity [1]. Inhibition of the F1-ATPase during ischemia would reduce the breakdown of ATP that occurs during ischemia by reverse mode of the F1-ATPase, which operates to consume ATP. Because SNO is a transient modification, inhibition of the F1-ATPase would be transient and on reperfusion, the F1-ATPase would operate to generate ATP again. Sun et al also showed that preconditioning results in the activation of the SR/ER Ca2+-ATPase (SERCA) [1]. The increase in SERCA activity, coupled with decreased Ca2+ entry via LTCC, would reduce cytosolic and hence mitochondrial Ca2+ levels, and would thereby reduce Ca2+ available to activate the mitochondrial permeability transition pore. Burwell et al [49] also showed that preconditioning results in an increase in SNO of complex I. They suggested that the increase in SNO of complex I results in inhibition of complex I, which they showed was associated with a decrease in ROS production during ischemia and reperfusion.
SNO protects from oxidation
As mentioned SNO has also been suggested to shield thiol groups from irreversible oxidation and this could play an important role in cardioprotection [15, 40–42]. The hypothesis is illustrated in Figure 3. It is proposed that ischemia and reperfusion result in the irreversible oxidation of protein thiol groups resulting in permanent alterations in their function. These proteins would have to be degraded and resynthesized to regain normal function. By increasing SNO of thiol groups, preconditioning can block irreversible oxidation of these thiols. Because we have shown SNO during preconditioning to be transient, with the modification lost during early reperfusion [15], we expect that the protein will return to normal function. To directly test this hypothesis we need to be able to identify sites of SNO and sites of oxidation since we predict that the sites of SNO during preconditioning would show less oxidation compared to non-preconditioned hearts. Kohr et al [15] used the SNO-RAC method to measure sites of SNO during preconditioning. They found 27 proteins with an increase in SNO in preconditioned hearts. Compared to non-preconditioned hearts, SNO proteins in preconditioned hearts showed a 76% decrease in oxidation at the same cysteine site after 5 minutes of reperfusion. To measure protein oxidation Kohr et al [15] used an Ox-RAC protocol, which combined procedures adapted from the SNO-RAC technique [13], and redox DIGE [52]. These data support the hypothesis that an increase in SNO can protect protein thiols from irreversible oxidation.
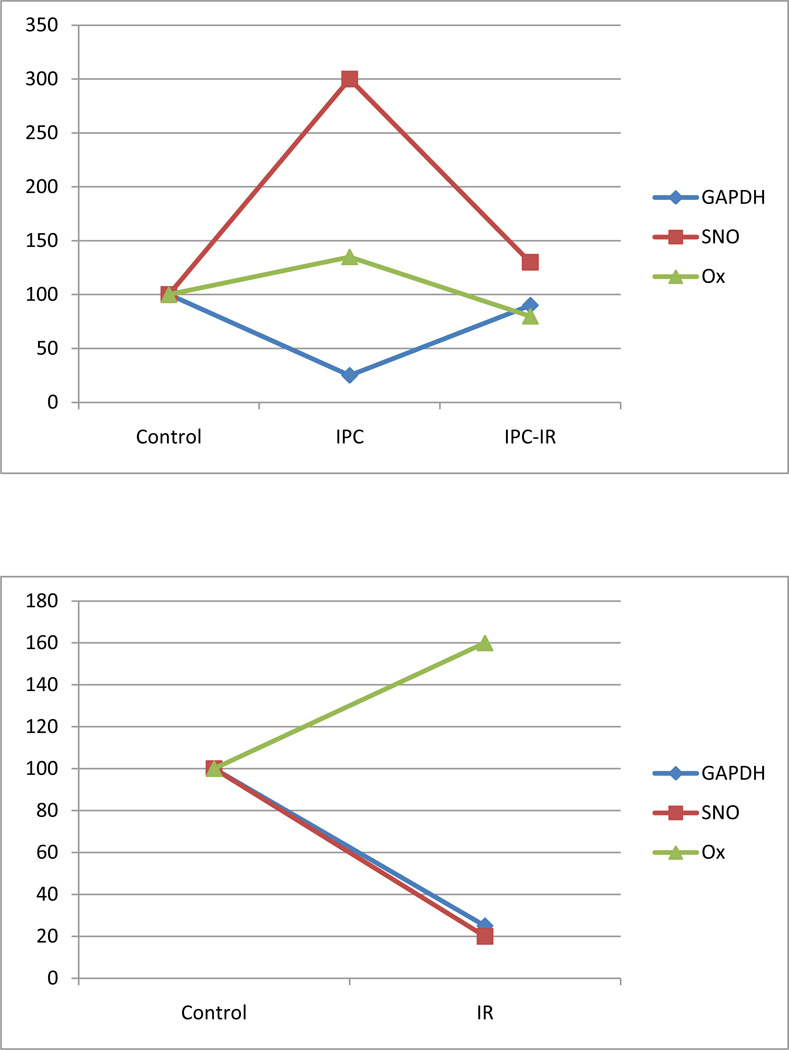
To further test this hypothesis, Kohr et al [15] monitored the activity of GAPDH in these samples and directly correlated this with SNO and oxidation levels. As shown in Figure 4 (adapted from Kohr et al [15]) preconditioning results in a decrease in GAPDH activity, presumably due to the increase in SNO. Ischemia followed by 5 minutes of reperfusion also results in a decrease in GAPDH activity, but in this case it appears to be due to an increase in oxidation as there is little or no increase in SNO. Interestingly in hearts that are preconditioned prior to 20 minutes of ischemia and 5 minutes of reperfusion, SNO which occurred during preconditioning is lost, and GAPDH activity is not significantly different than control; these hearts also show no significant increase in oxidation. Taken together these data support the hypothesis that SNO can protect thiols from oxidation thereby allowing recovery of protein activity during early reperfusion when the transient SNO is lost.
Figure 4.
SNO of GAPDH protects from oxidation.
In further support of a role for SNO in cardioprotection, Nadtochly et al [47] showed that IP injection of S-nitroso-2-mercaptopropionyl glycine, a mitochondrial targeted S-nitrosating agent, reduced infarct size in an in vivo mouse LAD occlusion model. Similarly, Prime et al [53] showed that addition of a mitochondrial targeted S-nitrosothiol (MitoSNO) reduced ischemia-reperfusion injury. Also consistent with a role for SNO in cardioprotection, Lima et al [23] showed that mice null for GSNO reductase, an enzyme involved in removing SNO, have reduced I/R injury. They also show SNO of hypoxic-inducible factor- 1α (HIF-1α) in GSNOR null mice and further show that SNO-HIF-1α binds to the VEGF gene resulting in increased capillary density.
5. Current and Future Issues Regarding SNO and Cardioprotection
SNO of a number of proteins has been shown to occur with cardioprotection. These proteins include the F1-ATPase, SERCA2a, LTCC, complex I, and PTEN. Altered activity or reduced oxidation of these proteins could contribute to cardioprotection. However, there are a number of questions regarding SNO and cardioprotection. Does SNO lead to cardioprotection by the summation of multiple pathways? In other words does SNO modulate many pathways which sum up to provide cardioprotection. Alternatively, is one or two of these SNO sites of critical importance in cardioprotection? In order to address this question we need to first identify the sites of SNO in key proteins with cardioprotection, select a group of proteins and mutate these key cysteine residues to determine if this blocks cardioprotection.
Approaches to Target Low Abundant Proteins
Another issue regarding the role of SNO and cardioprotection is that most of the SNO proteins that have been identified were found using broad based proteomic screens. These broad based proteomic screens are unbiased; however, they tend to select for highly abundant proteins [54]. It is likely that lower abundant signaling proteins might play a crucial role in cardioprotection. Thus we might be observing a change in SNO in abundant proteins that are not primarily responsible for cardioprotection, but which provide a marker for an SNO that is the key player in protection. Additional studies using a more targeted approach to examine SNO of candidate proteins will be needed.
Occupancy
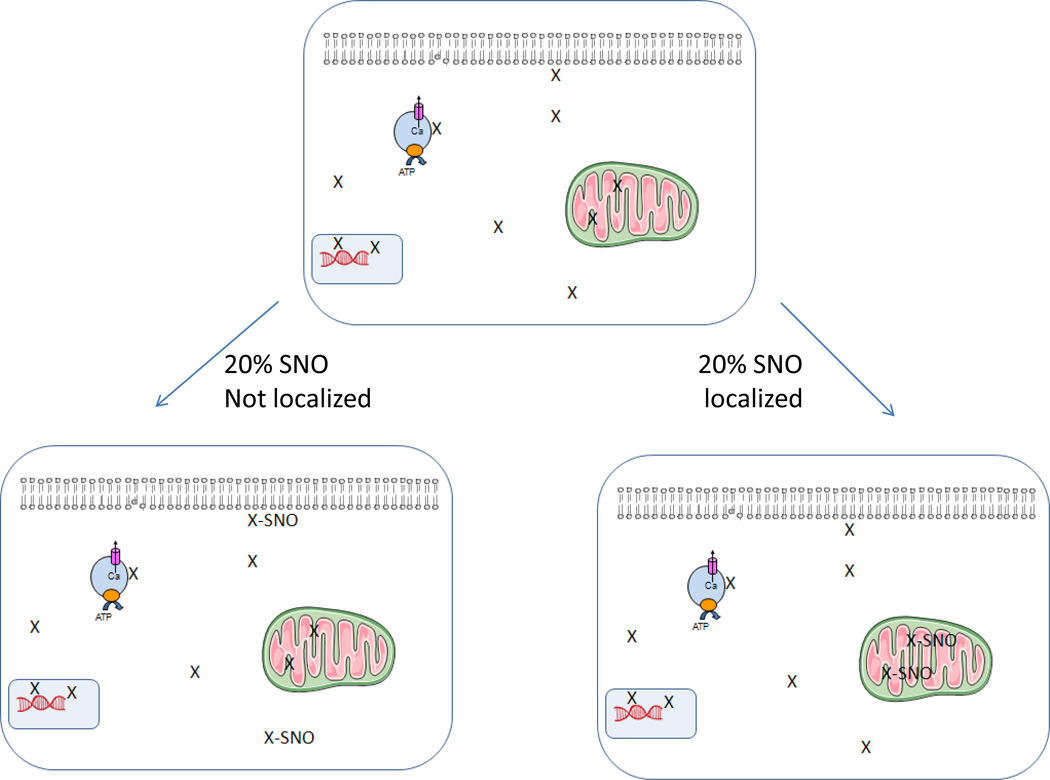
Although we have identified a large number of proteins that show an increase in SNO with preconditioning, the percentage of the total protein which is SNO (or the occupancy) is unknown. Sun et al [1] have found that preconditioning or GSNO perfusion of heart results in a 3 to 4 fold increase in SNO of many proteins. However, it is not clear whether SNO of these proteins increases from 10% to 40% occupancy, or whether this represents an increase from 2% to 8%. Clearly if SNO is going to protect a protein from oxidation, it is unlikely that an increase in SNO from 2 to 8% would have a major effect in reducing oxidation or preservation of the protein function. If SNO results in inhibition of an enzyme, assuming no compartmentalization of the enzyme, then one would expect that if 8% of the enzyme was SNO this would lead to only 8% inhibition of the activity. For most enzymes one would not expect this to have a major impact on cell function. If the enzyme is activated by SNO, going from 2% to 8% of the active enzyme could have a major impact on cell function. However, as illustrated in Table 2, SNO results in inhibition of the activity of many enzymes. Furthermore, as discussed, protection from oxidation would only be beneficial if a high percentage of the protein was SNO. Therefore it is important to determine the percentage of a protein that is SNO. It is also worth considering that the SNO modification is very labile and that it is likely that we are losing some of the modification during isolation and measurement of SNO. It would therefore also be important to have some way of estimating how much of the modification is lost. Lastly, one needs to consider, at least in some cases, that there could be localized compartments that are modified. As illustrated in figure 5, if only 20% of a protein (protein X) in the cell is inhibited by SNO this would not be expected to have a major impact on cell function. However, NO/SNO signaling is known to be localized and if there was more complete (e.g. ~100%) SNO and inhibition of localized protein X (e.g. mitochondrial, nuclear, SR) this could have important cellular effects. These are important issues for future studies
Figure 5.
Localized NOS signaling can result in targeted SNO. This is a hypothetical depiction of how localized signaling could play a role in signaling and why it is important to understand the localization of SNO as well as the total percentage of a protein that is SNO. For example if there is only 20% modification of a protein (protein X) and the modification is uniformly distributed, this would have a different effect on the cell than if for example 20% of the protein was localized in a compartment (e.g. mitochondria) and all of the protein in this compartment was SNO.
Uncoupled NOS
Cardioprotection is frequently lost with aging or in diseases such as diabetes and hypercholesterolemia [55–57]. Interestingly aging and many diseases are associated with an “uncoupled NOS” [55–58]. NOS converts the substrates oxygen and arginine into NO and citrulline. It also requires the cofactor tetrahydropterin (BH4). BH4 levels have been reported to decrease with aging and disease, which results in NOS uncoupling which causes the generation of high levels of superoxide and low levels of NO. [58, 59]. S-glutathionylation has been recently shown to uncouple eNOS [60]. Under conditions of low BH4, when NOS becomes uncoupled there would be lower levels of SNO. Thus, reduction in BH4 and uncoupling of NOS are likely to play an important role in the loss of protection with age and disease. In fact, stimulation of uncoupled NOS would be detrimental. The increased ROS and decreased NO will lead to further dysfunction by enhancing redox/nitrosative stress [41]. ROS can interact with NO to generate peroxynitrite. This reaction can lead to a decrease in cellular NO/SNO which can contribute to redox-nitroso imbalance. Activation of xanthine oxidase results in hyponitrosylation and oxidation of RyR2 resulting in Ca2+ leak [61].
SNO of Mitochondrial Proteins
It is also of interest that a large number of mitochondrial proteins have been shown to be SNO. This is interesting because mitochondria have been reported to be key regulators of cardioprotection. Mitochondria comprise ~30% of myocyte volume, however mitochondrial SNO proteins represented about 52% of the SNO proteins observed during preconditioning. One reason for the relative abundance of mitochondrial proteins showing SNO might be the large number of mitochondrial proteins containing cysteines. Also the mitochondrial matrix pH is alkaline which enhances reactivity of thiols, and mitochondria have many membrane proteins that are in a hydrophobic environment that can also enhance SNO.
Targeting of SNO
The abundance of SNO mitochondrial proteins raises the important issue of how SNO is targeted to the mitochondria? SNO appears to be targeted to selective proteins and selective cysteine residues, but the mechanisms responsible for targeting are poorly understood. Targeting appears to occur in several ways. NOS isoforms are targeted to different regions of the cells. There are scaffolding proteins that facilitate interaction of NOS and proteins. For example, CAPON (carboxy-terminal PDZ ligand of nNOS) has been shown to be an adaptor protein which binds nNOS and regulates its interaction with other proteins such as Dexras [62]. β-arrestin has also been shown to act as a scaffolding protein localizing eNOS, the β-adrenergic receptor and clathrin thereby promoting receptor endocytosis [30]. In addition, studies have reported that nNOS is targeted to the sarcoplasmic reticulum and eNOS is targeted to caveolae in the plasma membrane [63–65]. The mitochondrial source of NO is somewhat unclear. Some investigators have reported a mitochondrial NOS [66]; however, these findings have been debated and many question the presence of a mitochondrial NOS [67–69]. Given the close proximity of the SR and the mitochondria, nNOS localized to the SR could provide a source of NO to the mitochondria. Much of protein SNO is thought to occur as a result of transnitrosylation, a thiol to thiol transfer of NO. For example GSH can remove SNO from one protein and transnitrosylate or add it to another protein. Data are emerging suggesting that SNO-GAPDH may be targeted to selected nuclear proteins which become SNO by transnitrosylation with SNO-GAPDH as a donor [70]. Thioredoxin has also been shown to function as a transnitrosylase for specific proteins [21, 22]. Thioredoxin has been shown to specifically transnitrosylate peroxiredoxin and other proteins [22].
Thus, there appear to be multiple mechanisms by which SNO is targeted. NOS can be localized to different parts of the cell and this localization likely involves scaffolding proteins. A few scaffolding proteins such as CAPON and β-arrestin have been identified but there are likely to be others. Transnitrosylation also appears to be regulated by binding of SNO proteins such as SNO-GAPDH to selected proteins. It has been suggested that SNO-GAPDH mediated transnitrosylation might be a major mechanism for SNO of nuclear proteins. Interestingly GAPDH has also been shown to be present in the mitochondria [71] and it might be interesting to determine if it has a similar role in mitochondria.
It has been suggested that SNO might be targeted to cysteines with an acid-base motif. With the increasing number of SNO proteins it is now possible to use software to determine if SNO is targeted to specific motifs. At present there is no clear consensus [7, 14, 16, 72, 73]. It is possible that the lack of consensus is because we are lumping together a number of different transnitrosylating proteins that may recognize distinct motifs. This would be equivalent to attempting to find a consensus phosphorylation motif common among all kinases.
6. Summary
An increase in SNO occurs with cardioprotection. Agents or genetic alterations that increase SNO are cardioprotective. Cardioprotection also leads to an increase in SNO of many proteins and pathways that have been suggested to play a role in protection. SNO could protect by altering the activity of proteins (e.g. F1-ATPase, complex I, SERCA, LTCC). SNO can also protect by shielding thiol groups from oxidation and thereby allowing more rapid recovery of protein function. SNO has also been shown in other cell types to alter the localization of proteins such as Siah and thereby alter cell death signaling. The role for SNO in altering protein localization has not been well studied in heart. Whether SNO mediates protection by the sum of these multiple pathways or whether SNO of one or two proteins is of primary importance in mediating cardioprotection is unclear at this time. Addressing these questions has been hampered by the lack of methods for measuring sites of SNO. Now that methods, such as SNO-RAC, are available for measuring sites of SNO we should be able to address the role of specific SNO sites in cardioprotection.
Highlights.
Methods for measuring S-nitrosylation
S-nitrosylation regulates cardiovascular function
Role of S-nitrosylation in cardioprotection
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: None
Reference List
- 1.Sun J, Morgan M, Shen RF, Steenbergen C, Murphy E. Preconditioning results in S-nitrosylation of proteins involved in regulation of mitochondrial energetics and calcium transport. Circ Res. 2007;101:1155–1163. doi: 10.1161/CIRCRESAHA.107.155879. [DOI] [PubMed] [Google Scholar]
- 2.Seth D, Stamler JS. The SNO-proteome: causation and classifications. Curr Opin Chem Biol. 2011;15:129–136. doi: 10.1016/j.cbpa.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schulman IH, Hare JM. Regulation of cardiovascular cellular processes by S-nitrosylation. Biochim Biophys Acta. 2011 doi: 10.1016/j.bbagen.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun J, Murphy E. Protein S-nitrosylation and cardioprotection. Circ Res. 2010;106:285–296. doi: 10.1161/CIRCRESAHA.109.209452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Espey MG, Miranda KM, Thomas DD, Xavier S, Citrin D, Vitek MP, et al. A chemical perspective on the interplay between NO, reactive oxygen species, and reactive nitrogen oxide species. Ann N Y Acad Sci. 2002;962:195–206. doi: 10.1111/j.1749-6632.2002.tb04068.x. [DOI] [PubMed] [Google Scholar]
- 6.Jaffrey SR, Snyder SH. The biotin switch method for the detection of S-nitrosylated proteins. Sci STKE. 2001;2001 doi: 10.1126/stke.2001.86.pl1. pl1. [DOI] [PubMed] [Google Scholar]
- 7.Murray CI, Kane LA, Uhrigshardt H, Wang SB, Van Eyk JE. Site-Mapping of In Vitro S-nitrosation in Cardiac Mitochondria: Implications for Cardioprotection. Mol Cell Proteomics. 2010;10 doi: 10.1074/mcp.M110.004721. M110 004721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chouchani ET, Hurd TR, Nadtochiy SM, Brookes PS, Fearnley IM, Lilley KS, et al. Identification of S-nitrosated mitochondrial proteins by S-nitrosothiol difference in gel electrophoresis (SNO-DIGE): implications for the regulation of mitochondrial function by reversible S-nitrosation. Biochem J. 2010;430:49–59. doi: 10.1042/BJ20100633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu C, Hu J, Liu T, Ago T, Sadoshima J, Li H. Quantitative analysis of redox-sensitive proteome with DIGE and ICAT. J Proteome Res. 2008;7:3789–3802. doi: 10.1021/pr800233r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hao G, Derakhshan B, Shi L, Campagne F, Gross SS. SNOSID, a proteomic method for identification of cysteine S-nitrosylation sites in complex protein mixtures. Proc Natl Acad Sci U S A. 2006;103:1012–1017. doi: 10.1073/pnas.0508412103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Torta F, Elviri L, Bachi A. Direct and indirect detection methods for the analysis of S-nitrosylated peptides and proteins. Methods Enzymol. 2010;473:265–280. doi: 10.1016/S0076-6879(10)73014-7. [DOI] [PubMed] [Google Scholar]
- 12.Yang Y, Loscalzo J. S-nitrosoprotein formation and localization in endothelial cells. Proc Natl Acad Sci U S A. 2005;102:117–122. doi: 10.1073/pnas.0405989102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forrester MT, Thompson JW, Foster MW, Nogueira L, Moseley MA, Stamler JS. Proteomic analysis of S-nitrosylation and denitrosylation by resin-assisted capture. Nat Biotechnol. 2009;27:557–559. doi: 10.1038/nbt.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kohr MJ, Aponte AM, Sun J, Wang G, Murphy E, Gucek M, et al. Characterization of potential S-nitrosylation sites in the myocardium. Am J Physiol Heart Circ Physiol. 2011;300:H1327–H1335. doi: 10.1152/ajpheart.00997.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kohr MJ, Sun J, Aponte A, Wang G, Gucek M, Murphy E, et al. Simultaneous measurement of protein oxidation and S-nitrosylation during preconditioning and ischemia/reperfusion injury with resin-assisted capture. Circ Res. 2011;108:418–426. doi: 10.1161/CIRCRESAHA.110.232173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doulias PT, Greene JL, Greco TM, Tenopoulou M, Seeholzer SH, Dunbrack RL, et al. Structural profiling of endogenous S-nitrosocysteine residues reveals unique features that accommodate diverse mechanisms for protein S-nitrosylation. Proc Natl Acad Sci U S A. 2010;107:16958–16963. doi: 10.1073/pnas.1008036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Liu T, Wu C, Li H. A strategy for direct identification of protein S-nitrosylation sites by quadrupole time-of-flight mass spectrometry. J Am Soc Mass Spectrom. 2008;19:1353–1360. doi: 10.1016/j.jasms.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hao G, Xie L, Gross SS. Argininosuccinate synthetase is reversibly inactivated by S-nitrosylation in vitro and in vivo. J Biol Chem. 2004;279:36192–36200. doi: 10.1074/jbc.M404866200. [DOI] [PubMed] [Google Scholar]
- 19.Ying J, Clavreul N, Sethuraman M, Adachi T, Cohen RA. Thiol oxidation in signaling and response to stress: detection and quantification of physiological and pathophysiological thiol modifications. Free Radic Biol Med. 2007;43:1099–1108. doi: 10.1016/j.freeradbiomed.2007.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foster MW. Methodologies for the characterization, identification and quantification of S-nitrosylated proteins. Biochim Biophys Acta. 2011 doi: 10.1016/j.bbagen.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benhar M, Thompson JW, Moseley MA, Stamler JS. Identification of S-nitrosylated targets of thioredoxin using a quantitative proteomic approach. Biochemistry. 2010;49:6963–6969. doi: 10.1021/bi100619k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu C, Liu T, Chen W, Oka S, Fu C, Jain MR, et al. Redox regulatory mechanism of transnitrosylation by thioredoxin. Mol Cell Proteomics. 2010;9:2262–2275. doi: 10.1074/mcp.M110.000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lima B, Lam GK, Xie L, Diesen DL, Villamizar N, Nienaber J, et al. Endogenous S-nitrosothiols protect against myocardial injury. Proc Natl Acad Sci U S A. 2009;106:6297–6302. doi: 10.1073/pnas.0901043106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun J, Picht E, Ginsburg KS, Bers DM, Steenbergen C, Murphy E. Hypercontractile female hearts exhibit increased S-nitrosylation of the L-type Ca2+ channel alpha1 subunit and reduced ischemia/reperfusion injury. Circ Res. 2006;98:403–411. doi: 10.1161/01.RES.0000202707.79018.0a. [DOI] [PubMed] [Google Scholar]
- 25.Aracena-Parks P, Goonasekera SA, Gilman CP, Dirksen RT, Hidalgo C, Hamilton SL. Identification of cysteines involved in S-nitrosylation, S-glutathionylation, and oxidation to disulfides in ryanodine receptor type 1. J Biol Chem. 2006;281:40354–40368. doi: 10.1074/jbc.M600876200. [DOI] [PubMed] [Google Scholar]
- 26.Asada K, Kurokawa J, Furukawa T. Redox- and calmodulin-dependent S-nitrosylation of the KCNQ1 channel. J Biol Chem. 2009;284:6014–6020. doi: 10.1074/jbc.M807158200. [DOI] [PubMed] [Google Scholar]
- 27.Adachi T, Weisbrod RM, Pimentel DR, Ying J, Sharov VS, Schoneich C, et al. S-Glutathiolation by peroxynitrite activates SERCA during arterial relaxation by nitric oxide. Nat Med. 2004;10:1200–1207. doi: 10.1038/nm1119. [DOI] [PubMed] [Google Scholar]
- 28.Yoshida T, Inoue R, Morii T, Takahashi N, Yamamoto S, Hara Y, et al. Nitric oxide activates TRP channels by cysteine S-nitrosylation. Nat Chem Biol. 2006;2:596–607. doi: 10.1038/nchembio821. [DOI] [PubMed] [Google Scholar]
- 29.Eu JP, Sun J, Xu L, Stamler JS, Meissner G. The skeletal muscle calcium release channel: coupled O2 sensor and NO signaling functions. Cell. 2000;102:499–509. doi: 10.1016/s0092-8674(00)00054-4. [DOI] [PubMed] [Google Scholar]
- 30.Whalen EJ, Foster MW, Matsumoto A, Ozawa K, Violin JD, Que LG, et al. Regulation of beta-adrenergic receptor signaling by S-nitrosylation of G-protein-coupled receptor kinase 2. Cell. 2007;129:511–522. doi: 10.1016/j.cell.2007.02.046. [DOI] [PubMed] [Google Scholar]
- 31.Lu XM, Lu M, Tompkins RG, Fischman AJ. Site-specific detection of S-nitrosylated PKB alpha/Akt1 from rat soleus muscle using CapLC-Q-TOF(micro) mass spectrometry. J Mass Spectrom. 2005;40:1140–1148. doi: 10.1002/jms.885. [DOI] [PubMed] [Google Scholar]
- 32.Nogueira L, Figueiredo-Freitas C, Casimiro-Lopes G, Magdesian MH, Assreuy J, Sorenson MM. Myosin is reversibly inhibited by S-nitrosylation. Biochem J. 2009;424:221–231. doi: 10.1042/BJ20091144. [DOI] [PubMed] [Google Scholar]
- 33.Kapadia MR, Eng JW, Jiang Q, Stoyanovsky DA, Kibbe MR. Nitric oxide regulates the 26S proteasome in vascular smooth muscle cells. Nitric Oxide. 2009;20:279–288. doi: 10.1016/j.niox.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 34.Kwak YD, Ma T, Diao S, Zhang X, Chen Y, Hsu J, et al. NO signaling and S-nitrosylation regulate PTEN inhibition in neurodegeneration. Mol Neurodegener. 2010;5:49. doi: 10.1186/1750-1326-5-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burgoyne JR, Eaton P. Transnitrosylating nitric oxide species directly activate type I protein kinase A, providing a novel adenylate cyclase-independent cross-talk to beta-adrenergic-like signaling. J Biol Chem. 2009;284:29260–29268. doi: 10.1074/jbc.M109.046722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hara MR, Agrawal N, Kim SF, Cascio MB, Fujimuro M, Ozeki Y, et al. S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat Cell Biol. 2005;7:665–674. doi: 10.1038/ncb1268. [DOI] [PubMed] [Google Scholar]
- 37.Sen N, Hara MR, Ahmad AS, Cascio MB, Kamiya A, Ehmsen JT, et al. GOSPEL: a neuroprotective protein that binds to GAPDH upon S-nitrosylation. Neuron. 2009;63:81–91. doi: 10.1016/j.neuron.2009.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakamura T, Lipton SA. According to GOSPEL: filling in the GAP(DH) of NO-mediated neurotoxicity. Neuron. 2009;63:3–6. doi: 10.1016/j.neuron.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 39.Sen N, Hara MR, Kornberg MD, Cascio MB, Bae BI, Shahani N, et al. Nitric oxide-induced nuclear GAPDH activates p300/CBP and mediates apoptosis. Nat Cell Biol. 2008;10:866–873. doi: 10.1038/ncb1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun J, Steenbergen C, Murphy E. S-nitrosylation: NO-related redox signaling to protect against oxidative stress. Antioxid Redox Signal. 2006;8:1693–1705. doi: 10.1089/ars.2006.8.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hare JM, Stamler JS. NO/redox disequilibrium in the failing heart and cardiovascular system. J Clin Invest. 2005;115:509–517. doi: 10.1172/JCI200524459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen YY, Chu HM, Pan KT, Teng CH, Wang DL, Wang AH, et al. Cysteine S-nitrosylation protects protein-tyrosine phosphatase 1B against oxidation-induced permanent inactivation. J Biol Chem. 2008;283:35265–35272. doi: 10.1074/jbc.M805287200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones SP, Bolli R. The ubiquitous role of nitric oxide in cardioprotection. J Mol Cell Cardiol. 2006;40:16–23. doi: 10.1016/j.yjmcc.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 44.Murphy E, Steenbergen C. Mechanisms underlying acute protection from cardiac ischemia-reperfusion injury. Physiol Rev. 2008;88:581–609. doi: 10.1152/physrev.00024.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Costa AD, Pierre SV, Cohen MV, Downey JM, Garlid KD. cGMP signalling in pre- and post-conditioning: the role of mitochondria. Cardiovasc Res. 2008;77:344–352. doi: 10.1093/cvr/cvm050. [DOI] [PubMed] [Google Scholar]
- 46.Shiva S, Sack MN, Greer JJ, Duranski M, Ringwood LA, Burwell L, et al. Nitrite augments tolerance to ischemia/reperfusion injury via the modulation of mitochondrial electron transfer. J Exp Med. 2007;204:2089–2102. doi: 10.1084/jem.20070198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nadtochiy SM, Burwell LS, Brookes PS. Cardioprotection and mitochondrial S-nitrosation: effects of S-nitroso-2-mercaptopropionyl glycine (SNO-MPG) in cardiac ischemia-reperfusion injury. J Mol Cell Cardiol. 2007;42:812–825. doi: 10.1016/j.yjmcc.2007.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin J, Steenbergen C, Murphy E, Sun J. Estrogen receptor-beta activation results in S-nitrosylation of proteins involved in cardioprotection. Circulation. 2009;120:245–254. doi: 10.1161/CIRCULATIONAHA.109.868729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Burwell LS, Nadtochiy SM, Tompkins AJ, Young S, Brookes PS. Direct evidence for S-nitrosation of mitochondrial complex I. Biochem J. 2006;394:627–634. doi: 10.1042/BJ20051435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Campbell DL, Stamler JS, Strauss HC. Redox modulation of L-type calcium channels in ferret ventricular myocytes. Dual mechanism regulation by nitric oxide and S-nitrosothiols. J Gen Physiol. 1996;108:277–293. doi: 10.1085/jgp.108.4.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murry CE, Jennings RB, Reimer KA. Preconditioning with ischemia: a delay of lethal cell injury in ischemic myocardium. Circulation. 1986;74:1124–1136. doi: 10.1161/01.cir.74.5.1124. [DOI] [PubMed] [Google Scholar]
- 52.Hurd TR, Prime TA, Harbour ME, Lilley KS, Murphy MP. Detection of reactive oxygen species-sensitive thiol proteins by redox difference gel electrophoresis: implications for mitochondrial redox signaling. J Biol Chem. 2007;282:22040–22051. doi: 10.1074/jbc.M703591200. [DOI] [PubMed] [Google Scholar]
- 53.Prime TA, Blaikie FH, Evans C, Nadtochiy SM, James AM, Dahm CC, et al. A mitochondria-targeted S-nitrosothiol modulates respiration, nitrosates thiols, and protects against ischemia-reperfusion injury. Proc Natl Acad Sci U S A. 2009;106:10764–10769. doi: 10.1073/pnas.0903250106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gucek M, Murphy E. What can we learn about cardioprotection from the cardiac mitochondrial proteome? Cardiovasc Res. 88:211–218. doi: 10.1093/cvr/cvq277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tang XL, Stein AB, Shirk G, Bolli R. Hypercholesterolemia blunts NO donor-induced late preconditioning against myocardial infarction in conscious rabbits. Basic Res Cardiol. 2004;99:395–403. doi: 10.1007/s00395-004-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Settergren M, Bohm F, Malmstrom RE, Channon KM, Pernow J. L-arginine and tetrahydrobiopterin protects against ischemia/reperfusion-induced endothelial dysfunction in patients with type 2 diabetes mellitus and coronary artery disease. Atherosclerosis. 2009;204:73–78. doi: 10.1016/j.atherosclerosis.2008.08.034. [DOI] [PubMed] [Google Scholar]
- 57.Landmesser U, Dikalov S, Price SR, McCann L, Fukai T, Holland SM, et al. Oxidation of tetrahydrobiopterin leads to uncoupling of endothelial cell nitric oxide synthase in hypertension. J Clin Invest. 2003;111:1201–1209. doi: 10.1172/JCI14172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dumitrescu C, Biondi R, Xia Y, Cardounel AJ, Druhan LJ, Ambrosio G, et al. Myocardial ischemia results in tetrahydrobiopterin (BH4) oxidation with impaired endothelial function ameliorated by BH4. Proc Natl Acad Sci U S A. 2007;104:15081–15086. doi: 10.1073/pnas.0702986104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moens AL, Leyton-Mange JS, Niu X, Yang R, Cingolani O, Arkenbout EK, et al. Adverse ventricular remodeling and exacerbated NOS uncoupling from pressure-overload in mice lacking the beta3-adrenoreceptor. J Mol Cell Cardiol. 2009;47:576–585. doi: 10.1016/j.yjmcc.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen CA, Wang TY, Varadharaj S, Reyes LA, Hemann C, Talukder MA, et al. S-glutathionylation uncouples eNOS and regulates its cellular and vascular function. Nature. 2010;468:1115–1118. doi: 10.1038/nature09599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gonzalez DR, Treuer AV, Castellanos J, Dulce RA, Hare JM. Impaired S-nitrosylation of the ryanodine receptor caused by xanthine oxidase activity contributes to calcium leak in heart failure. J Biol Chem. 2010;285:28938–28945. doi: 10.1074/jbc.M110.154948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fang M, Jaffrey SR, Sawa A, Ye K, Luo X, Snyder SH. Dexras1: a G protein specifically coupled to neuronal nitric oxide synthase via CAPON. Neuron. 2000;28:183–193. doi: 10.1016/s0896-6273(00)00095-7. [DOI] [PubMed] [Google Scholar]
- 63.Feron O, Balligand JL. Caveolins and the regulation of endothelial nitric oxide synthase in the heart. Cardiovasc Res. 2006;69:788–797. doi: 10.1016/j.cardiores.2005.12.014. [DOI] [PubMed] [Google Scholar]
- 64.Barouch LA, Harrison RW, Skaf MW, Rosas GO, Cappola TP, Kobeissi ZA, et al. Nitric oxide regulates the heart by spatial confinement of nitric oxide synthase isoforms. Nature. 2002;416:337–339. doi: 10.1038/416337a. [DOI] [PubMed] [Google Scholar]
- 65.Lim G, Venetucci L, Eisner DA, Casadei B. Does nitric oxide modulate cardiac ryanodine receptor function? Implications for excitation-contraction coupling. Cardiovasc Res. 2008;77:256–264. doi: 10.1093/cvr/cvm012. [DOI] [PubMed] [Google Scholar]
- 66.Kanai AJ, Pearce LL, Clemens PR, Birder LA, VanBibber MM, Choi SY, et al. Identification of a neuronal nitric oxide synthase in isolated cardiac mitochondria using electrochemical detection. Proc Natl Acad Sci U S A. 2001;98:14126–14131. doi: 10.1073/pnas.241380298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Venkatakrishnan P, Nakayasu ES, Almeida IC, Miller RT. Absence of nitric-oxide synthase in sequentially purified rat liver mitochondria. J Biol Chem. 2009;284:19843–19855. doi: 10.1074/jbc.M109.003301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.French S, Giulivi C, Balaban RS. Nitric oxide synthase in porcine heart mitochondria: evidence for low physiological activity. Am J Physiol Heart Circ Physiol. 2001;280:H2863–H2867. doi: 10.1152/ajpheart.2001.280.6.H2863. [DOI] [PubMed] [Google Scholar]
- 69.Brookes PS. Mitochondrial nitric oxide synthase. Mitochondrion. 2004;3:187–204. doi: 10.1016/j.mito.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 70.Kornberg MD, Sen N, Hara MR, Juluri KR, Nguyen JV, Snowman AM, et al. GAPDH mediates nitrosylation of nuclear proteins. Nat Cell Biol. 2010;12:1094–1100. doi: 10.1038/ncb2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tristan C, Shahani N, Sedlak TW, Sawa A. The diverse functions of GAPDH: views from different subcellular compartments. Cell Signal. 23:317–323. doi: 10.1016/j.cellsig.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Marino SM, Gladyshev VN. Structural analysis of cysteine S-nitrosylation: a modified acid-based motif and the emerging role of trans-nitrosylation. J Mol Biol. 395:844–859. doi: 10.1016/j.jmb.2009.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xue Y, Liu Z, Gao X, Jin C, Wen L, Yao X, et al. GPS-SNO: computational prediction of protein S-nitrosylation sites with a modified GPS algorithm. PLoS One. 5:e11290. doi: 10.1371/journal.pone.0011290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Straub AC, Billaud M, Johnstone SR, Best AK, Yemen S, Dwyer ST, et al. Compartmentalized connexin 43 s-nitrosylation/denitrosylation regulates heterocellular communication in the vessel wall. Arterioscler Thromb Vasc Biol. 31:399–407. doi: 10.1161/ATVBAHA.110.215939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Secondo A, Molinaro P, Pannaccione A, Esposito A, Cantile M, Lippiello P, et al. Nitric oxide stimulates NCX1 and NCX2 but inhibits NCX3 isoform by three distinct molecular determinants. Mol Pharmacol. 2011;79:558–568. doi: 10.1124/mol.110.069658. [DOI] [PubMed] [Google Scholar]
- 76.Xu L, Eu JP, Meissner G, Stamler JS. Activation of the cardiac calcium release channel (ryanodine receptor) by poly-S-nitrosylation. Science. 1998;279:234–237. doi: 10.1126/science.279.5348.234. [DOI] [PubMed] [Google Scholar]
- 77.Sun J, Yamaguchi N, Xu L, Eu JP, Stamler JS, Meissner G. Regulation of the cardiac muscle ryanodine receptor by O(2) tension and S-nitrosoglutathione. Biochemistry. 2008;47:13985–13990. doi: 10.1021/bi8012627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Moon KH, Kim BJ, Song BJ. Inhibition of mitochondrial aldehyde dehydrogenase by nitric oxide-mediated S-nitrosylation. FEBS Lett. 2005;579:6115–6120. doi: 10.1016/j.febslet.2005.09.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ozawa K, Whalen EJ, Nelson CD, Mu Y, Hess DT, Lefkowitz RJ, et al. S-nitrosylation of beta-arrestin regulates beta-adrenergic receptor trafficking. Mol Cell. 2008;31:395–405. doi: 10.1016/j.molcel.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Maejima Y, Adachi S, Morikawa K, Ito H, Isobe M. Nitric oxide inhibits myocardial apoptosis by preventing caspase-3 activity via S-nitrosylation. J Mol Cell Cardiol. 2005;38:163–174. doi: 10.1016/j.yjmcc.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 81.Cleeter MW, Cooper JM, Darley-Usmar VM, Moncada S, Schapira AH. Reversible inhibition of cytochrome c oxidase, the terminal enzyme of the mitochondrial respiratory chain, by nitric oxide. Implications for neurodegenerative diseases. FEBS Lett. 1994;345:50–54. doi: 10.1016/0014-5793(94)00424-2. [DOI] [PubMed] [Google Scholar]
- 82.Atar S, Ye Y, Lin Y, Freeberg SY, Nishi SP, Rosanio S, et al. Atorvastatin-induced cardioprotection is mediated by increasing inducible nitric oxide synthase and consequent S-nitrosylation of cyclooxygenase-2. Am J Physiol Heart Circ Physiol. 2006;290:H1960–H1968. doi: 10.1152/ajpheart.01137.2005. [DOI] [PubMed] [Google Scholar]
- 83.Arstall MA, Bailey C, Gross WL, Bak M, Balligand JL, Kelly RA. Reversible S-nitrosation of creatine kinase by nitric oxide in adult rat ventricular myocytes. J Mol Cell Cardiol. 1998;30:979–988. doi: 10.1006/jmcc.1998.0662. [DOI] [PubMed] [Google Scholar]
- 84.Ito G, Ariga H, Nakagawa Y, Iwatsubo T. Roles of distinct cysteine residues in S-nitrosylation and dimerization of DJ-1. Biochem Biophys Res Commun. 2006;339:667–672. doi: 10.1016/j.bbrc.2005.11.058. [DOI] [PubMed] [Google Scholar]
- 85.Cho DH, Nakamura T, Fang J, Cieplak P, Godzik A, Gu Z, et al. S-nitrosylation of Drp1 mediates beta-amyloid-related mitochondrial fission and neuronal injury. Science. 2009;324:102–105. doi: 10.1126/science.1171091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ravi K, Brennan LA, Levic S, Ross PA, Black SM. S-nitrosylation of endothelial nitric oxide synthase is associated with monomerization and decreased enzyme activity. Proc Natl Acad Sci U S A. 2004;101:2619–2624. doi: 10.1073/pnas.0300464101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Erwin PA, Mitchell DA, Sartoretto J, Marletta MA, Michel T. Subcellular targeting and differential S-nitrosylation of endothelial nitric-oxide synthase. J Biol Chem. 2006;281:151–157. doi: 10.1074/jbc.M510421200. [DOI] [PubMed] [Google Scholar]
- 88.Garban HJ, Marquez-Garban DC, Pietras RJ, Ignarro LJ. Rapid nitric oxide-mediated S-nitrosylation of estrogen receptor: regulation of estrogen-dependent gene transcription. Proc Natl Acad Sci U S A. 2005;102:2632–2636. doi: 10.1073/pnas.0409854102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Martinez-Ruiz A, Villanueva L, Gonzalez de Orduna C, Lopez-Ferrer D, Higueras MA, Tarin C, et al. S-nitrosylation of Hsp90 promotes the inhibition of its ATPase and endothelial nitric oxide synthase regulatory activities. Proc Natl Acad Sci U S A. 2005;102:8525–8530. doi: 10.1073/pnas.0407294102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Reynaert NL, Ckless K, Korn SH, Vos N, Guala AS, Wouters EF, et al. Nitric oxide represses inhibitory kappaB kinase through S-nitrosylation. Proc Natl Acad Sci U S A. 2004;101:8945–8950. doi: 10.1073/pnas.0400588101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yao D, Gu Z, Nakamura T, Shi ZQ, Ma Y, Gaston B, et al. Nitrosative stress linked to sporadic Parkinson's disease: S-nitrosylation of parkin regulates its E3 ubiquitin ligase activity. Proc Natl Acad Sci U S A. 2004;101:10810–10814. doi: 10.1073/pnas.0404161101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Barrett DM, Black SM, Todor H, Schmidt-Ullrich RK, Dawson KS, Mikkelsen RB. Inhibition of protein-tyrosine phosphatases by mild oxidative stresses is dependent on S-nitrosylation. J Biol Chem. 2005;280:14453–14461. doi: 10.1074/jbc.M411523200. [DOI] [PubMed] [Google Scholar]
- 93.Haendeler J, Hoffmann J, Tischler V, Berk BC, Zeiher AM, Dimmeler S. Redox regulatory and antiapoptotic functions of thioredoxin depend on S-nitrosylation at cysteine 69. Nat Cell Biol. 2002;4:743–749. doi: 10.1038/ncb851. [DOI] [PubMed] [Google Scholar]
- 94.Lai TS, Hausladen A, Slaughter TF, Eu JP, Stamler JS, Greenberg CS. Calcium regulates S-nitrosylation, denitrosylation, and activity of tissue transglutaminase. Biochemistry. 2001;40:4904–4910. doi: 10.1021/bi002321t. [DOI] [PubMed] [Google Scholar]