Abstract
Na+,K+ ATPase pumps Na+ out of and K+ into the cytosol, maintaining a resting potential that is essential for the function of excitable tissues like cardiac muscle. In addition to its well-characterized physiological role in the heart, Na+,K+ ATPase also regulates the morphogenesis of the embryonic zebrafish heart via an as yet unknown mechanism. Here, we describe a novel non-cell autonomous function of Na+,K+ ATPase/Atp1a1 in the elongation of the zebrafish heart tube. Embryos lacking Atp1a1 function exhibit abnormal migration behavior of cardiac precursors, defects in the elongation of the heart tube, and a severe reduction in ECM/Fibronectin deposition around the myocardium, despite the presence of normal cell polarity and junctions in the myocardial epithelium prior to the timeframe of heart tube elongation. Interestingly, we found that Atp1a1 is not present in the myocardium at the time when cardiac morphogenesis defects first become apparent, but is expressed in an extraembryonic tissue, the yolk syncytial layer (YSL), at earlier stages. Knockdown of Atp1a1 activity specifically in the YSL using morpholino oligonucleotides produced heart tube elongation defects like those found in atp1a1 mutants, indicating that Atp1a1 function in the YSL is necessary for heart tube elongation. Furthermore, atp1a1 expression in the YSL was regulated by the homeobox transcription factor mxtx1. Together, these data reveal a new non-cell autonomous role for Atp1a1 in cardiac morphogenesis and establish Na+,K+ ATPase as a major player in the genetic pathway by which the YSL regulates embryonic ECM deposition.
Keywords: zebrafish, heart development, Na+, K+ ATPase, yolk syncytial layer, fibronectin, extracellular matrix
Introduction
The vertebrate heart forms from two populations of progenitor cells in the anterior lateral plate mesoderm. These cardiac progenitors first migrate medially to fuse at the midline, and then rearrange to form a simple, linear heart tube. In zebrafish, the migration of cardiac progenitors to the midline results in the formation of the cardiac cone. The cardiac cone is essentially a flattened version of the primitive heart tube with the ventricular and atrial cells arranged along the dorsoventral axis (Glickman and Yelon, 2002). In roughly a five hour window, the cardiac cone elongates anteriorly and leftward, generating a primitive heart tube with ventricular and atrial cells arranged along the anteroposterior axis.
Recent studies in zebrafish have greatly expanded our understanding of the molecular mechanisms governing primitive heart tube elongation. While migrating to the midline, cardiomyocyte progenitors epithelialize, establishing clear apicobasal polarity upon formation of the cardiac cone (Trinh and Stainier, 2004). The cardiomyocytes of the cardiac cone also display basolateral ZO1-positive cell-cell junctions (Trinh and Stainier, 2004). Disruption of cardiomyocyte apicobasal polarity by mutation of atypical protein kinase C (aPKC) prevents heart tube elongation, resulting in an extremely abnormal heart in which the ventricular cells are engulfed by the cells of the atrium (Horne-Badovinac et al., 2001; Peterson et al., 2001). Loss of function of another important regulator of cell polarity, MAGUK p55 subfamily member 5 (Mpp5), also compromises the polarity and junctions of cardiomyocytes and disrupts heart tube elongation in zebrafish (Cibrian-Uhalte et al., 2007; Rohr et al., 2006).
Cardiac cell polarity and junctions during heart tube morphogenesis are also regulated by the presence of extracellular matrix (ECM) proteins. Migrating cardiomyocytes travel on a basement membrane containing the ECM proteins Fibronectin and Laminin (Sakaguchi et al., 2006; Trinh and Stainier, 2004). Absence of Fibronectin protein in the zebrafish natter mutant prevents the complete medial migration of cardiomyocytes, indicating the important role of ECM in cardiac morphogenesis (Trinh and Stainier, 2004).
Recent studies have revealed that expression of ECM proteins in the embryo is regulated by the yolk syncytial layer (YSL), an extra-embryonic tissue consisting of a syncytium of nuclei near the surface of the yolk (Kimmel and Law, 1985). YSL-specific knockdown of mxtx1 in zebrafish demonstrates that this mix-type homeobox transcription factor is required for Fibronectin protein expression, ECM assembly, and myocardial migration (Sakaguchi et al., 2006). Syndecan 2, a transmembrane heparin sulfate proteoglycan, also functions in the YSL to regulate ECM deposition and cardiac development (Arrington and Yost, 2009). ECM deposition and heart morphogenesis are similarly defective when the activity of the sphingosine-1-phosphate transporter Spinster is eliminated by morpholino knockdown (Osborne et al., 2008). Spinster is primarily expressed in the YSL during early development, and knockdown of spinster specifically in the YSL disrupts the migration of the cardiomyocyte precursors to the midline (Kawahara et al., 2009; Osborne et al., 2008). Interestingly, knockdown of Retinol binding protein 4 (Rbp4) in the YSL causes a reduction in the posterior expression of fibronectin 1 without affecting its anterior expression level or myocardial migration, suggesting that signals from the YSL can regulate anterior and posterior ECM deposition independently (Li et al., 2007).
Another critical regulator of heart tube morphogenesis is Na+,K+ ATPase. Na+,K+ ATPase is a pump that generates the Na+ and K+ gradients necessary for the physiology of living cells and has well characterized roles in excitatory cells of the heart, skeletal muscle, and nervous system (Therien and Blostein, 2000). By maintaining the Na+ gradient, Na+,K+ ATPase also indirectly regulates intracellular Ca2+ levels (McDonough et al., 2002; Therien and Blostein, 2000; Tian and Xie, 2008). Mutation in atp1a1, an isoform of the Na+,K+ ATPase alpha subunit, results in a severe delay in heart tube elongation in zebrafish (Shu et al., 2003). Atp1a1 mutants exhibit a small heart positioned at the midline. The small size of the atp1a1 mutant heart is not a result of decreased cardiomyocyte number, but instead a failure of these cells to spread out as they normally would do during heart tube elongation. Later, atp1a1 mutants do generate a shortened heart tube, but display functional defects including reduced heart rate and contractility (Shu et al., 2003). Atp1a1 also regulates the maintenance of myocardial tight junctions via a genetic interaction with the cell polarity protein Mpp5 (Cibrian-Uhalte et al., 2007). While the physiological role of the sodium pump in the heart has been explored extensively, the mechanisms underlying the requirement of atp1a1 in cardiac morphogenesis have not previously been elucidated.
Here, we report a novel non-cell autonomous role for Atp1a1 in cardiac morphogenesis. Our data demonstrate that Atp1a1 activity in the YSL regulates the elongation of the zebrafish heart tube, and that loss of Atp1a1 function results in a profound reduction in ECM deposition around the zebrafish heart. Furthermore, atp1a1 expression in the YSL is regulated by the homeobox transcription factor mxtx1. These findings highlight a new function for Na+,K+ ATPase in cardiac development that is separate from its cell autonomous role in cardiac function.
Materials and Methods
Zebrafish lines and husbandry
Zebrafish colonies were cared for and bred under standard conditions (Westerfield, 2000). Developmental stages of zebrafish embryos were determined using standard morphological features of fish raised at 28.5°C (Westerfield, 2000). The atp1a1m883 allele (Ellertsdottir et al., 2006) was crossed into the Tg(myl7:EGFP) transgenic background (Huang et al., 2003) to fluorescently label cardiomyocytes. The Tg(myl7:mCherry)chb1 transgenic line (from J. Mably) was used when injecting fluorescein-labeled morpholino oligonucleotides.
Time-lapse confocal microscopy
Tg(myl7:EGFP) transgenic (Huang et al., 2003) wild type and atp1a1m883 mutant embryos were embedded in 1% low melt agarose at 18.5 hpf and confocal z-stack images were acquired every 5 minutes using a Zeiss LSM 510 confocal microscope equipped with a heated stage. Zeiss LSM 510 imaging software was used to acquire time-lapse movies and generate zstack projections. Manual tracking of cardiomyocyte migration was performed using ImageJ software (http://rsbweb.nih.gov/ij/).
Transfection and Western blotting
HEK293T cells were transfected with HisMyc-Atp1a1 (Cibrian-Uhalte et al., 2007) and Flag-Atp1a2a expression vectors using Lipofectamine 2000 reagent (Invitrogen). After 24 hours, cells were lysed in RIPA lysis buffer and total protein concentrations were obtained by DC Protein assay (Bio-Rad). 20 ug of protein lysate per lane was separated on a 10% polyacrylamide gel and transferred to a nitrocellulose membrane for Western analysis.
Atp1a1-specific antibody was produced by BioSource and raised against the following peptide: Ac-GKKSKSKGKKEKDKDMC-amide. Rabbit polyclonal anti-Atp1a1 was used at a concentration of 1:20,000, and was detected with an HRP-conjugated secondary antibody (1:50,000, 81-6120, Zymed). Mouse anti-Myc 9E10 (1:10,000, 05-419, Upstate), mouse anti-FLAG M2 (1:10,000, F1804, Sigma), and mouse anti-β-Actin were used to detect Myc-tagged, Flag-tagged, and β-Actin proteins, respectively. HRP-conjugated rabbit anti-mouse secondary antibody (1:20,000, 61-6020, Invitrogen) was used to detect Myc, Flag, and β-Actin primary antibodies.
Vibratome Sectioning and Antibody Staining
Embryos were fixed in 2% PFA/PBS overnight at 4 °C, embedded in 4% low melt agarose, and sectioned at 200 µm using a Vibratome 1000 Plus. Sections were stained for β-catenin, aPKC, ZO-1, Fibronectin, and Actin as previously described (Trinh and Stainier, 2004). Anti-Atp1a1 (BioSource) was used at a concentration of 1:200. Fluorescent secondary antibodies were used at a concentration of 1:200 and include anti-mouse IgG1-R (sc-2084, Santa Cruz Biotechnology) and anti-rabbit IgG-Cy5 (81-6116, Invitrogen). Confocal images were acquired with a Zeiss LSM 510 confocal microscope.
Histology
Fixed embryos were dehydrated, embedded in plastic blocks (JB-4, Polysciences), sectioned at 8-12.5 µm on a Leica RM 2065 microtome, and stained with 0.1% methylene blue as previously described (Chen and Fishman, 1996) or with 0.1% neutral red in 7.7 mM sodium acetate solution at pH 4.8. Sections were imaged on a Zeiss Axioplan 2 microscope equipped with a Zeiss Axiocam camera.
Morpholino injections to yolk syncytial layer
Fluorescein-tagged atp1a1 morpholinos (8 ng atp1a1MO1, 5’-CTGCCAGCTCATATTGTTCTCGGCC-3’; 2 ng atp1a1MO2, 5’-GCCTTCTCCTCGTCCCATTTTGCTG-3’) (Blasiole et al., 2006; Shu et al., 2003) were coinjected with p53 morpholino (8 ng or 2 ng, 5’- GCGCCATTGCTTTGCAAGAATTG -3’) into the yolks of 1000-cell stage embryos. 8 ng of standard control morpholino oligo (5’-CCTCTTACCTCAGTTACAATTTATA-3’) or slc8a1a morpholino (5’-GATGAAGTCCCAGATTGGCCCATGT-3’) (Langenbacher et al., 2005) was coinjected with rhodamine-dextran into the yolks of 1000-cell stage embryos as negative controls. Injection of mxtx1 morpholino was performed as described previously (Sakaguchi et al., 2006). During gastrulation, embryos with successful uptake of morpholino to the yolk syncytial layer were selected based on fluorescence and grown until the appropriate stages for in situ hybridization analysis.
Whole-mount in situ hybridization and antibody staining
Embryos for in situ hybridization or antibody staining were raised in embryo medium supplemented with 0.2 mM 1-phenyl-2-thiourea to maintain optical transparency (Westerfield, 2000). Whole-mount in situ hybridization was performed as described previously (Chen and Fishman, 1996). The antisense RNA probes used in this study include cmlc2 (myl7), atp1a1, and fn1 (Trinh and Stainier, 2004). Whole-mount antibody staining was performed using anti-Atp1a1 antibody (1:200, BioSource) followed by incubation with anti-rabbit HRP-conjugated secondary antibody (Zymed) and detection with 3,3'-Diaminobenzidine (Sigma).
Results
Atp1a1 regulates cardiomyocyte migration during heart tube elongation
In wild type zebrafish embryos, the cardiac cone elongates into the primitive heart tube within a 5 hour window from 19 – 24 hours post fertilization (hpf). Heart tube elongation is severely delayed in atp1a1 mutants, resulting in a clump of cells at the midline at 24 hpf. Despite this severe defect in morphogenesis, atp1a1 mutants have a normal number of cardiomyocytes at 24 hpf (Shu et al., 2003).
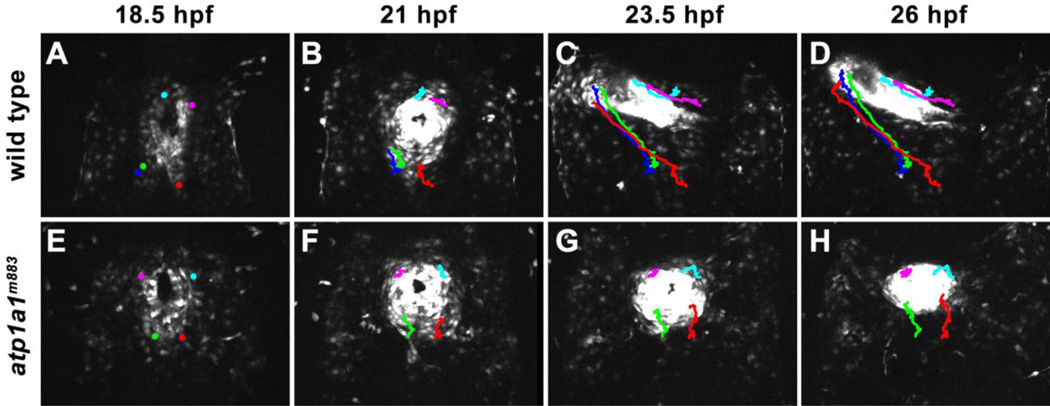
To better characterize the cellular mechanism underlying the atp1a1 heart tube elongation defect, we used time-lapse confocal microscopy to track cardiomyocyte migration beginning at 18.5 hpf (prior to heart tube elongation) in wild type and atp1a1m883 mutant embryos. Wild type cardiomyocytes exhibit dramatic migration between 18.5 hpf and 26 hpf (Figure 1A–D). Notably, between 18.5 and 23.5 hpf, cardiomyocytes in the posterior portion of the wild type heart field rapidly migrate anteriorly in order to assume a similar anteroposterior position to cardiomyocytes that originated in the anterior portion of the heart field (Figure 1A–C, Movie S1). Conversely, atp1a1m883 cardiomyocytes exhibit limited migration. Cardiomyocytes in the posterior portion of the atp1a1m883 heart field migrate anteriorly, but fail to reach the same anteroposterior position as cells that originated in the anterior region of the heart field. Furthermore, cardiomyocytes of the anterior heart field exhibit no net anterior migration (Figure 1E–H). Instead of elongating into a tube, this limited anterior migration of cardiomyocytes causes the atp1a1m883 heart to become increasingly clump-like (Figure 1F–H, Movie S2), suggesting that a defect in migration behavior is the underlying cellular cause of the atp1a1 mutant phenotype.
Figure 1. Sodium pump function is important for cardiomyocyte migration and heart tube elongation.
(A–D) Time-lapse images showing a dorsal view (anterior to top) of heart morphogenesis in a wild type Tg(myl7:EGFP) embryo. After arriving at the midline, wild type cardiomyocytes rapidly migrate anteriorly and leftward between 18.5 and 26 hpf, generating the heart tube. (E–H) Time-lapse images showing a dorsal view (anterior to top) of heart morphogenesis in a atp1a1m883 mutant Tg(myl7:EGFP) embryo. Cardiomyocytes migrate to the midline normally in the majority of atp1a1m883 mutants (E), but only exhibit limited migration from 18.5 to 26 hpf. At 26 hpf, atp1a1m883 mutants have a clump of cardiomyocytes at the midline (H).
Initial establishment of myocardial cell polarity and junctions does not require Atp1a1 function
The migration of cardiomyocytes, in particular their medial migration prior to cardiac cone formation, requires the establishment of cell-cell junctions and apicobasal polarity (Cibrian-Uhalte et al., 2007; Rohr et al., 2006; Trinh and Stainier, 2004). To determine if the migration defect in atp1a1 mutants was caused by a defect in cell polarity or junctions, we examined the distribution of proteins known to localize to junctions or specific apicobasal membrane domains.
Myocardial cells in wild type embryos are well polarized at the 20 somite (20S) stage with basal/basolateral β-catenin and apical/apicolateral aPKC protein distributions (Figure 2A). Furthermore, basolateral Zona occludens-1 (ZO-1) positive junctions are present at the boundaries between individual wild type myocardial cells (Figure 2B). In atp1a1 mutants, β-catenin and aPKC proteins are localized to the appropriate membrane domains, and ZO-1 positive junctions are present between cells of the myocardium (Figure 2C,D). These data suggest that atp1a1 function is not required for the establishment of cell-cell junctions or cell polarity in the heart, and that the cardiomyocyte migration defects in atp1a1 mutants are not a result of disrupted cell polarity or junctions.
Figure 2. Cell polarity and tight junctions are not disrupted by loss of Atp1a1 function.
(A–D) Transverse vibratome sections of 20S stage wild type and atp1a1m883 Tg(myl7:EGFP) embryos. (A,C) The polarized localizations of β-catenin (red, basolateral) and aPKC (green, apicolateral) are indistinguishable between wild type (A) and atp1a1m883 mutant (C) embryos at the 20S stage. (B,D) Tight junctions are present basolaterally in both wild type (B) and atp1a1m883 mutant (D) embryos based on the localization of the tight junction protein ZO-1 (green) at the 20S stage.
Atp1a1 is required for ECM deposition around the myocardium of the cardiac cone
Another factor that is critical for cardiomyocyte migration is ECM deposition. Wild type embryos display thickened ECM between the myocardium and neighboring cells at the 20S stage (Figure 3A). This thickened matrix stains pink with methylene blue dye, indicating the presence of negatively charged moieties such as the sulfated proteoglycans typically found in ECM (Walsh and Stainier, 2001). Remarkably, in atp1a1 mutants, this thickened ECM is strongly reduced and the myocardium is almost in direct contact with the neighboring cells (Figure 3B).
Figure 3. Sodium pump activity is required for extracellular matrix deposition around the cardiac cone.
(A,B) Transverse microtome sections of 20S stage wild type and atp1a1m883 embryos stained with methylene blue. In wild type embryos, sulfated proteoglycans are stained pink by methylene blue, allowing visualization of the abundant extracellular matrix present around the cardiomyocytes of the cardiac cone (*) at the 20S stage (A). In atp1a1m883 mutants, the cardiac cone forms normally (*), but thickened extracellular matrix is completely absent (B). (C,D) Transverse vibratome sections of 20S stage wild type and atp1a1m883 Tg(myl7:EGFP) embryos. Wild type embryos (C) have Fibronectin protein (green) surrounding the cardiomyocytes of the cardiac cone (blue), while atp1a1m883 mutants (D) have extremely reduced Fibronectin protein deposition. (E,F) Dorsal views of fn1 expression in 20S stage wild type and atp1a1m883 embryos. Fn1 is expressed in the cardiac region of wild type embryos (E), but is dramatically reduced in atp1a1m883 mutants (F). (G,H) Lateral views of fn1 expression in 20S stage wild type and atp1a1m883 embryos. Both wild type (G) and atp1a1m883 mutant (H) embryos exhibit fn1 expression in posterior structures including the yolk extension and tail.
Fibronectin protein is one important component of the ECM surrounding the myocardium (Trinh and Stainier, 2004). Wild type embryos exhibit strong Fibronectin protein localization (100%, n=13) to the basal surface of cardiomyocytes at the 20S stage (Figure 3C). On the other hand, most atp1a1 mutants (79%, n=14) exhibited a reduction in Fibronectin deposition (Figure 3D). A reduction in Fibronectin deposition was detected in the hypomorphic atp1a1 allele atp1a1la1 as well (52%, n=21) (Figure S1). We also examined the expression of fn1, the gene encoding the predominant isoform of Fibronectin expressed in the cardiac region. At the 20S stage, fn1 expression was reduced in the atp1a1 mutant cardiac mesoderm (Figure 3E,F), indicating that atp1a1 regulates Fibronectin production at the transcriptional level. Interestingly, the posterior expression domain of fn1 was maintained in atp1a1 mutants (Figure 3G,H), indicating that loss of Atp1a1 activity does not cause a general shutdown in embryonic production of Fn1. These data show that atp1a1 is critical for deposition of significant amounts of ECM proteins, specifically Fibronectin, around the developing myocardium.
Atp1a1 protein is not present in the myocardial membrane prior to formation of the heart tube
To better analyze the function of atp1a1 during cardiac development, we generated an antibody that recognizes a unique sequence in the N-terminal intracellular domain of the Atp1a1 protein. By Western blotting, we found that this anti-Atp1a1 antibody specifically recognizes zebrafish Atp1a1 protein and does not cross react with another highly homologous zebrafish Na+,K+ ATPase α subunit isoform, Atp1a2a (Figure 4A). At 2 days post fertilization (dpf), whole mount staining using the anti-Atp1a1 antibody detected abundant protein in the heart (Figure 4B), brain, and pronephros (Figure S2). This expression domain is identical to the expression pattern of atp1a1 detected by in situ hybridization (Canfield et al., 2002; Serluca et al., 2001).
Figure 4. Endogenous Atp1a1 protein is not detected in the heart at stages when morphogenesis defects are first observed in atp1a1 mutants.
(A) Western blot visualizing Myc-tagged, Flag-tagged, Atp1a1, and β-Actin proteins in HEK293T cell lysates. Mock transfected cell lysates have no reactivity with Myc, Flag, or Atp1a1 antibodies. Lysates from cells transfected with a Myc-tagged zebrafish Atp1a1 construct contain a protein that reacts with both Myc and Atpa1a antibodies. Lysates from cells transfected with a Flag-tagged zebrafish Atp1a2a construct contain a protein that reacts only with the Flag antibody. The presence of β-Actin protein in mock, Myc-Atp1a1, and Flag-Atp1a2a lysates demonstrates equal loading of total protein. (B) Ventral view of Atp1a1 antibody staining on 48 hpf wild type embryo. Atp1a1 protein is strongly detected in the heart. (C,D) Transverse vibratome sections of 20S stage (C) and 24 hpf (D) wild type embryos. At the 20S stage (C), Actin (red) is primarily basolateral around the cardiomyocytes of the cardiac cone (blue), whereas Atp1a1 protein (green) is not detectable in the cardiomyocyte membranes. By 24 hpf (D), polymerized Actin (red) is present on the ventral and lumenal surfaces of the primitive heart tube. Atp1a1 protein (green) is strongly localized to the membrane of cardiomyocytes in a punctate manner by this stage. (E,F) Microtome section of atp1a1 expression in 90% epiboly wild type embryo. (E) At 90% epiboly, atp1a1 is expressed ubiquitously. (F) Magnified portion of image in (E) shows that yolk syncytial layer nuclei strongly express atp1a1 (arrows).
We used the anti-Atp1a1 antibody to analyze the distribution of Atp1a1 protein at the 20S stage, immediately before the onset of the atp1a1 mutant heart tube elongation phenotype. In contrast to what we observed at 2 dpf, no Atp1a1 protein was detectable in cardiomyocytes at the 20S stage (Figure 4C). However, by 24 hpf, when the heart tube begins to contract, Atp1a1 was easily detectable in the membrane of cardiomyocytes (Figure 4D). These data suggest that heart tube elongation is unlikely to depend on a function of Atp1a1 in cardiomyocytes.
Yolk syncytial layer activity of Atp1a1 regulates heart tube elongation in zebrafish
During the gastrula period, at 90% epiboly, we found that atp1a1 is strongly expressed throughout the embryo (Figure 4E). Interestingly, this strong level of expression is also present in an extra-embryonic layer of the embryo, the yolk syncytial layer (YSL). We found that nuclei of the YSL were surrounded by atp1a1 transcripts (Figure 4F).
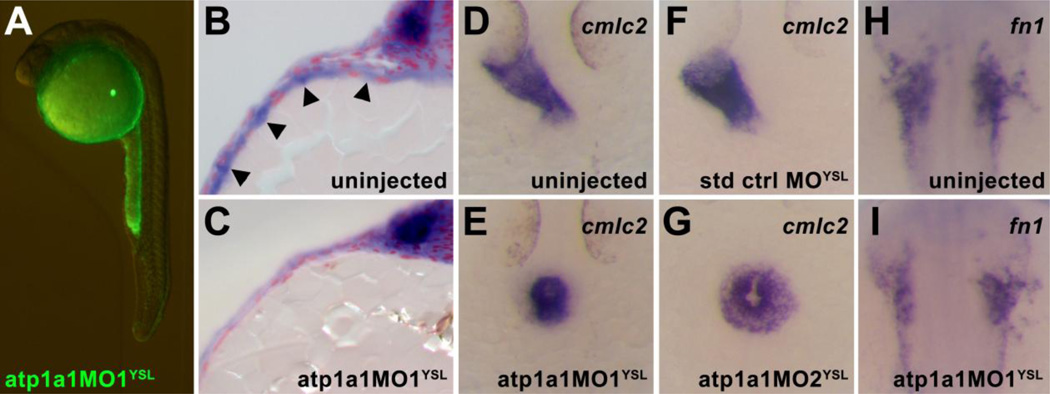
Because the YSL plays an important role in embryonic ECM deposition (Arrington and Yost, 2009; Kawahara et al., 2009; Li et al., 2007; Sakaguchi et al., 2006), we decided to test whether Atp1a1 function in the YSL regulated heart tube elongation in zebrafish. In zebrafish, knockdown of gene expression specifically in the YSL is possible by injecting morpholino oligonucleotides into the yolk at the 1000-cell stage, when the marginal blastomeres fuse with the yolk and generate the YSL (Sakaguchi et al., 2006; Sakaguchi et al., 2001). We chose to analyze the YSL requirement of atp1a1 using two independent and previously characterized translation blocking morpholinos, atp1a1MO1 (Shu et al., 2003) and atp1a1MO2 (Blasiole et al., 2006), which are known to phenocopy the atp1a1 mutant phenotype (atp1a1MO1 and atp1a1MO2) and reduce the endogenous levels of Atp1a1 protein (atp1a1MO2). When a fluorescently tagged atp1a1 morpholino was injected into the yolk at the 1000-cell stage, fluorescent signal was present in the yolk and was excluded from the body of the embryo at 24 hpf (Figure 5A). Furthermore, embryos in which atp1a1 morpholino was targeted to the YSL exhibited a strong reduction in YSL expression of atp1a1 without a reduction in embryonic expression (Figure 5B,C), indicating that this morpholino modulates YSL gene expression without directly affecting the expression of genes in the embryo.
Figure 5. Atp1a1 is required in the yolk syncytial layer for heart tube elongation.
(A) Embryos injected at the 512 to 1000-cell stage with fluorescein-tagged atp1a1MO1 exhibit a yolk-specific fluorescence pattern at 24 hpf, indicating that the morpholino’s effects are restricted to the yolk syncytial layer. (B,C) Microtome sections of atp1a1 expression in 12S stage wild type and atp1a1MO1YSL embryos. Strong expression of atp1a1 is visible in the YSL of wild type embryos (arrowheads, B), whereas atp1a1MO1YSL embryos display a specific loss of YSL expression of atp1a1 while maintaining normal embryonic expression (C). (D–G) Dorsal views of cmlc2 expression in 24 hpf uninjected and YSL-morphant embryos. Uninjected and standard control oligo YSL-injected (std ctrl MOYSL) embryos have an elongated heart tube at 24 hpf visualized by cmlc2 expression (D,F) whereas most atp1a1MO1YSL and atp1a1MO2YSL embryos have a clump of cardiomyocytes at the midline at 24 hpf (E,G). (H,I) Dorsal views of fn1 expression in the cardiac region of 15S stage wild type and YSL-morphant embryos. Expression of fn1 is reduced in atp1a1MO1YSL embryos (I) compared to wild type embryos (H).
The majority of uninjected control embryos generate an elongated heart tube by 24 hpf (81%, n=132; Figure 5D, Table S1). Most embryos in which 8 ng of atp1a1MO1 was injected into the YSL (atp1a1MOYSL) were not able to generate an elongated heart tube (12% with elongated tube, n=42), but instead exhibited a clump of cardiomyocytes at the midline reminiscent of the atp1a1 mutant phenotype (60%, n=42; Figure 5E, Table S1). Similarly, most embryos in which 2 ng of atp1a1MO2 was injected into the YSL (atp1a1MO2YSL) had defective heart tube elongation (89%, n=79; Figure 5G, Table S1). As a negative control, we injected 8 ng of a standard negative control morpholino (std ctrl MO) targeting the human β-globin pre-mRNA (Eisen and Smith, 2008) or 8ng of a morpholino targeting slc8a1a, a zebrafish gene not expressed at detectable levels in the YSL (Langenbacher et al., 2005). Injection of these negative control morpholinos at the 1000-cell stage to target them to the YSL did not alter the process of heart tube formation, and the majority of injected embryos were able to generate normal elongated heart tubes by 24 hpf (81% of std ctrl MO injected, n=99; 78% of slc8a1a MO injected, n=79; Figure 5F, Table S1). These data indicate that the heart tube elongation defect induced by injection of atp1a1 morpholinos into the YSL is a specific result of Atp1a1 deficiency and is not likely to be a general defect caused by injection of morpholinos into the yolk.
We next tested if loss of Atp1a1 function specifically in the YSL could replicate the ECM defects that we identified in atp1a1m883 mutants. We found that fn1 expression was reduced in the cardiac region of atp1a1MO1YSL embryos (Figure 5I) compared to wild type uninjected controls (Figure 5H), suggesting that the deposition of Fibronectin around the developing heart depends on expression of atp1a1 in the extra-embryonic YSL. Interestingly, we found that Atp1a1 function in the YSL was not required for contraction of the heart. Embryos in which atp1a1 was knocked down in the YSL, like atp1a1m883 mutants, have a shortened heart tube at 32 hpf. However, unlike atp1a1m883 mutants, the hearts of atp1a1MO1YSL embryos were able to contract (Movies S3–S6). These data indicate that atp1a1 plays a dual role during cardiac development, functioning non-cell autonomously to regulate heart tube elongation and cell autonomously to control cardiac contraction.
Expression of atp1a1 in the YSL is regulated by mxtx1
One critical YSL-specific transcription factor that regulates the embryonic deposition of ECM is mxtx1 (Sakaguchi et al., 2006). Downstream targets of mxtx1 that directly regulate the expression of fn1 and other ECM genes have not yet been identified. Phenotypes characteristic of atp1a1 mutants and morphants, including a flattened hindbrain, cardiac defects, and a loss of ECM deposition around the developing heart are also present in mxtx1 YSL-morphant (mxtx1MOYSL) embryos (Sakaguchi et al., 2006). Because of these phenotypic similarities, we questioned whether atp1a1 might be one of the transcriptional targets of mxtx1.
To test this, we examined the expression of atp1a1 in wild type and mxtx1MOYSL embryos. In wild type embryos, atp1a1 expression can be detected adjacent to YSL nuclei during both epiboly and somitogenesis (Figure 6A,C). YSL nuclei were visible in mxtx1MOYSL embryos, but expression of atp1a1 in the YSL was dramatically reduced throughout development (Figure 6B,D). This indicates that extra-embryonic expression of atp1a1 depends on Mxtx1 activity, and that atp1a1 may be one of the downstream effectors of Mxtx1 during embryonic ECM deposition.
Figure 6. Atp1a1 expression by the YSL is regulated by mxtx1.
(A–D) Microtome sections of atp1a1 expression in 95% epiboly (A,B) and 19S stage (C,D) wild type and mxtx1MOYSL embryos. Nuclei are labeled red by neutral red staining. Expression of atp1a1 (purple staining, arrowheads) is present in the YSL of wild type embryos at 95% epiboly (A) and the 19S stage (C). The expression of atp1a1 in the YSL is strongly reduced in mxtx1MOYSL embryos at the same stages (B,D). (G) Summary of Atp1a1 function. Atp1a1 expression in the YSL is regulated by mxtx1, and is required for the deposition of Fn/ECM around the developing heart. This ECM deposition is critical for normal heart tube elongation. Atp1a1 also has a cell autonomous role within cardiomyocytes to regulate the contractility of the heart.
Discussion
The hearts of zebrafish atp1a1 mutants exhibit severe defects in primitive heart tube elongation (Shu et al., 2003), indicating that Na+,K+ ATPase is required for cardiac morphogenesis in addition to its well-defined role in cardiac function. The cellular and molecular mechanisms by which Na+,K+ ATPase regulates cardiac morphogenesis were previously unknown. In this study, we demonstrate a novel non-cell autonomous function of Na+,K+ ATPase in heart tube elongation. We found that knockdown of Atp1a1 function in the extra-embryonic yolk syncytial layer interferes with the deposition of ECM in the heart field and disrupts heart tube elongation, suggesting that the atp1a1 mutant heart phenotype may be the result of loss of Atp1a1 activity in the YSL.
From this study and previous studies by our lab (Shu et al., 2003), we propose that Atp1a1 regulates at least two aspects of cardiac development in zebrafish (Figure 6E). Prior to the formation of a functioning heart, Atp1a1 activity in the YSL is required for the deposition of ECM in the heart field, providing a substrate for the myocardial migration that drives elongation of the primitive heart tube. Upon formation of the primitive heart tube, Atp1a1 is expressed in cardiomyocytes and is required for maintaining cardiac function in a cell autonomous manner. Embryos lacking Atp1a1 activity have no heart beat at 24 hpf and exhibit a slow heart beat, reduced ventricular contractility, and cardiac arrhythmia at 2 dpf (Shu et al., 2003). We have also previously identified a genetic interaction with mpp5 by which atp1a1 regulates the maintenance of ZO-1 positive cell-cell junctions. Simultaneous loss of mpp5 and atp1a1 causes a disruption of ZO-1 junctions at 36 hpf, a later developmental stage than was examined in this study (Cibrian-Uhalte et al., 2007). Future studies analyzing the cellular requirements of atp1a1 and mpp5 will reveal whether this genetic interaction depends upon the role of atp1a1 in ECM deposition or another yet undiscovered cell autonomous function.
Loss of ECM deposition in several previous zebrafish studies resulted in defective medial migration of cardiomyocytes and cardia bifida at 24 hpf (Arrington and Yost, 2009; Osborne et al., 2008; Sakaguchi et al., 2006; Trinh and Stainier, 2004). However, we have not detected cardia bifida in atp1a1m883 mutants and have only observed a small percentage of embryos with cardia bifida in another atp1a1 mutant allele, atp1a1la1. Based on immunostaining, low levels of Fibronectin protein are present in atp1a1m883 mutants, while fn1 mutants and mxtx1 YSL-morphants have a much more dramatic loss of Fibronectin deposition (Sakaguchi et al., 2006; Trinh and Stainier, 2004). This suggests that cardiac morphogenesis may be sensitive to the dosage of Fibronectin. A reduced level of Fibronectin/ECM may still permit robust migration of the cardiomyocytes to the midline, but be insufficient for heart tube elongation. In agreement with this hypothesis are the findings that excessive Fibronectin deposition in the zebrafish hand2 mutant blocks the medial migration of cardiac progenitors and that this migration can be rescued by genetically reducing Fibronectin levels (Garavito-Aguilar et al., 2010). Thus the migration of cardiomyocytes during zebrafish cardiac development is highly sensitive to Fibronectin levels and may occur in two phases: migration to the midline requiring at least some, but not excessive, Fibronectin/ECM deposition and migration to form the primitive heart tube requiring normal levels of Fibronectin/ECM deposition.
Our data reveals that atp1a1 mutants have a thinning of the ECM between the cardiac cone and surrounding tissues, suggesting that multiple ECM components are reduced upon loss of Atp1a1 function. In this study we examined the deposition of Fibronectin, a critical modulator of cardiomyocyte migration (Arrington and Yost, 2009; Osborne et al., 2008; Sakaguchi et al., 2006; Trinh and Stainier, 2004), but a combinatorial loss of multiple ECM components may result in an array of cardiac migration defects in zebrafish. Laminin protein is also present around the cardiac cone in zebrafish (Arrington and Yost, 2009; Sakaguchi et al., 2006). Although loss of function of any single laminin gene product does not induce significant cardiac migration or tube elongation defects in zebrafish, simultaneous loss of laminin c1 and fibronectin 1 produces a severely enhanced phenotype (Sakaguchi et al., 2006), suggesting that interactions between individual ECM components are important for cardiac morphogenesis.
Regulation of heart tube elongation by Atp1a1 activity in the YSL highlights the important roles played by extra-embryonic signals during zebrafish development. The YSL signal or signals that directly modulate embryonic ECM deposition have not yet been elucidated. Based on YSL knockdown experiments, some potential regulators may include sphingosine-1-phosphate signaling, retinoid signaling, and the ECM-interacting transmembrane protein syndecan (Arrington and Yost, 2009; Li et al., 2007; Osborne et al., 2008). The dramatic reduction of ECM deposition in mxtx1 YSL-morphants suggests that this transcription factor is likely to regulate a number of genes that are critical for embryonic ECM deposition (Sakaguchi et al., 2006). Our study indicates that atp1a1 is one such gene. Identifying more targets of Mxtx1 will undoubtedly reveal additional members of the genetic pathway controlling embryonic ECM deposition.
Loss of Atp1a1 activity results in a strong reduction in anterior, but not posterior, Fibronectin expression, suggesting that the YSL regulates anterior and posterior deposition of Fibronectin/ECM independently. Retinoid signaling appears to play a critical role in posterior ECM deposition, since only posterior Fibronectin expression is affected by loss of Rbp4 (Li et al., 2007). In the case of rbp4, spatially restricted gene expression may be responsible for this posterior-specific effect (Li et al., 2007). However, atp1a1 expression is not anteriorly-biased. Functional redundancies in the posterior YSL or anteriorly localized downstream effectors may be responsible for the anterior-specific effect of Atp1a1 on Fibronectin expression.
Further studies are required to determine how Atp1a1 influences Fibronectin/ECM production and deposition. Na+,K+ ATPase is known to possess structural and signaling roles in addition to its pump function. In some cell types, Na+,K+ ATPase can regulate MAPK and PKC signaling via interaction with Src (Xie and Cai, 2003). However, since cardiac development in atp1a1 mutants can be rescued with wild type Atp1a1 but not with a catalytically inactive form of Atp1a1 (Cibrian-Uhalte et al., 2007), the pump function and not a structural or signaling function of Atp1a1 is likely to be necessary in the YSL. Na+,K+ ATPase directly regulates the concentration of Na+ and K+ in the cytosol, and indirectly modulates Ca2+ levels and pH (Casey et al., 2010; McDonough et al., 2002; Swift et al., 2010; Therien and Blostein, 2000; Tian and Xie, 2008). This regulation of ion homeostasis by Atp1a1 may serve as a critical permissive factor for signaling from the extra-embryonic YSL to the embryo.
Research Highlights.
-
>
Zebrafish atp1a1 mutants exhibit defects in cardiomyocyte migration.
-
>
Abnormal cardiomyocyte migration in atp1a1 is caused by a loss of ECM/Fibronectin.
-
>
Atp1a1 is expressed in the extra-embryonic yolk syncytial layer (YSL).
-
>
Loss of Atp1a1 function in the YSL phenocopies the atp1a1 heart morphogenesis defect.
-
>
Atp1a1 functions cell autonomously and non-cell autonomously in cardiac development.
Supplementary Material
Movie of wild type heart morphogenesis from 18.5 to 26 hpf. The migration paths of 5 cardiomyocytes have been manually traced.
Movie of atp1a1m883 mutant heart morphogenesis from 18.5 to 32 hpf. The migration paths of 4 cardiomyocytes have been manually traced.
Movie of beating heart in wild type Tg(myl7:EGFP) embryo at 1 day post fertilization.
Movie of non-beating heart in atp1a1m883 mutant Tg(myl7:EGFP) embryo at 1 day post fertilization.
Movie of beating heart in wild type Tg(myl7:mCherry)chb1 embryo at 1 day post fertilization.
Movie of beating heart in atp1a1MO1YSL Tg(myl7:mCherry)chb1 embryo at 1 day post fertilization. Atp1a1MO1 is tagged with fluorescein (green), confirming restriction of morpholino to the yolk.
(A,B) Transverse vibratome sections of 20S stage wild type and atp1a1la1 Tg(myl7:EGFP) embryos. Wild type embryos (A) have Fibronectin protein (green) surrounding the cardiomyocytes of the cardiac cone (blue), while atp1a1la1 mutants (B) have reduced Fibronectin protein deposition.
(A) Lateral view of Atp1a1 protein in 2 days post fertilization embryo. Atp1a1 protein is detected in the brain (b), heart (h), and pronephros (p). (B) A magnified image of the pronephros expression (p) detected by the anti-Atp1a1 antibody.
Acknowledgements
We are indebted to John Mably for allowing us to use his Tg(myl7:mCherry)chb1 transgenic line before publication. We also thank K. Mouillesseaux, H. Shimizu, and X. Shu for providing reagents and experimental advice. This work was supported by grants from the NIH (HL081700 and HL096980) to JNC and an NSF Graduate Research Fellowship to ADL.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arrington CB, Yost HJ. Extra-embryonic syndecan 2 regulates organ primordia migration and fibrillogenesis throughout the zebrafish embryo. Development. 2009;136:3143–3152. doi: 10.1242/dev.031492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasiole B, Canfield VA, Vollrath MA, Huss D, Mohideen MA, Dickman JD, Cheng KC, Fekete DM, Levenson R. Separate Na,K-ATPase genes are required for otolith formation and semicircular canal development in zebrafish. Dev Biol. 2006;294:148–160. doi: 10.1016/j.ydbio.2006.02.034. [DOI] [PubMed] [Google Scholar]
- Canfield VA, Loppin B, Thisse B, Thisse C, Postlethwait JH, Mohideen MA, Rajarao SJ, Levenson R. Na,K-ATPase alpha and beta subunit genes exhibit unique expression patterns during zebrafish embryogenesis. Mech Dev. 2002;116:51–59. doi: 10.1016/s0925-4773(02)00135-1. [DOI] [PubMed] [Google Scholar]
- Casey JR, Grinstein S, Orlowski J. Sensors and regulators of intracellular pH. Nat Rev Mol Cell Biol. 2010;11:50–61. doi: 10.1038/nrm2820. [DOI] [PubMed] [Google Scholar]
- Chen JN, Fishman MC. Zebrafish tinman homolog demarcates the heart field and initiates myocardial differentiation. Development. 1996;122:3809–3816. doi: 10.1242/dev.122.12.3809. [DOI] [PubMed] [Google Scholar]
- Cibrian-Uhalte E, Langenbacher A, Shu X, Chen JN, Abdelilah-Seyfried S. Involvement of zebrafish Na+,K+ ATPase in myocardial cell junction maintenance. J Cell Biol. 2007;176:223–230. doi: 10.1083/jcb.200606116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen JS, Smith JC. Controlling morpholino experiments: don't stop making ntisense. Development. 2008;135:1735–1743. doi: 10.1242/dev.001115. [DOI] [PubMed] [Google Scholar]
- Ellertsdottir E, Ganz J, Durr K, Loges N, Biemar F, Seifert F, Ettl AK, Kramer-Zucker AK, Nitschke R, Driever W. A mutation in the zebrafish Na,K-ATPase subunit atp1a1a.1 provides genetic evidence that the sodium potassium pump contributes to left-right asymmetry downstream or in parallel to nodal flow. Dev Dyn. 2006;235:1794–1808. doi: 10.1002/dvdy.20800. [DOI] [PubMed] [Google Scholar]
- Garavito-Aguilar ZV, Riley HE, Yelon D. Hand2 ensures an appropriate environment for cardiac fusion by limiting Fibronectin function. Development. 2010 doi: 10.1242/dev.052225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman NS, Yelon D. Cardiac development in zebrafish: coordination of form and function. Semin Cell Dev Biol. 2002;13:507–513. doi: 10.1016/s1084952102001040. [DOI] [PubMed] [Google Scholar]
- Horne-Badovinac S, Lin D, Waldron S, Schwarz M, Mbamalu G, Pawson T, Jan Y, Stainier DY, Abdelilah-Seyfried S. Positional cloning of heart and soul reveals multiple roles for PKC lambda in zebrafish organogenesis. Curr Biol. 2001;11:1492–1502. doi: 10.1016/s0960-9822(01)00458-4. [DOI] [PubMed] [Google Scholar]
- Huang CJ, Tu CT, Hsiao CD, Hsieh FJ, Tsai HJ. Germ-line transmission of a myocardium-specific GFP transgene reveals critical regulatory elements in the cardiac myosin light chain 2 promoter of zebrafish. Dev Dyn. 2003;228:30–40. doi: 10.1002/dvdy.10356. [DOI] [PubMed] [Google Scholar]
- Kawahara A, Nishi T, Hisano Y, Fukui H, Yamaguchi A, Mochizuki N. The sphingolipid transporter spns2 functions in migration of zebrafish myocardial precursors. Science. 2009;323:524–527. doi: 10.1126/science.1167449. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Law RD. Cell lineage of zebrafish blastomeres. II. Formation of the yolk syncytial layer. Dev Biol. 1985;108:86–93. doi: 10.1016/0012-1606(85)90011-9. [DOI] [PubMed] [Google Scholar]
- Langenbacher AD, Dong Y, Shu X, Choi J, Nicoll DA, Goldhaber JI, Philipson KD, Chen JN. Mutation in sodium-calcium exchanger 1 (NCX1) causes cardiac fibrillation in zebrafish. Proc Natl Acad Sci U S A. 2005;102:17699–17704. doi: 10.1073/pnas.0502679102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Korzh V, Gong Z. Localized rbp4 expression in the yolk syncytial layer plays a role in yolk cell extension and early liver development. BMC Dev Biol. 2007;7:117. doi: 10.1186/1471-213X-7-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonough AA, Velotta JB, Schwinger RH, Philipson KD, Farley RA. The cardiac sodium pump: structure and function. Basic Res Cardiol. 2002;97 Suppl 1:I19–I24. doi: 10.1007/s003950200024. [DOI] [PubMed] [Google Scholar]
- Osborne N, Brand-Arzamendi K, Ober EA, Jin SW, Verkade H, Holtzman NG, Yelon D, Stainier DY. The spinster homolog, two of hearts, is required for sphingosine 1- phosphate signaling in zebrafish. Curr Biol. 2008;18:1882–1888. doi: 10.1016/j.cub.2008.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson RT, Mably JD, Chen JN, Fishman MC. Convergence of distinct pathways to heart patterning revealed by the small molecule concentramide and the mutation heart-andsoul. Curr Biol. 2001;11:1481–1491. doi: 10.1016/s0960-9822(01)00482-1. [DOI] [PubMed] [Google Scholar]
- Rohr S, Bit-Avragim N, Abdelilah-Seyfried S. Heart and soul/PRKCi and nagie oko/Mpp5 regulate myocardial coherence and remodeling during cardiac morphogenesis. Development. 2006;133:107–115. doi: 10.1242/dev.02182. [DOI] [PubMed] [Google Scholar]
- Sakaguchi T, Kikuchi Y, Kuroiwa A, Takeda H, Stainier DY. The yolk syncytial layer regulates myocardial migration by influencing extracellular matrix assembly in zebrafish. Development. 2006;133:4063–4072. doi: 10.1242/dev.02581. [DOI] [PubMed] [Google Scholar]
- Sakaguchi T, Kuroiwa A, Takeda H. A novel sox gene, 226D7, acts downstream of Nodal signaling to specify endoderm precursors in zebrafish. Mech Dev. 2001;107:25–38. doi: 10.1016/s0925-4773(01)00453-1. [DOI] [PubMed] [Google Scholar]
- Serluca FC, Sidow A, Mably JD, Fishman MC. Partitioning of tissue expression accompanies multiple duplications of the Na+/K+ ATPase alpha subunit gene. Genome Res. 2001;11:1625–1631. doi: 10.1101/gr.192001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu X, Cheng K, Patel N, Chen F, Joseph E, Tsai HJ, Chen JN. Na,K-ATPase is essential for embryonic heart development in the zebrafish. Development. 2003;130:6165–6173. doi: 10.1242/dev.00844. [DOI] [PubMed] [Google Scholar]
- Swift F, Tovsrud N, Sjaastad I, Sejersted OM, Niggli E, Egger M. Functional coupling of alpha(2)-isoform Na(+)/K(+)-ATPase and Ca(2+) extrusion through the Na(+)/Ca(2+)-exchanger in cardiomyocytes. Cell Calcium. 2010;48:54–60. doi: 10.1016/j.ceca.2010.06.006. [DOI] [PubMed] [Google Scholar]
- Therien AG, Blostein R. Mechanisms of sodium pump regulation. Am J Physiol Cell Physiol. 2000;279:C541–C566. doi: 10.1152/ajpcell.2000.279.3.C541. [DOI] [PubMed] [Google Scholar]
- Tian J, Xie ZJ. The Na-K-ATPase and calcium-signaling microdomains. Physiology (Bethesda) 2008;23:205–211. doi: 10.1152/physiol.00008.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh LA, Stainier DY. Fibronectin regulates epithelial organization during myocardial migration in zebrafish. Dev Cell. 2004;6:371–382. doi: 10.1016/s1534-5807(04)00063-2. [DOI] [PubMed] [Google Scholar]
- Walsh EC, Stainier DY. UDP-glucose dehydrogenase required for cardiac valve formation in zebrafish. Science. 2001;293:1670–1673. doi: 10.1126/science.293.5535.1670. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The zebrafish book. A guide for the laboratory use of zebrafish (Danio rerio), 4th ed. University of Oregon Press, Eugene; 2000. [Google Scholar]
- Xie Z, Cai T. Na+-K+--ATPase-mediated signal transduction: from protein interaction to cellular function. Mol Interv. 2003;3:157–168. doi: 10.1124/mi.3.3.157. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie of wild type heart morphogenesis from 18.5 to 26 hpf. The migration paths of 5 cardiomyocytes have been manually traced.
Movie of atp1a1m883 mutant heart morphogenesis from 18.5 to 32 hpf. The migration paths of 4 cardiomyocytes have been manually traced.
Movie of beating heart in wild type Tg(myl7:EGFP) embryo at 1 day post fertilization.
Movie of non-beating heart in atp1a1m883 mutant Tg(myl7:EGFP) embryo at 1 day post fertilization.
Movie of beating heart in wild type Tg(myl7:mCherry)chb1 embryo at 1 day post fertilization.
Movie of beating heart in atp1a1MO1YSL Tg(myl7:mCherry)chb1 embryo at 1 day post fertilization. Atp1a1MO1 is tagged with fluorescein (green), confirming restriction of morpholino to the yolk.
(A,B) Transverse vibratome sections of 20S stage wild type and atp1a1la1 Tg(myl7:EGFP) embryos. Wild type embryos (A) have Fibronectin protein (green) surrounding the cardiomyocytes of the cardiac cone (blue), while atp1a1la1 mutants (B) have reduced Fibronectin protein deposition.
(A) Lateral view of Atp1a1 protein in 2 days post fertilization embryo. Atp1a1 protein is detected in the brain (b), heart (h), and pronephros (p). (B) A magnified image of the pronephros expression (p) detected by the anti-Atp1a1 antibody.