Abstract
We report a systems genetics analysis of high density lipoproteins (HDL) levels in an F2 intercross between inbred strains CAST/EiJ and C57BL/6J. We previously showed that there are dramatic differences in HDL metabolism in a cross between these strains, and we now report co-expression network analysis of HDL that integrates global expression data from liver and adipose with relevant metabolic traits. Using data from a total of 293 F2 intercross mice, we constructed weighted gene co-expression networks and identified modules (subnetworks) associated with HDL and clinical traits. These were examined for genes implicated in HDL levels based on large human genome-wide associations studies (GWAS) and examined with respect to conservation between tissue and sexes in a total of 9 data sets. We identify genes that are consistently ranked high by association with HDL across the 9 data sets. We focus in particular on two genes, Wfdc2 and Hdac3, that are located in close proximity to HDL QTL peaks where causal testing indicates that they may affect HDL. Our results provide a rich resource for studies of complex metabolic interactions involving HDL.
Introduction
HDLs, classically defined as a plasma fraction of lipoproteins at a density range 1.063–1.21mg/ml, include a wide range of circulating particles that are highly heterogeneous in terms of shape, size, and lipid composition. As with other lipoproteins, the basic structure of HDL consists of a lipid core, surrounded by a surface containing a phospholipid bilayer, free cholesterol, and a number of apolipoproteins. In addition to the major apolipoproteins of mammalian HDL, apolipoprotein AI (APOAI) and apolipoprotein AII (APOAII), HDL also contains a number of other apolipoproteins as well as enzymes such as lecithin cholesterol acyl transferase (LCAT) and phospholipid transfer protein (PLTP). Cholesteryl ester transferase (CETP), an important constituent of human HDL, is missing in mice. Shotgun proteomic studies have shown that HDL are associated with dozens of proteins with many different functions, including host defense (1–4). The assembly of HDL occurs largely extracellularly in the circulation following secretion of APOAI by liver and intestine (5–8).
HDL is a well-documented negative risk factor for coronary heart disease (CHD) (1, 2, 9, 10). In addition to human epidemiologic studies, experimental studies in animal models indicate that elevation of HDL cholesterol is protective (11–16). One mechanism by which HDL protects against atherosclerosis is by removing cholesterol from artery wall macrophages in vascular cells (7, 17, 18). The anti-inflammatory or anti-oxidant properties of HDL also appear to contribute to its cardioprotective effects (19–21).
Human studies with rare HDL disorders, such as Tangier disease, and association studies have been particularly informative in identifying genes and pathways contributing to HDL levels and functions. Also, several recent genome-wide association studies (GWAS), each involving tens of thousands of individuals typed for hundreds of thousands of single nucleotide polymorphisms (SNPs) revealed several novel HDL loci and confirmed the roles of a dozen previously identified genes such as CETP, LIPG, LIPC, ABCA1, LCAT, LPL, and APOAI. However, altogether, these explained only about 5–10% of the variation in HDL levels (2, 3, 22–26). Since estimates of the heritability of HDL in human populations have ranged from 40%–70%, there clearly remains a great deal to be discovered. Little is known about the factors contributing to functional HDL differences in the human populations. In addition to human studies, extensive studies on the regulation of HDL cholesterol have been carried out in animal models, particularly the mouse. Altogether, over 40 different loci have been mapped using quantitative trait locus analyses in mice and shown to affect HDL cholesterol levels. Many of these loci do not contain any known genes contributing to HDL metabolism, suggesting strongly that there are many novel genes and factors remaining to be identified regulating HDL metabolism (27–35). Recently, microRNAs have been recognized as having a regulatory role in cholesterol metabolism and transport (36). Moreover, Vickers and colleagues have shown that microRNAs are transported in plasma by HDL (37). This uncovers a previously unknown role of HDL in cell-cell communication and genetic regulation.
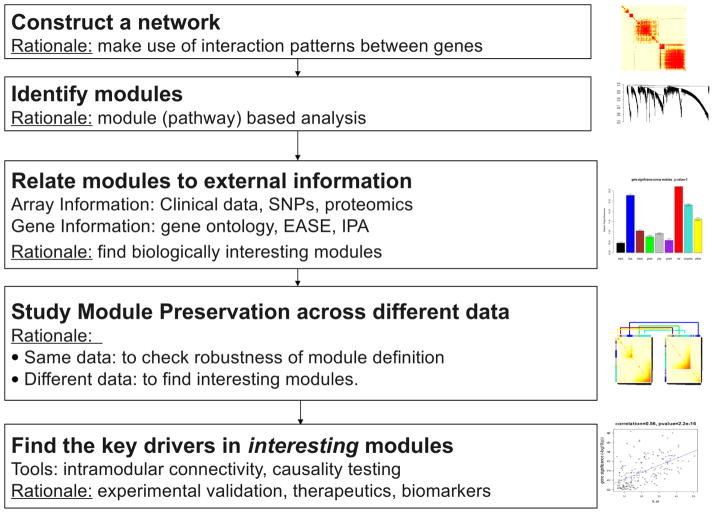
The strain CAST/EiJ, derived from a castaneus subspecies of musculus, differs dramatically in HDL cholesterol metabolism from common laboratory strains of mouse such as C57BL/6J. In previous studies, we have examined parameters relating to HDL metabolism in CAST/EiJ (CAST) and C57BL/6J (B6) and have identified several loci contributing to HDL metabolism in crosses between the two strains (38). We now report a systems-level study of variations between the strains. For this, we characterized HDL metabolism in a large intercross between the strains and carried out global expression array analysis of tissues relevant to HDL metabolism, including liver and adipose. These data were used to identify candidate genes contributing to variation in HDL metabolism and also to model biologic networks associated with HDL levels using Weighted Gene Co-expression Network Analysis (WGCNA, Figure 1) (39).
Figure 1. Overview of Weighted Gene Co-expression Network Analysis (WGCNA).
WGCNA starts from the level of tens of thousands of genes, constructs a correlation-based gene network, identifies interesting gene modules by a clustering analysis, and finally uses gene significance (e.g., based on the correlation of a gene expression profile with HDL) and intramodular connectivity to identify key genes for further validation. WGCNA alleviates the multiple testing problem inherent in microarray data analysis. Instead of relating tens of thousands of genes to a microarray sample trait, it focuses on the relationship between a few tens (typically less than 100) modules and the sample trait. Toward this end, it represents each module by a summary profile, the module eigengene. Correlation between the sample trait (HDL) and the eigengene of a module is referred to as module eigengene significance (MES) (40). MES also serves as a measure of module significance for the trait. Because module eigengene significance is defined as a correlation, the corresponding p-value can be used to measure the statistical significance of the association between the module and HDL. The module definition does not make use of a priori defined gene sets. Instead, modules are constructed from the expression data using hierarchical clustering. Although it is advisable to relate the resulting modules to gene ontology information to assess their biological plausibility, it is not required. Since the modules may correspond to biological pathways, focusing the analysis on the modules amounts to a biologically motivated data reduction scheme.
Materials and Methods
Mice, breeding, and diets
CAST/EiJ and C67BL/6J mice were purchased from the Jackson Laboratory, Bar Harbor, Maine. Female CAST/EiJ were bred to C67BL/6 mice to generate F1 hybrids. The F1 hybrid mice were subsequently intercrossed to generate F2 mice, referred to as CASTxB6. All mice were fed ad libitum and maintained on a 12-hour light/dark cycle. The mice were fed Purina Chow (Ralston-Purina Co., St. Louis, MO) containing 4% fat until 10 weeks of age, and then transferred to a Western diet (Teklad 88137, Harland Teklad, Madison, WI), containing 42% fat and 0.5% cholesterol for 8 weeks until euthanization at 18 weeks of age. Lipid measurements and biochemical analyses. Mice were fasted overnight and bled retro-orbitally under isoflurane anesthesia. Enzymatic assays for total cholesterol, HDL cholesterol and triglycerides were carried out as previously described (41). Plasmas were stored at −80°C.
Body composition
At 10 and 18 weeks of age, body mass parameters were determined using NMR spectroscopy (42). The NMR instrument (Bruker Biospin, Billerica, MA) and software from Echo Medical Systems was used to determine fat mass (g), lean mass (g), and free fluid (g). Percent fat and lean mass was calculated by dividing each value by total body mass. Fat-to-muscle ratio was calculated by dividing fat mass by lean mass. At euthanasia, the epididymal, retroperitoneal, visceral, and subcutaneous fat pads were removed and weighed. The sum of these weights is referred to as total fat pad depot.
Linkage and genotype data analysis
A 1.5 cM dense SNP map was constructed using the multiple inversion probe technology (parALLELE, San Francisco, CA). The map was created according to the Celera and NCBI Public Databases. The QTL analysis was performed using R/qtl software (http://www.rqtl.org) (43, 44). We tested two models, one with sex as a covariate and one with sex as an interactive trait. Data were permuted 10,000 times to determine genome wide significant (p<0.05) and suggestive (p<0.20) LOD scores. We removed outlier trait values, defined as values further than 3 standard deviations from the mean.
Global gene transcript studies
RNA was isolated from the livers, and gonadal fat pads, of F2 mice using the Trizol method. Microanalyses were performed on RNA essentially as previously described (45). Briefly, 60mer oligonucleotide chips were utilized (Agilent Technologies) and all hybridizations were done in duplicate with Fluor reversal. Each individual sample was hybridized against a pool of F2 samples. Expression data in the form of mean log ratios (MLratios) were treated as quantitative trait in eQTL analysis, while taking genotype sex interactions into account as described above. Correlations between gene expression measured as MLratios, and each quantitative trait were calculated using biweight midcorrelation (a robust correlation).
Weighted Gene Co-expression Network Analysis (WGCNA)
WGCNA has the advantage of preserving the continuous nature of co-expression that is lost in unweighted networks (39). The analysis starts by constructing a matrix of pairwise correlations (we use the robust biweight midcorrelation) between all pairs of probes across the measured samples. To construct a “signed hybrid” weighted network, we first create a pairwise probe co-expression similarity that equals the probe-probe correlation if the correlation is positive, and equals zero otherwise. Next the co-expression similarity is raised to the power β=4 to arrive at the network adjacency. This has the effect of suppressing low correlations that may be due to noise. Thus, the network adjacency is zero for negatively correlated probes and it is positive for positively correlated probes. Adjacency of weakly correlated probes is nearly zero due to the power transformation.
To identify modules of co-expressed genes, we construct the Topological Overlap-based dissimilarity (39, 46) and use it as input to average linkage hierarchical clustering (47) that results in a clustering tree (dendrogram) whose branches are identified using the Dynamic Hybrid Tree Cut algorithm (48). Modules whose summary profiles (eigengenes) were too similar (correlation above 0.8) were merged. Modules were further pruned of probes whose module membership (described below) was below 0.3. The statistical analysis software (WGCNA R package) and R tutorials for constructing a weighted gene co-expression network can be found in (49).
Summarizing expression profiles of a module using the module eigengene
The module identification procedure results in modules containing genes with highly correlated expression profiles. It is useful to summarize such modules using a single expression profile. We use the module eigengene E, defined as the first principal component of the standardized expression matrix.
Continuous measure of module membership
Module eigengenes lead to a natural measure of similarity (membership) of all individual genes to all modules. We define a fuzzy measure of module membership of gene i in module q, as , where xi is the expression profile of gene i and E q is the eigengene of module q. The value of module membership lies between −1 and 1. The higher , the more similar the expression profile of gene i is to the summary profile of module q. Since we use signed networks here, we consider module membership near −1 low. The advantage of using a correlation to quantify module membership is that the corresponding statistical significance (p-values) can be easily computed. Genes represented by probes with highest module membership are called hub genes. Hub genes are centrally located inside the module and represent the expression profiles of the entire module.
Module preservation statistics
To assess the preservation of CASTxB6 female liver modules in other expression data sets, we use the network module preservation statistics described in (50) and implemented in the function modulePreservation in the WGCNA R package. Network module preservation statistics assess whether the density and connectivity patterns of modules defined in a reference data set are preserved in a test data set. Unlike traditional cross-tabulation statistics that rely on module matching between reference and test data sets, network preservation statistics do not require that modules be identified in the test data set. This makes them independent of the ambiguities associated with module identification in the test data set.
Although it is useful to study all statistics to determine which module properties are preserved, it is practical to consider summary statistics that summarize the evidence for preservation of each module. We concentrate on the Zsummary statistic that summarizes evidence that a module is preserved more significantly than a random sample of all network genes. We use the thresholds proposed by Langfelder et al: Zsummary<2 implies no evidence for module preservation, 2<Zsummary<10 implies weak to moderate evidence, and Zsummary<10 implies strong evidence for module preservation. Thus, we report Zsummary for each CASTxB6 female liver module in each of the test data sets.
Calculation of the module preservation statistics described above requires that the sets of measured variables (probes or genes) in the compared sets be the same. Since the probes on different microarray platforms are typically different, we first converted probe-level measurements into gene-level measurements using the function collapseRows in the WGCNA package. This function selects one representative probe per gene. We used the MaxMean option for probe selection; this option selects the probe with the highest mean expression. We then matched genes using their Entrez codes. For each pair of reference-test data set in the preservation calculations we retained all common genes between the two data sets. Hence, the number of genes in different reference-test pairs is in general different.
Independent test data sets
We study module preservation as well as preservation of gene and module eigengene significance for HDL in the following test data sets: CAST x B6 male liver, CASTxB6 female adipose, female and male liver of a reciprocal C57BL/6J x C3H/HeJ F2 mouse cross (denoted BxH and HxB) (51), female and male liver of a C57BL/6J x C3H/HeJ cross on an ApoE null background (denoted BxH ApoE or ApoE for short) (52), and male liver of the Hybrid Mouse Diversity Panel (denoted HMDP) (53).
Identifying genes consistently associated with HDL
Our aim is to identify genes that relate to HDL consistently more strongly (positively or negatively) than other genes. A standard approach for selecting HDL-related genes is based on the marginal associations between gene expression levels and HDL. An advantage of this approach is that it is straightforward to generalize it to a meta analysis involving 9 data sets. By definition, the resulting genes reproducibly correlate with HDL in multiple independent data sets. Toward this end, we measure the association of each gene and HDL in each data set by robust correlation. Next, for each data set, we standardize the gene-HDL associations by scaling the vector of gene-HDL associations in this data set to mean 0 and variance 1. This results in an approximately normally distributed Zia statistic for each gene i in each data set a. We next form a meta-analysis Zi statistic for each gene as
The meta-analysis statistic Zi is approximately normally distributed with mean 0 and variance 1 and can be used to calculate the corresponding two-sided p-value using standard normal distribution. To calculate the corresponding local False Discovery Rate (FDR), also known as the q-value, we use the R software package qvalue.
Causal testing using NEO software
To identify candidate genes whose expression causally affects HDL and genes whose expression is affected by HDL, we use the Network Edge Orienting (NEO) causal testing software (54) to test for evidence of causal effect between each gene and HDL, using peak QTL SNPs for HDL as causal anchors. NEO uses genotypes as causal anchors and test each pair of variables (in our case one gene and HDL) independently of all other variables. For each pair of variables (gene G, HDL), NEO outputs Local Edge Orienting (LEO) scores for the two models “expression of G causally affects HDL” and “HDL causally affects expression of G”. Values greater than 0 indicate that the causal model fits the data better than alternative (reactive and confounded) models. We use the LEO threshold 1 (recommended in (53)) for reporting genes that may be causal for HDL.
Using module information to screen for HDL-related genes
Network information (including information about module membership) can be used in several ways to gain insight into the genomics of a complex trait such as HDL levels. First, co-expression networks provide insights into the organization of the gene expression and how that organization relates to a trait such as HDL. Preservation of (individual) modules across different conditions suggests that regulatory mechanisms responsible for each preserved module are universal (at least across the studied conditions). Further, robust module significance for HD) across different conditions further suggests that regulatory mechanisms underlying the module expression are associated with HDL.
There are several possible strategies for using HDL related co-expression modules for screening for HDL related genes. First, if the module is known to be enriched with biologically relevant pathways then it makes sense to make use of intramodular connectivity (kME) since intramodular hub genes are centrally located inside the module (55). The resulting gene screening strategy has been successfully used for a number of complex diseases, e.g. brain cancer and atherosclerosis (56, 57)
Second, gene information on individual module genes can be used to screen for transcriptional regulators of the module (e.g. transcription factors or splicing factors). For example, an intramodular hub genes in an autism related module was identified given its role as splicing factor (58).
Third, a systems genetic screening strategy can be implemented based on one more more trait related SNP(s) since it can be used as causal anchor in a causal testing procedure (54), (59) In this case, one can prioritize module genes according to the evidence of their causal effect on the complex trait. The resulting systems biologic screening strategies have been applied to chronic fatigue syndrome (60), hyperlipidemia (61), and conditional fear (62). In our article, we focus on this third strategy.
Results
Complex inheritance pattern of HDL-C in CAST x B6 Intercross: Gene-by-sex and gene-by-diet interactions
For this study we used 293 mice of a CAST x B6 F2 intercross. F2 male (n=111) and female F2 (n=182) mice were placed on a standard chow diet until 10 weeks of age, and then switched to a high fat “Western” diet for 8 weeks to generate a diet induced inflammatory state where genetic interactions could be analyzed. During this period a number of traits pertaining to lipoprotein metabolism, obesity and diabetes were collected. The mice were genotyped with 1375 SNPs at an average genome wide density of ~1.5 MB. Expression profiles for 23,623 genes were determined for liver and adipose, and treated as quantitative traits (eQTL) (45).
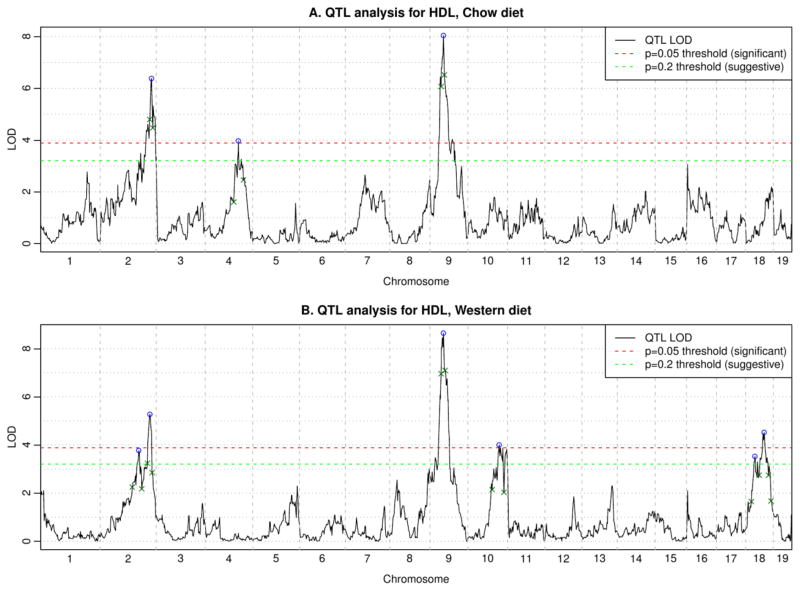
HDL levels differed significantly between chow (45 ± 14 mg/dl) and Western (73 ± 23 mg/dl) diets (t-test p-value 2e-56). Treating sex as a covariate, we identified suggestive or significant QTL loci associated with HDL cholesterol levels on either a chow or a Western diet on 5 chromosomes: 2, 4, 9, 10, 18 (Figure 2). Permutation tests using R/qtl indicated suggestive (p<0.2) LOD = 3.2 and significant (p<0.05) LOD = 3.9. QTL analysis suggests that HDL associated loci are sensitive to dietary interactions as was observed in our previous cross (38). LOD scores for all genotyped markers with sex as a covariate are listed in Supplementary Table 1, and with sex as an interactive trait in Supplementary Table 2. All supplementary materials (tables and figures) are posted at the following URL: http://genetics.ucla.edu/labs/horvath/CoexpressionNetwork/CASTxB6-HDL/.
Figure 2. QTL analysis for HDL.
Here we combined male and female mice together and treated sex as a covariate. Graphs show the LOD scores as well as significant (p=0.05) and suggestive (p=0.20) thresholds. Peaks are indicated by blue circles; small green crosses indicate boundaries of peak regions (LOD drop of 1.5 from the peak).
ANOVA analysis for HDL cholesterol levels at the QTL peak SNP markers (Table 1a and 1b) confirmed differences due to genotype distribution on Chrs 2, 4, and 9 for the mice on the chow diet, and Chrs 2, 9, 10 and 18 on the Western diet. The highly significant Chr 4 locus at ~108 MB for mice on a chow diet has not been previously observed (29). The identified significant and suggestive peaks explain 16% of the variance of HDL for chow diet and 25% for Western diet.
Table 1a.
Male and female mice on a chow diet. Median HDL levels by genotype at cQTL peak markers with half-range of 95% confidence interval indicated in brackets, Kruskal-Wallis p-value, the corresponding R/qtl LOD score, and the genome-wide significance (permutation test p-value).
| SNP | Chr | Bp | BB | BC | CC | p-value | LOD | GW signif. |
|---|---|---|---|---|---|---|---|---|
| rs6219107 | 2 | 167167187 | 46 (4) | 46 (2) | 36 (3) | 1E-05 | 6.4 | 3E-04 |
| rs3664637 | 4 | 108062431 | 48 (3) | 45 (2) | 38 (2) | 3E-04 | 4 | 0.04 |
| rs3700385 | 9 | 44710804 | 39 (3) | 44 (2) | 53 (3) | 2E-07 | 8 | 1E-04 |
Table 1b.
Male and female mice on a Western diet. Median HDL levels by genotype at cQTL peak markers, with half-range of 95% confidence interval indicated in brackets, Kruskal-Wallis p-value, the corresponding R/qtl LOD score, and the genome-wide significance (permutation test p-value).
| SNP | Chr | Bp | BB | BC | CC | p-value | LOD | GW signif. |
|---|---|---|---|---|---|---|---|---|
| rs3708635 | 2 | 125570240 | 76 (6) | 74 (4) | 65 (5) | 0.003 | 3.8 | 0.06 |
| rs3692104 | 2 | 161902891 | 78 (5) | 73 (4) | 64 (5) | 6.00E-05 | 5.3 | 0.003 |
| rs3700385 | 9 | 44710804 | 61 (4) | 74 (4) | 81 (5) | 5.00E-08 | 8.7 | 1E-04 |
| rs6382566 | 10 | 102218998 | 76 (7) | 75 (3) | 64 (5) | 5.00E-04 | 4 | 0.04 |
| rs6174809 | 18 | 31312897 | 78 (6) | 68 (4) | 69 (6) | 0.003 | 3.5 | 0.10 |
| rs6381950 | 18 | 61521178 | 78 (6) | 72 (4) | 62 (4) | 1.00E-04 | 4.5 | 0.01 |
The chow QTL on Chrs 2, 4, and 9 exhibited dominant inheritance, with that on Chr 9 being dominant for the CAST allele (Table 1a). HDL levels were significantly higher on the Western diet, where a dominant allele effect was seen on Chrs 2, 9, 10 and 18. The B6 allele was associated with increased HDL on chromosomes 2, 10 and 18 (Table 1b).
The differences between HDL QTL on chow and Western diets may be the result of acute-phase or inflammatory responses due to the high fat dietary intervention. In our previous cross (38) the mice were analyzed for HDL levels on either a chow or a cholic acid containing atherosclerotic (Ath) diet, which differs considerably from the Western diet. The change from an Ath to a Western diet could account for the variation in HDL QTL loci observed as well as in the LOD scores.
QTL analysis of HDL with sex as an interactive trait (Supplementary Figure 1 posted with other supplementary material at our web page) identified several significant and suggestive loci that are detailed in Supplementary Tables 3 and 4 for chow and Western diets, respectively. For mice on chow diet we observed significant sex-dependent effects on chomosomes 1 and 2, while on Western diet the sex interaction peaks are located on chromosomes 4, 5, 10. The peaks on chromosomes 1, 4, and 5 were not observed in the analysis with sex as a covariate.
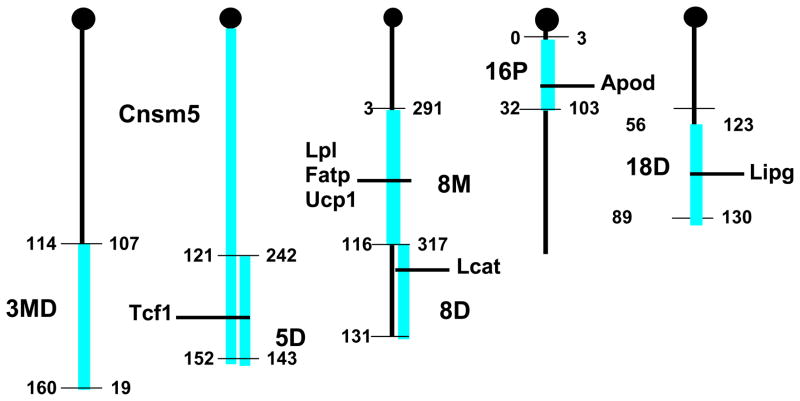
We have constructed congenic strains for a number of the loci identified in (38) by introgressing the chromosomal region spanning the QTL segment from CAST/EiJ onto the background of B6, using a speed congenic, marker assisted protocol (63). Following 10 generations of backcrossing, the F1 mice were brother/sister mated to produce homozygous congenics. Phenotyping of these twelve congenic strains for variation in HDL levels compared to B6 mice confirmed most of the original QTLs identified in the cross. For further study, we chose congenic strains for HDL cQTL loci on chrs 3 (CON3md), 5 (CON5 and Consomic5 (Cnsmc5), 8 (CON8m and CON8d), 16 (CON16p) and 18 (CON18d) (Figure 3). These strains were shown to vary in HDL and other lipid levels as compared to the control B6 mice for both males and females (Table 2a and 2b). B6 mice carrying the CAST loci showed significant reduction in HDL cholesterol as compared with B6 wild type. Many of the loci had a substantial impact on HDL levels, for example, CON8m, CON8d and CON18d loci all decrease HDL cholesterol by about 25%. Of the various congenics analyzed, only the CON16p strain did not exhibit a significant difference in HDL levels, although this congenic exhibited approximately two-fold higher TG levels in both males and females. A surprising finding was the observation of significant variations in glucose levels between the congenics, particularly in females. Metabolic syndrome is characterized by low HDL, high TG and insulin resistance (64). From epidemiological and genome wide association studies (GWAS) it is clear that there is overlap of genetic factors contributing to insulin resistance and HDL/TG metabolism (25). These lipid profiles are reflected in the congenics and may lead to identification of genes with pleiotropic metabolic effects. Interestingly, the results also demonstrate a sexual bias for HDL levels in CON3md and CON8d, where the trait was significantly associated with the male congenics and not the females.
Figure 3. Congenic Intervals for B6.CAST.3MD, B6.CAST.5, B6.CAST.5D, B6.CAST.8M, B6.CAST.8D, B6.CAST.16P and B6.CAST.18D.
Megabase distances are shown on the left and MIT markers on the right of the cartoon chromosomes. Genes known to affect HDL metabolism are indicated.
Table 2a.
Comparison of mean lipid levels (mg/dL) among male HDL congenics (15wks old) on a chow diet (mean ±SE).
| Strain (N) | Tchol | HDLc | VLDL/LDL | TG | UC | FFA | Glucose |
|---|---|---|---|---|---|---|---|
| B6 (50) | 95.8±1.7 | 83.4±1.4 | 12.4±0.6 | 40.0±1.7 | 19.3±0.5 | 62.0±1.3 | 120.6±3.5 |
| B6.CAST.3MD (12) | 91.1±1.8 | 75.3±2.0** | 15.8±0.7 | 52.2±3.8 | 23.6±0.8** | 53.8±3.0 | 119.8±5.9 |
| B6.CAST.5 (19) | 103.9±6.0* | 78.9±3.1 | 25.0±3.8**** | 84.8±13.5**** | 28.4±1.6**** | 81.7±7.7**** | 125.4±4.5 |
| B6.CAST.5D (9) | 87.2±3.3 | 73.0±3.4** | 14.2±0.6 | 63.8±13.5* | 24.9±3.9*** | 66.1±3.1 | 112.1±4.0 |
| B6.CAST.8M (4) | 76.2±4.0** | 62.5±3.8**** | 13.7±0.4 | 83.7±17.6** | 13.0±0.8** | 58.0±4.7 | 130.0±7.2 |
| B6.CAST.8D (24) | 78.4±2.0**** | 64.5±1.1**** | 18.9±1.3 | 65.1±7.1*** | 18.6±0.7 | 70.5±3.8* | 112.5±4.6 |
| B6.CAST.16P (19) | 102.0±3.3 | 82.5±2.0 | 19.5±1.6*** | 106.7±8.2**** | 24.6±0.5**** | 75.8±5.6*** | 116.7±5.0 |
| B6.CAST.18D (19) | 70.8±1.6**** | 62.5±1.1**** | 8.3±0.6* | 38.7±2.3 | 17.0±0.6* | 44.7±2.3*** | 117.8±5.0 |
p ≤0.05;
p≤0.01;
p≤0.001;
p≤0.0001 compared to B6
Table 2b.
Comparison of mean lipid levels (mg/dL) among female HDL congenics (15wks old) on a chow diet(mean ±SE).
| Strain (N) | Tchol | HDLc | VLDL/LDL | TG | UC | FFA | Glucose |
|---|---|---|---|---|---|---|---|
| B6 (51) | 85.1±2.0 | 67.2±1.7 | 18.1±0.7 | 32.8±2.1 | 15.7±0.5 | 65.7±1.7 | 124.8±5.0 |
| B6.CAST.3MD (9) | 69.5±4.2*** | 52.7±4.2 | 16.9±1.4 | 45.7±5.8 | 18.9±1.2 | 49.9±2.8* | 103.3±10.0* |
| B6.CAST.5 (24) | 79.7±3.2 | 50.2±2.0** | 20.5±1.8 | 59.9±6.1*** | 16.2±1.2 | 83.5±5.8**** | 103.4±3.7*** |
| B6.CAST.5D (12) | 70.5±1.9*** | 54.5±1.6*** | 16.0±1.0 | 57.0±5.6** | 30.1±8.2**** | 70.8±5.8 | 107.3±6.8* |
| B6.CAST.8D (16) | 75.5±3.2** | 60.6±3.0* | 14.8±1.0 | 40.0±4.8 | 15.5±1.1 | 74.7±2.7 | 103.2±3.5*** |
| B6.CAST.16P (24) | 86.5±2.8 | 66.6±1.7 | 20.0±1.8 | 64.9±10.9**** | 17.0±1.0 | 68.9±4.4 | 120.3±5.6 |
| B6.CAST.18D (12) | 65.5±1.7**** | 51.5±1.2**** | 13.9±0.8* | 41.7±3.7 | 15.2±0.7 | 50.3±3.2** | 93.2±4.1*** |
p ≤0.05;
p≤0.01;
p≤0.001;
p≤0.0001 compared to B6
Relationship of HDL and other metabolic traits
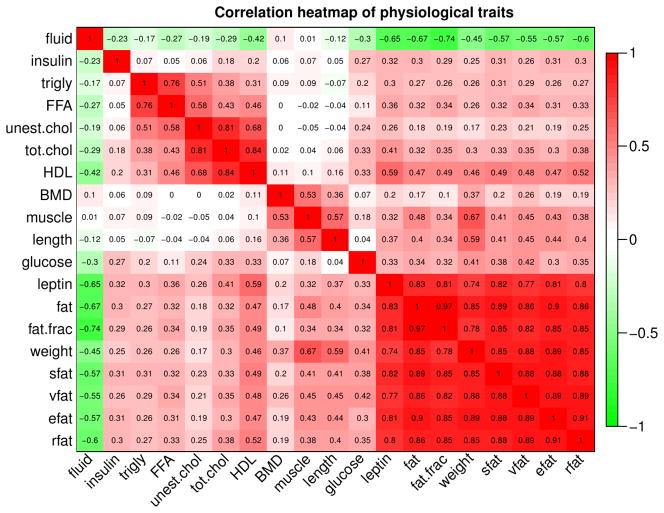
In addition to HDL, several other physiological traits were measured in the crosses. In Figure 4 we present an overview of the statistical associations among the measured traits. In this work we concentrate mostly on HDL, but it is worth noting that HDL is strongly correlated with total cholesterol (robust correlation r = 0.88) and unesterified cholesterol (r = 0.68), as well as leptin (r = 0.59) and adiposity traits (for example, body fat fraction, r = 0.49). For reasons that are not yet understood, the relationship between HDL and certain metabolic syndrome traits, including triglycerides and body fat, tends to be opposite between mice and humans, although this finding is also to some degree cross-, diet-, and/or perturbation-dependent. For example, while human HDL and triglyceride (TG) levels tend to be anti-correlated (26), (65), we observe a positive correlation of 0.31. This is concordant with results of other studies (e.g., (66) (67)) that have found positive HDL-TG correlations of varying strength. Increased HDL accompanying increases in weight, BMI and fat pad weigh in mice has been reported, for example, in (68). This implies some fundamental differences in the regulation of HDL. For example, mice lack CETP. Despite these differences, it is expected that many aspects of HDL regulation are similar between humans and mice (this is indirectly confirmed by multiple genes that have been shown to affect HDL in both humans and mice).
Figure 4. Correlation heatmap of physiological traits measured in the CASTxB6 cross.
Statistical association, as measured by robust correlation, of physiological traits in female mice of the CASTxB6 cross. Each row and each column corresponds to a trait. Numbers within the table represent robust correlations. Cells in the table are color coded using correlation values according to color scale on the right, that is high positive correlations are denoted by strong red color, and high negative correlations by strong green color.
Similarly, the relationship between HDL and plasma glucose levels in mice is sometimes opposite to that in humans. We observe a strong positive relationship in the female congenics but an non-significant (and negative) correlation in the male congenics (Supplementary Figure 2). Across our studied data sets, the HDL-glucose correlations range from moderate (r = 0.41, p=2e-5 in CASTxB6 male) to non-significant (BxH ApoE, Supplementary Figure 3). On the other hand, negative correlation between HDL and plasma glucose has also been reported in multiple studies of Type 2 diabetes (see, e.g., the review (69) and references therein). It appears that the HDL-glucose association in mice depends strongly on genetic background (e.g., strain or cross), diet, and experimental intervention and presents an interesting topic of future research.
Standard analysis of expression data: genes associated with HDL
The expression data consist of 23623 probes measured in 141 female liver samples and 165 female adipose samples that were retained after quality control. Since expression data is only available for the Western diet, we restrict our analysis to HDL on Western diet. Statistical association of each individual probe set with HDL and other quantitative traits was assessed using robust correlation, its associated p value, and the corresponding local false discovery rate (q-value). Supplementary Tables 5 and 6 contain the associations of all probe sets with the measured quantitative traits.
In liver we find 340 probes strongly positively correlated with HDL (Bonferroni-corrected p-value < 0.05) and 134 probes strongly negatively correlated with HDL. The highest enriched GO terms for positively-correlated probes included “catalytic activity” (Bonferroni-corrected p-value 1e-20), “oxidoreductase activity” (p=2e-17) and related terms, “mitochondrion” (p=2e-9), “cytoplasm” (p=2e-8), “metabolic process” (p=5e-6), “lipid metabolic process” (p=2e-4). Negatively-correlated probes exhibited much less significant enrichment (Supplementary Table 7). Enrichment of genes whose adipose expression levels were associated with HDL is detailed in Supplementary Table 8.
Network modules statistically associated with HDL and other clinical traits
We used Weighted Gene Co-expression Network Analysis (Methods) to construct co-expression networks and identify modules of co-expressed probes. The module analysis identified 42 modules with sizes ranging from 25 to 2821 genes. Of all probes, 13713 were assigned to a module, and 9910 were not assigned to a module (Supplementary Figure 4). Supplementary Figure 5 shows the results of a similar network analysis of the adipose data.
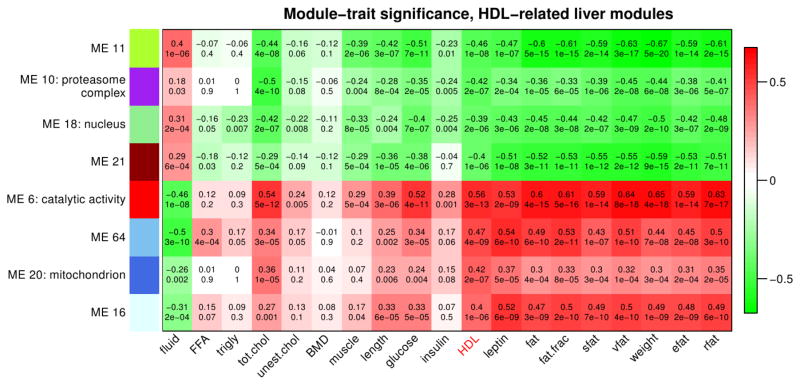
We find several modules with strong statistical association with selected traits (robust correlation between module eigengene and trait above 0.35, corresponding to significant Bonferroni-corrected p-values). Eight modules relate strongly to HDL (Figure 5 and Supplementary Table 9). The module most highly associated with HDL is module 6 (666 probes, r = 0.56, p = 3e-13). As for other traits, we find strong associations of multiple modules, including the HDL-related modules, with adiposity traits (fat weight, fat percentage, subcutaneous fat, etc) and leptin (Figure 5 and Supplementary Figure 6). For example, liver module 6 and fat percentage are highly correlated (r = 0.61, p=5e-16). We note that the correlation of HDL-related modules with adiposity traits is even stronger in adipose, reaching r=0.8 (Supplementary Figure 7 and Supplementary Table 10).
Figure 5. Association of HDL-related liver modules with physiological traits.
Each row corresponds to a module (more precisely, its eigengene), and each column corresponds to a trait. In each cell, we display the robust correlation (top) and the corresponding asymptotic p-value (bottom). Cells in the table are color coded using correlation values according to color scale on the right, that is high positive correlations are denoted by strong red color, and high negative correlations by strong green color.
Functional enrichment analysis of identified co-expression modules
We used the GO enrichment calculation implemented in the WGCNA R package to study enrichment of the found modules in GO categories (GO2000) (70). Supplementary tables 11 and 12 summarize the full results of the enrichment analysis in liver and adipose, respectively. Liver module 6 is enriched in “catalytic activity” (Bonferroni corrected p = 2e-28), “oxidoreductase activity” (p = 4e-27) as well as “lipid metabolic process” (p = 2e-10). Liver module 10 is enriched in terms “cytosol” and “intracellular” (p = 5e-10); liver module 11 shows weak enrichment in “cellular lipid metabolic process” (p = 1e-1); liver module 18 is enriched in “nucleus” (p = 9e-11) and related terms; module 20 is enriched in “mitochondrion” (p = 1e-3); and modules 16, 21, 64 do not exhibit significant GO enrichment.
Annotating all genes by their fuzzy module membership
The WGCNA methodology allows one to annotate all genes (probes) on the microarray by a continuous (fuzzy) measure of membership in all identified modules. In Supplementary Tables 13 and 14 we provide annotation tables for all genes in the analysis that list the gene significance for all traits as well as module membership values in all modules.
HDL-related genes implicated in previous genome-wide association studies
Past genome-wide association studies of HDL levels have implicated a number of genes whose genetic variation has a measurable effect on HDL levels. Here we look at genes identified in the review (71) and a more recent meta-analysis of multiple GWA studies (40). In liver, module 6 contains the following corresponding mouse genes: Angptl3, Fads3. Gckr, Lipc, Mmab; module 10 contains genes Galnt2, Pgs1; module 11 contains genes Apoa1, Scarb4, Fads2; module 16 contains Stard3, Ube2l3; module 21 contains Angptl4, Hnf4a, Pabpc4; and module 64 contains Apoa2.
Preservation of liver modules in adipose
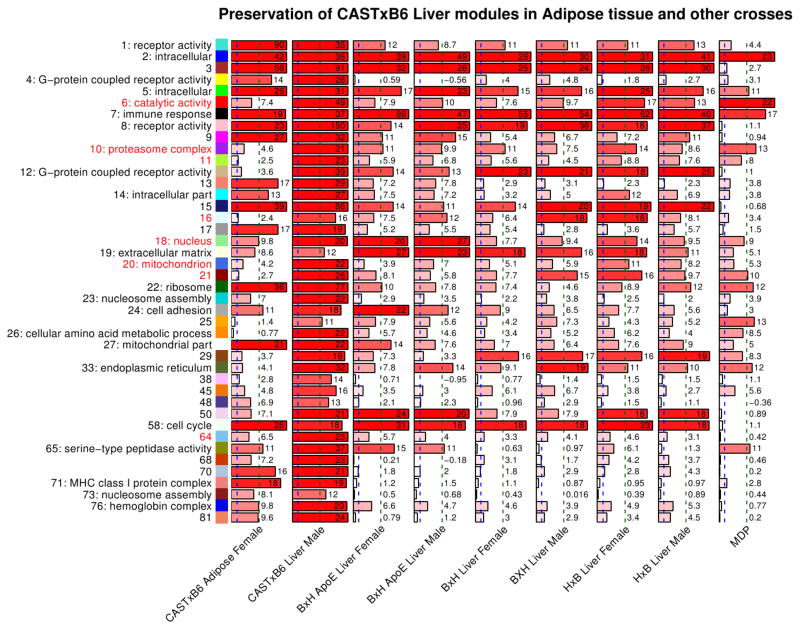
We used the network module preservation statistics (Methods) to study the preservation of liver modules in adipose. We find that multiple liver modules show strong evidence for preservation in adipose (first column in Figure 6). In general, modules whose GO annotation suggests basic cellular function are strongly preserved, for example the ribosomal module 22 exhibits a high preservation statistics Zsummary=36. On the other hand, the HDL-related modules have lower preservation scores. These results suggest that the co-expression patterns of HDL-related modules vary strongly between liver and adipose. In contrast, genes active in basic cellular and molecular functions show much stronger preservation of co-expression between liver and adipose. Supplementary Figure 8 shows the overlap of liver and adipose modules.
Figure 6. Preservation of CASTxB6 female liver modules in the corresponding adipose female, liver male, and other crosses expression data.
Each column represents a single barplot and corresponds to a test set indicated below each column. Each row corresponds to a CASTxB6 female liver module. The modules are indicated on the left side by module color, module number, and the most significant GO annotation. The barplots represent the summary preservation statistics Zsummary for the corresponding module in the test data sets. The scores are also indicated by the numbers next to the bars. The blue and green vertical dashed lines indicate the thresholds Zsummary=2 and 10 for weak and strong evidence of preservation, respectively.
Preservation of female liver network in male data
Although the expression data contain samples from both male and female mice, we restricted the network analysis to female mice only since significant sex differences in expression patterns have been reported in the literature (51). We now use the module preservation statistics to study whether the module-level organization of gene expression is preserved between the female and male CASTxB6 samples. Further, we check whether module eigengene significances for HDL are similar in the female and male data.
We find that all female liver modules show strong evidence of preservation in the male data, with Zsummary≥11 (second column of Figure 6). The HDL-related modules exhibit high preservation scores between 16 and 49. We thus conclude that the module-level organization of female expression is preserved in male samples.
Association of individual probes and modules with HDL are also strongly preserved between the female and male co-expression networks. The module eigengene-HDL correlations are strongly preserved (r = 0.84, p=4e-12, Supplementary Figure 9). Similarly, female and male probe-HDL correlations, when viewed as two vectors, are highly correlated (r = 0.62, p<1e-200).
Studies of network module preservation in other crosses
Thus far we have studied the large-scale organization of gene expression in the CASTxB6 cross. We now apply the network module preservation statistics to study whether the CASTxB6 female liver modules are also present in female and male co-expression networks other liver data sets that include 3 crosses and a more genetically diverse Hybrid Mouse diversity Panel (see Methods for a more detailed description).
Recall that all modules identified in the female CAST x B6 data were strongly preserved in the male CAST x B6 data. In contrast, we find that the preservation in other crosses varies markedly (Figure 6). For example, modules 2 and 5 (GO term “intracellular”) and module 7 (“immune response”) are strongly preserved in all data sets (Zsummary>10). On the other hand, several mostly smaller modules, for example 71 (MHC class I protein complex), 73 (nucleosome) and some modules without strong GO enrichment, are preserved very weakly or not at all (Zsummary<3) in all other data sets.
Most HDL-related modules are moderately to strongly preserved in most or all test data sets. For example, module 6 (catalytic activity) is strongly preserved in MDP (Zsummary=22) and the HxB cross (Zsummary=17 in female and 13 in male). One exception is module 64 that only shows weak evidence of preservation in the crosses (Zsummary between 3.1 and 5.7), and no evidence of preservation in the MDP (Zsummary=0.4). This indicates that module 64 may be unique to the CASTxB6 cross while all other HDL-related modules appear to be more universal.
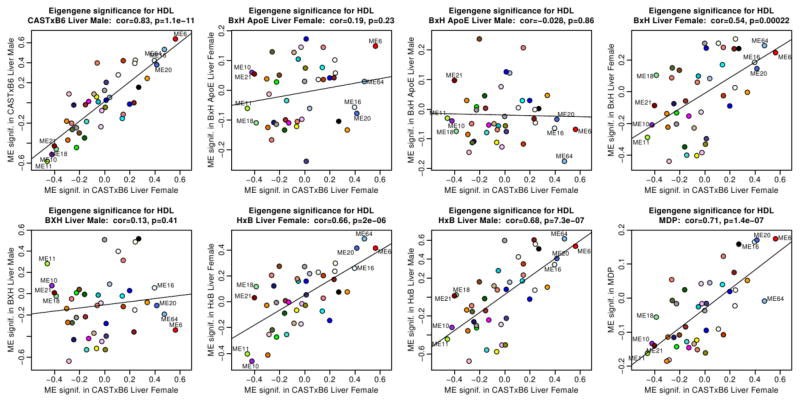
Preservation of module-HDL associations in other crosses
Recall that in the male CAST x B6 liver data, module associations with HDL were strongly preserved. We find (Figure 7) that module-HDL associations are overall preserved in BxH female (r = 0.54, p=2e-4), HxB female (r=0.66, p=2e-6), HxB male (r=0.68, p=7e-7) and HMDP (r=0.71, p=1e-7). In contrast, the module-HDL associations are overall not preserved in BxH ApoE −/− female (r = 0.19, p = 0.2) and male (r = −0.03, p=0.9) and BxH male (r = 0.13, p=0.4). The non-preservation in the ApoE −/− cross may be due to the fact that in this cross the major gene for HDL (Apoa2) accounted for a large fraction of the HDL variance. In the Supplementary Figure 10 we provide evidence that the module significance tends to be better preserved in those test data sets that in which module is more strongly preserved.
Figure 7. Preservation of module eigengene significance for HDL between the CASTxB6 female liver data and liver data from other crosses and the MDP.
Each panel presents a scatterplot of module significance for HDL in one test set (y-axis) vs. module significance for HDL in the female CASTxB6 data (x-axis). The corresponding test data set is indicated in the title. Each point corresponds to a module represented by its eigengene. HDL-related module eigengenes are labeled by their number. These plots indicate that module significance for HDL is preserved in the CASTxB6 male, BxH female, HxB male and female, and MDP data. Module significance for HDL is not preserved in the BxH ApoE −/− female and male data, and in the BxH male data.
Genes consistently associated with HDL
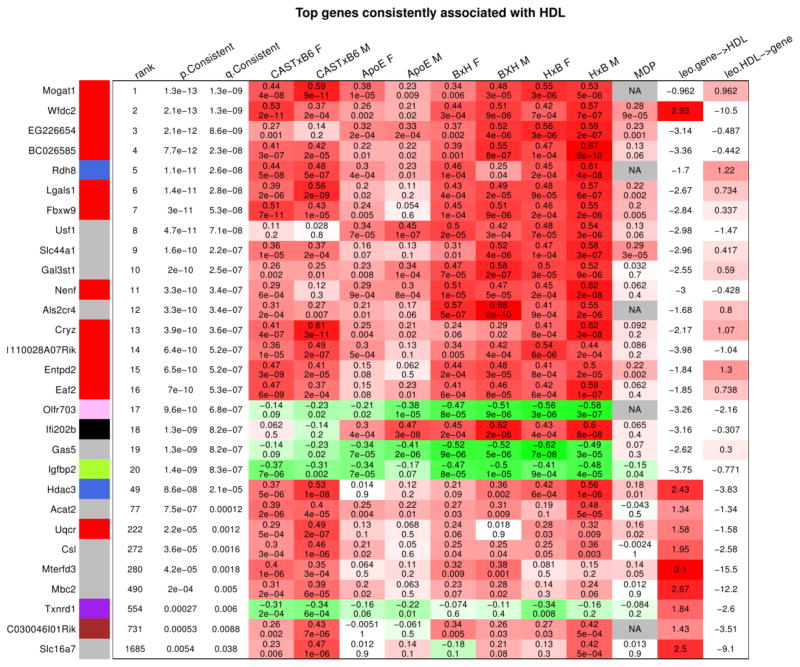
Starting from gene significance for (that is, robust correlation with) HDL, we study genes that relate to HDL most strongly and consistently across the studied data sets (including the reference CASTxB6 female liver and all test data sets). Toward this end, we rank all genes by a p-value derived from standardized gene-HDL correlations across all studied data sets (Methods). The top identified genes are summarized in Figure 8; detailed results are provided in Supplementary Tables 15 and 16. Twelve of the top 20 genes are members of module 6, the most strongly related module in the CASTxB6 female liver.
Figure 8. Top genes consistently associated with HDL across all studied data sets.
Each row corresponds to one gene indicated on the left. The color indicator next to the gene symbol indicates module membership (grey corresponds to genes not assigned to any of the modules). The first two columns give the p- and q-value (local FDR) for the null hypothesis of no consistent association. The following columns show the association (as measured by robust correlation) and the corresponding p-value of the gene and HDL in each of our data sets. The table shows the 20 genes with most significant p-value for consistent association with HDL. We note that 12 of the top 20 genes belong to module 6 (red color)
We used the NEO causal testing software (54) to identify candidate genes whose expression causally affects HDL and vide-versa. Genes with LEO score (Methods) greater than 1 and whose FDR for consistent association with HDL is below 0.05 are also shown in Figure 8. Of the top 20 genes, 4 are reactive candidates (LEO score in the last column of nearly or above 1), and 1 is a causal candidate. We now briefly discuss the top 3 causal candidate genes: Wfdc2, Hdac3, and Acat2.
The WAP four-disulfide core domain 2 (Wfdc2) gene is located on chromosome 2 at approximately 164MB, near the QTL peak at 162MB. In the CAST x B6 cross it has a strong cis-eQTL peak with LOD score of 15.9 which essentially coincides with the HDL QTL peak on chromosome 2 (Supplementary Figure 11). To the best of our knowledge this gene has not been associated with HDL, although it has been observed that its expression increases with higher cholesterol levels in aging canine livers (72). Interestingly, the human WDC2 gene is located on chromosome 20 at base The next causal candidate gene is histone deacetylase 3 (Hdac3). Hdac3 is located on chromosome 18 at approximately 38.1 MB and has a strong cis-eQTL peak with LOD score of 15.3 there. This gene has been implicated in the regulation of hepatic lipid metabolism (73). The third causal gene is acetyl-coenzyme A acetyltransferase 2 (Acat2), a member of the superpathway of cholesterol biosynthesis that catalyzes the synthesis of cholesteryl esters from cholesterol. This gene is located on chromosome 17 where we have not observed a QTL for HDL. It has a moderately strong trans-eQTL (LOD score 5.9) on chromosome 10 near the observed HDL peak.
Discussion
In previous studies, we have examined parameters relating to HDL metabolism and obesity in crosses between CAST and B6 mice (74). Whereas variations among the “classic” inbred strains of mice exhibit relatively subtle variations in HDL levels and functions, we observed tremendous variation in the cross with CAST mice. This undoubtedly reflects the much greater evolution divergence of CAST mice from the set of classic inbred strains. Numerous loci contributing to HDL-C levels were mapped but, altogether, these explained a small fraction of the total HDL variance in the cross. Thus, it is likely that dozens, or perhaps hundreds of loci, control HDL levels and functions in this cross. We also observed that HDL levels exhibited correlations with a variety of metabolic traits, such as body fat and insulin levels, and differ strikingly between sexes. The present study was designed to examine these complex interactions affecting HDL using a systems biology approach. For this, we have analyzed transcript levels globally in liver and adipose tissue of a large cross between the two strains and used this to model biologic networks related to HDL levels.
It is noteworthy that whereas our previous study was performed using mice maintained on an “atherosclerotic” (15% fat, 1.25% cholesterol, and 0.05% cholate) diet, the present study was conducted on a high fat (42% fat, 0.5% cholesterol) diet. This had a striking impact on the HDL-cholesterol QTLs identified, as only the loci on chromosomes 2, 9, and 18 were preserved. In an effort to confirm and fine map the loci from our first study, we have created congenic strains in which the chromosomal regions harboring the QTL loci from strain CAST were transferred by a series of genetic crosses onto the background of strain C57BL/6J. In each case, the analysis of the congenic phenotypes confirmed the original mapping.
These studies have revealed striking correlations, both positive and negative, between HDL and various metabolic parameters. In particular, measures of body fat were strongly associated with HDL cholesterol levels. A number of candidate genes consistently associated with HDL levels in liver and adipose were also identified. Interestingly, most of these genes have not been associated with HDL cholesterol levels in GWAS studies published to date. One likely explanation is that these genes do not exhibit common variations in the human populations studied. Also, these genes could be reactive rather than causal for HDL cholesterol levels. In the latter case, they provide interesting candidates for mechanisms by which HDL protects against atherosclerosis or other inflammatory disorders.
Several co-expression modules in liver were strongly correlated with HDL levels and these also exhibited correlations with various physiologic traits, including insulin levels, leptin levels, and body fat. The modules were found to be enriched in various gene ontology categories such as proteosome complex and mitochondrion. Some of the HDL modules were highly conserved between tissues, sexes, and other crosses. This suggests that they represent fundamental relationships in genes influencing HDL metabolism. Thus, the genes in these modules are good candidates for the regulation of HDL metabolism and the edges between the gene nodes in the modules provide hypotheses for mechanistic interactions.
Human studies have observed that HDL and triglyceride levels (TG) tend to be anti-correlated (26), and at loci that are associated with both HDL and TG the two associations tend to go in the opposite direction. As mentioned earlier, in mice HDL and TG tend to be positively correlated; in our study HDL and TG show a moderate positive correlation of 0.31. For the top 20 genes consistently associated with HDL, we observe strong positive correlations between the gene significance for HDL and gene significance for TG (Supplementary Figure 12). Nine of the 20 genes pass the FDR threshold of 0.01 for consistent association with TG: Wfdc2, EG226654, BC026585, Usf1, Slc44a1, Nenf, Olfr703, Gas5, Igfbp2.
Our study presents several resources for future experimental as well as computational studies. First, we provide a ranking of genes in terms of their consistent association with HDL across several independent data sets. Second, we provide fuzzy membership of all genes in all modules determined from the CAST x B6 cross. Third, by studying the preservation of the identified modules in other data sets we provide an indication how universal (or, conversely, specific to our cross) each found module is. Because our test gene expression data sets were measured on different microarray platforms (in the case of the MDP, using a different technology, Affymetrix 25-mers vs. Agilent 60-mers for the current cross), our findings are less likely to reflect possible array-specific technical artifacts.
Supplementary Material
Highlights.
We investigate genetic factors affecting HDL in a CASTxB6 F2 mouse cross.
Network analysis identifies gene co-expression modules associated with HDL.
Studies across independent data sets confirm robustness of identified modules.
Using meta-analysis techniques we identify genes consistently associated with HDL.
Causal testing implicates Wfdc2 and Hdac3 as novel genes affecting HDL levels.
Acknowledgments
This work was carried out with funds from National Institutes of Health grants HL28481, HL30568 HL60030 and HL70526 (AJL) and the American Heart Association (0655108Y), the Laubish Fund, UCLA, and support from Rosetta Inpharmatics/Merck Inc. We thank Ms. Rosa Chen for help in the preparation of this manuscript.
Footnotes
Availability: Data, R code for statistical analysis, Supplementary Tables, and Supplementary Figures are posted at http://www.genetics.ucla.edu/labs/horvath/CoexpressionNetwork/CASTxB6-HDL/.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gordon DJ, Rifkind BM. High-density lipoprotein--the clinical implications of recent studies. N Engl J Med. 1989;321:1311–1316. doi: 10.1056/NEJM198911093211907. [DOI] [PubMed] [Google Scholar]
- 2.Rader DJ. Molecular regulation of HDL metabolism and function: implications for novel therapies. J Clin Invest. 2006;116:3090–3100. doi: 10.1172/JCI30163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tall AR, Costet P, Wang N. Regulation and mechanisms of macrophage cholesterol efflux. J Clin Invest. 2002;110:899–904. doi: 10.1172/JCI16391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vaisar T, Pennathur S, Green PS, Gharib SA, Hoofnagle AN, Cheung MC, Byun J, Vuletic S, Kassim S, Singh P, et al. Shotgun proteomics implicates protease inhibition and complement activation in the antiinflammatory properties of HDL. J Clin Invest. 2007;117:746–756. doi: 10.1172/JCI26206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chau P, Nakamura Y, Fielding CJ, Fielding PE. Mechanism of prebeta-HDL formation and activation. Biochemistry. 2006;45:3981–3987. doi: 10.1021/bi052535g. [DOI] [PubMed] [Google Scholar]
- 6.Chisholm JW, Burleson ER, Shelness GS, Parks JS. ApoA-I secretion from HepG2 cells: evidence for the secretion of both lipid-poor apoA-I and intracellularly assembled nascent HDL. J Lipid Res. 2002;43:36–44. [PubMed] [Google Scholar]
- 7.Fielding CJ, Fielding PE. Molecular physiology of reverse cholesterol transport. J Lipid Res. 1995;36:211–228. [PubMed] [Google Scholar]
- 8.Tsujita M, Wu CA, Abe-Dohmae S, Usui S, Okazaki M, Yokoyama S. On the hepatic mechanism of HDL assembly by the ABCA1/apoA-I pathway. J Lipid Res. 2005;46:154–162. doi: 10.1194/jlr.M400402-JLR200. [DOI] [PubMed] [Google Scholar]
- 9.Wilson PW, Abbott RD, Castelli WP. High density lipoprotein cholesterol and mortality. The Framingham Heart Study. Arteriosclerosis. 1988;8:737–741. doi: 10.1161/01.atv.8.6.737. [DOI] [PubMed] [Google Scholar]
- 10.Navab M, Reddy ST, Van Lenten BJ, Fogelman AM. HDL and cardiovascular disease: atherogenic and atheroprotective mechanisms. Nat Rev Cardiol. 2011;8:222–232. doi: 10.1038/nrcardio.2010.222. [DOI] [PubMed] [Google Scholar]
- 11.Badimon JJ, Badimon L, Fuster V. Regression of atherosclerotic lesions by high density lipoprotein plasma fraction in the cholesterol-fed rabbit. J Clin Invest. 1990;85:1234–1241. doi: 10.1172/JCI114558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plump AS, Scott CJ, Breslow JL. Human apolipoprotein A-I gene expression increases high density lipoprotein and suppresses atherosclerosis in the apolipoprotein E-deficient mouse. Proc Natl Acad Sci U S A. 1994;91:9607–9611. doi: 10.1073/pnas.91.20.9607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rong JX, Li J, Reis ED, Choudhury RP, Dansky HM, Elmalem VI, Fallon JT, Breslow JL, Fisher EA. Elevating high-density lipoprotein cholesterol in apolipoprotein E-deficient mice remodels advanced atherosclerotic lesions by decreasing macrophage and increasing smooth muscle cell content. Circulation. 2001;104:2447–2452. doi: 10.1161/hc4501.098952. [DOI] [PubMed] [Google Scholar]
- 14.Rubin EM, Krauss RM, Spangler EA, Verstuyft JG, Clift SM. Inhibition of early atherogenesis in transgenic mice by human apolipoprotein AI. Nature. 1991;353:265–267. doi: 10.1038/353265a0. [DOI] [PubMed] [Google Scholar]
- 15.Oram JF. HDL apolipoproteins and ABCA1: partners in the removal of excess cellular cholesterol. Arterioscler Thromb Vasc Biol. 2003;23:720–727. doi: 10.1161/01.ATV.0000054662.44688.9A. [DOI] [PubMed] [Google Scholar]
- 16.Osorio J. Vascular disease: cholesterol-efflux capacity might be the key to the protective effects of HDL. Nat Rev Cardiol. 2011;8:122. doi: 10.1038/nrcardio.2011.9. [DOI] [PubMed] [Google Scholar]
- 17.Fielding PE, Nagao K, Hakamata H, Chimini G, Fielding CJ. A two-step mechanism for free cholesterol and phospholipid efflux from human vascular cells to apolipoprotein A-1. Biochemistry. 2000;39:14113–14120. doi: 10.1021/bi0004192. [DOI] [PubMed] [Google Scholar]
- 18.Khera AV, Cuchel M, de la Llera-Moya M, Rodrigues A, Burke MF, Jafri K, French BC, Phillips JA, Mucksavage ML, Wilensky RL, et al. Cholesterol efflux capacity, high-density lipoprotein function, and atherosclerosis. N Engl J Med. 2011;364:127–135. doi: 10.1056/NEJMoa1001689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ansell BJ, Fonarow GC, Fogelman AM. The paradox of dysfunctional high-density lipoprotein. Curr Opin Lipidol. 2007;18:427–434. doi: 10.1097/MOL.0b013e3282364a17. [DOI] [PubMed] [Google Scholar]
- 20.Barter PJ, Nicholls S, Rye KA, Anantharamaiah GM, Navab M, Fogelman AM. Antiinflammatory properties of HDL. Circ Res. 2004;95:764–772. doi: 10.1161/01.RES.0000146094.59640.13. [DOI] [PubMed] [Google Scholar]
- 21.Kontush A, Chapman MJ. Antiatherogenic function of HDL particle subpopulations: focus on antioxidative activities. Curr Opin Lipidol. 2010;21:312–318. doi: 10.1097/MOL.0b013e32833bcdc1. [DOI] [PubMed] [Google Scholar]
- 22.Bodzioch M, Orso E, Klucken J, Langmann T, Bottcher A, Diederich W, Drobnik W, Barlage S, Buchler C, Porsch-Ozcurumez M, et al. The gene encoding ATP-binding cassette transporter 1 is mutated in Tangier disease. Nat Genet. 1999;22:347–351. doi: 10.1038/11914. [DOI] [PubMed] [Google Scholar]
- 23.Kathiresan S, Melander O, Guiducci C, Surti A, Burtt NP, Rieder MJ, Cooper GM, Roos C, Voight BF, Havulinna AS, et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 2008;40:189–197. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kooner JS, Chambers JC, Aguilar-Salinas CA, Hinds DA, Hyde CL, Warnes GR, Gomez Perez FJ, Frazer KA, Elliott P, Scott J, et al. Genome-wide scan identifies variation in MLXIPL associated with plasma triglycerides. Nat Genet. 2008;40:149–151. doi: 10.1038/ng.2007.61. [DOI] [PubMed] [Google Scholar]
- 25.Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, Heath SC, Timpson NJ, Najjar SS, Stringham HM, et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Demirkan A, Amin N, Isaacs A, Jarvelin MR, Whitfield JB, Wichmann HE, Kyvik KO, Rudan I, Gieger C, Hicks AA, et al. Genetic architecture of circulating lipid levels. Eur J Hum Genet. 2011 doi: 10.1038/ejhg.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.LeBoeuf RC, Puppione DL, Schumaker VN, Lusis AJ. Genetic control of lipid transport in mice. I. Structural properties and polymorphisms of plasma lipoproteins. J Biol Chem. 1983;258:5063–5070. [PubMed] [Google Scholar]
- 28.Machleder D, Ivandic B, Welch C, Castellani L, Reue K, Lusis AJ. Complex genetic control of HDL levels in mice in response to an atherogenic diet. Coordinate regulation of HDL levels and bile acid metabolism. J Clin Invest. 1997;99:1406–1419. doi: 10.1172/JCI119300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rollins J, Chen Y, Paigen B, Wang X. In search of new targets for plasma high-density lipoprotein cholesterol levels: promise of human-mouse comparative genomics. Trends Cardiovasc Med. 2006;16:220–234. doi: 10.1016/j.tcm.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 30.Wang X, Ishimori N, Korstanje R, Rollins J, Paigen B. Identifying novel genes for atherosclerosis through mouse-human comparative genetics. Am J Hum Genet. 2005;77:1–15. doi: 10.1086/431656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X, Le Roy I, Nicodeme E, Li R, Wagner R, Petros C, Churchill GA, Harris S, Darvasi A, Kirilovsky J, et al. Using advanced intercross lines for high-resolution mapping of HDL cholesterol quantitative trait loci. Genome Res. 2003;13:1654–1664. doi: 10.1101/gr.1185803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X, Paigen B. Quantitative trait loci and candidate genes regulating HDL cholesterol: a murine chromosome map. Arterioscler Thromb Vasc Biol. 2002;22:1390–1401. doi: 10.1161/01.atv.0000030201.29121.a3. [DOI] [PubMed] [Google Scholar]
- 33.Wang X, Paigen B. Genetics of variation in HDL cholesterol in humans and mice. Circ Res. 2005;96:27–42. doi: 10.1161/01.RES.0000151332.39871.13. [DOI] [PubMed] [Google Scholar]
- 34.Warden CH, Hedrick CC, Qiao JH, Castellani LW, Lusis AJ. Atherosclerosis in transgenic mice overexpressing apolipoprotein A-II. Science. 1993;261:469–472. doi: 10.1126/science.8332912. [DOI] [PubMed] [Google Scholar]
- 35.Leduc MS, Lyons M, Darvishi K, Walsh K, Sheehan S, Amend S, Cox A, Orho-Melander M, Kathiresan S, Paigen B, et al. The mouse QTL map helps interpret human genome-wide association studies for HDL cholesterol. J Lipid Res. 2011 doi: 10.1194/jlr.M009175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moore KJ, Rayner KJ, Suarez Y, Fernandez-Hernando C. microRNAs and cholesterol metabolism. Trends Endocrinol Metab. 2010;21:699–706. doi: 10.1016/j.tem.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13:423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mehrabian M, Castellani LW, Wen PZ, Wong J, Rithaporn T, Hama SY, Hough GP, Johnson D, Albers JJ, Mottino GA, et al. Genetic control of HDL levels and composition in an interspecific mouse cross (CAST/Ei x C57BL/6J) J Lipid Res. 2000;41:1936–1946. [PubMed] [Google Scholar]
- 39.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4:Article17. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- 40.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mehrabian M, Wong J, Wang X, Jiang Z, Shi W, Fogelman AM, Lusis AJ. Genetic locus in mice that blocks development of atherosclerosis despite extreme hyperlipidemia. Circ Res. 2001;89:125–130. doi: 10.1161/hh1401.093458. [DOI] [PubMed] [Google Scholar]
- 42.Taicher GZ, Tinsley FC, Reiderman A, Heiman ML. Quantitative magnetic resonance (QMR) method for bone and whole-body-composition analysis. Anal Bioanal Chem. 2003;377:990–1002. doi: 10.1007/s00216-003-2224-3. [DOI] [PubMed] [Google Scholar]
- 43.Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- 44.Lander ES, Botstein D. Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics. 1989;121:185–199. doi: 10.1093/genetics/121.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schadt EE, Monks SA, Drake TA, Lusis AJ, Che N, Colinayo V, Ruff TG, Milligan SB, Lamb JR, Cavet G, et al. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- 46.Ravasz E, Somera AL, Mongru DA, Oltvai ZN, Barabasi AL. Hierarchical organization of modularity in metabolic networks. Science. 2002;297:1551–1555. doi: 10.1126/science.1073374. [DOI] [PubMed] [Google Scholar]
- 47.Kaufman L, Rousseeuw PJ. Finding Groups in Data: An Introduction to Cluster Analysis. New York: John Wiley and Sons, Inc; 1990. [Google Scholar]
- 48.Langfelder P, Horvath S. Eigengene networks for studying the relationships between co-expression modules. BMC Syst Biol. 2007;1:54. doi: 10.1186/1752-0509-1-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Langfelder P, Luo R, Oldham MC, Horvath S. Is my network module preserved and reproducible? PLoS Comput Biol. 2011;7:e1001057. doi: 10.1371/journal.pcbi.1001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Nas A, Guhathakurta D, Wang SS, Yehya N, Horvath S, Zhang B, Ingram-Drake L, Chaudhuri G, Schadt EE, Drake TA, et al. Elucidating the role of gonadal hormones in sexually dimorphic gene coexpression networks. Endocrinology. 2009;150:1235–1249. doi: 10.1210/en.2008-0563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang SS, Schadt EE, Wang H, Wang X, Ingram-Drake L, Shi W, Drake TA, Lusis AJ. Identification of pathways for atherosclerosis in mice: integration of quantitative trait locus analysis and global gene expression data. Circ Res. 2007;101:e11–30. doi: 10.1161/CIRCRESAHA.107.152975. [DOI] [PubMed] [Google Scholar]
- 53.Bennett BJ, Farber CR, Orozco L, Kang HM, Ghazalpour A, Siemers N, Neubauer M, Neuhaus I, Yordanova R, Guan B, et al. A high-resolution association mapping panel for the dissection of complex traits in mice. Genome Res. 2010;20:281–290. doi: 10.1101/gr.099234.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aten JE, Fuller TF, Lusis AJ, Horvath S. Using genetic markers to orient the edges in quantitative trait networks: the NEO software. BMC Syst Biol. 2008;2:34. doi: 10.1186/1752-0509-2-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Horvath S, Dong J. Geometric interpretation of gene coexpression network analysis. PLoS Comput Biol. 2008;4:e1000117. doi: 10.1371/journal.pcbi.1000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gargalovic PS, Imura M, Zhang B, Gharavi NM, Clark MJ, Pagnon J, Yang WP, He A, Truong A, Patel S, et al. Identification of inflammatory gene modules based on variations of human endothelial cell responses to oxidized lipids. Proc Natl Acad Sci U S A. 2006;103:12741–12746. doi: 10.1073/pnas.0605457103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Horvath S, Zhang B, Carlson M, Lu KV, Zhu S, Felciano RM, Laurance MF, Zhao W, Qi S, Chen Z, et al. Analysis of oncogenic signaling networks in glioblastoma identifies ASPM as a molecular target. Proc Natl Acad Sci U S A. 2006;103:17402–17407. doi: 10.1073/pnas.0608396103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, Mill J, Cantor RM, Blencowe BJ, Geschwind DH. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schadt EE, Lamb J, Yang X, Zhu J, Edwards S, Guhathakurta D, Sieberts SK, Monks S, Reitman M, Zhang C, et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nat Genet. 2005;37:710–717. doi: 10.1038/ng1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Presson AP, Sobel EM, Papp JC, Suarez CJ, Whistler T, Rajeevan MS, Vernon SD, Horvath S. Integrated weighted gene co-expression network analysis with an application to chronic fatigue syndrome. BMC Syst Biol. 2008;2:95. doi: 10.1186/1752-0509-2-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Plaisier CL, Horvath S, Huertas-Vazquez A, Cruz-Bautista I, Herrera MF, Tusie-Luna T, Aguilar-Salinas C, Pajukanta P. A systems genetics approach implicates USF1, FADS3, and other causal candidate genes for familial combined hyperlipidemia. PLoS Genet. 2009;5:e1000642. doi: 10.1371/journal.pgen.1000642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Park CC, Gale GD, de Jong S, Ghazalpour A, Bennett BJ, Farber CR, Langfelder P, Lin A, Khan AH, Eskin E, et al. Gene networks associated with conditional fear in mice identified using a systems genetics approach. BMC Syst Biol. 2011;5:43. doi: 10.1186/1752-0509-5-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Davis RC, Jin A, Rosales M, Yu S, Xia X, Ranola K, Schadt EE, Lusis AJ. A genome-wide set of congenic mouse strains derived from CAST/Ei on a C57BL/6 background. Genomics. 2007;90:306–313. doi: 10.1016/j.ygeno.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 64.Watts GF, Barrett PH, Chan DC. HDL metabolism in context: looking on the bright side. Curr Opin Lipidol. 2008;19:395–404. doi: 10.1097/MOL.0b013e328306596d. [DOI] [PubMed] [Google Scholar]
- 65.Demirkan A, Amin N, Isaacs A, Jarvelin MR, Whitfield JB, Wichmann HE, Kyvik KO, Rudan I, Gieger C, Hicks AA, et al. Genetic architecture of circulating lipid levels. Eur J Hum Genet. 2011;19:813–819. doi: 10.1038/ejhg.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Su Z, Ishimori N, Chen Y, Leiter EH, Churchill GA, Paigen B, Stylianou IM. Four additional mouse crosses improve the lipid QTL landscape and identify Lipg as a QTL gene. J Lipid Res. 2009;50:2083–2094. doi: 10.1194/jlr.M900076-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wergedal JE, Ackert-Bicknell CL, Beamer WG, Mohan S, Baylink DJ, Srivastava AK. Mapping genetic loci that regulate lipid levels in a NZB/B1NJxRF/J intercross and a combined intercross involving NZB/B1NJ, RF/J, MRL/MpJ, and SJL/J mouse strains. J Lipid Res. 2007;48:1724–1734. doi: 10.1194/jlr.M700015-JLR200. [DOI] [PubMed] [Google Scholar]
- 68.Warden CH, Fisler JS, Shoemaker SM, Wen PZ, Svenson KL, Pace MJ, Lusis AJ. Identification of four chromosomal loci determining obesity in a multifactorial mouse model. J Clin Invest. 1995;95:1545–1552. doi: 10.1172/JCI117827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.von Eckardstein A, Sibler RA. Possible contributions of lipoproteins and cholesterol to the pathogenesis of diabetes mellitus type 2. Curr Opin Lipidol. 22:26–32. doi: 10.1097/MOL.0b013e3283412279. [DOI] [PubMed] [Google Scholar]
- 70.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lusis AJ, Pajukanta P. A treasure trove for lipoprotein biology. Nat Genet. 2008;40:129–130. doi: 10.1038/ng0208-129. [DOI] [PubMed] [Google Scholar]
- 72.Kil DY, Vester Boler BM, Apanavicius CJ, Schook LB, Swanson KS. Age and diet affect gene expression profiles in canine liver tissue. PLoS One. 2010;5:e13319. doi: 10.1371/journal.pone.0013319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Feng D, Liu T, Sun Z, Bugge A, Mullican SE, Alenghat T, Liu XS, Lazar MA. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011;331:1315–1319. doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mehrabian M, Wen PZ, Fisler J, Davis RC, Lusis AJ. Genetic loci controlling body fat, lipoprotein metabolism, and insulin levels in a multifactorial mouse model. J Clin Invest. 1998;101:2485–2496. doi: 10.1172/JCI1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.