Abstract
The rapid accumulation of neuroproteomics data in recent years has prompted the emergence of novel antibody-based imaging methods which aim to understand the anatomical and functional context of the multitude of identified proteins. The pioneering field of ultrastructural multiplexed proteomic imaging now includes a number of high resolution methods, such as array tomography, stimulated emission depletion microscopy, stochastic optical reconstruction microscopy and automated transmission electron microscopy, which allow a detailed molecular characterization of individual synapses and subsynaptic structures within brain tissues for the first time. While all of these methods still face considerable limitations, a combined complementary approach building on the respective strengths of each method is possible and will enable fascinating research into the proteomic diversity of the nervous system.
Introduction
A major challenge for neuroproteomics is the great diversity of the nervous system building blocks. For example, there are at least 60 different neuronal types in the mammalian retina [1] and 24 neuronal types in the CA1 region of hippocampus alone [2]. The number of different synapse types is undoubtedly even higher, at the very least because a synapse is usually made by two neurons. And while neuroproteomics has made great advances in recent years by identifying the majority of proteins at the synapse (~2000), including about 410 proteins of synaptic vesicles [3] and 200 to 1000 proteins of the postsynaptic density [4–7], we still don’t know the make up of a real-life synapse. The structure of the ‘average’ synapse is now documented in great detail, but which of all the identified proteins are found at a particular individual synapse? What different combinations of these proteins exist at different types of synapses? How do we define a synapse type anyway? These are the kinds of questions that we ultimately need to answer to make sense of the proteomic information that is quickly accumulating. The pioneering field of ultrastructural multiplexed proteomic imaging is now beginning to grapple with these questions.
Antibody-based proteomic imaging in situ: What is where and what else is there?
Antibodies have been an invaluable tool for protein localization in situ ever since their introduction for biological imaging [8]. In recent years, the rapid accumulation of proteomics data has prompted the emergence of novel antibody-based imaging methods that aim to address the new challenges (Table 1). A variety of approaches have been explored to achieve multiplexed proteomic imaging of tissues. As a general rule, however, the higher the resolution of the imaging method is, the lower its capability for multiplexing. This is a new variation of the old conundrum faced by immunoelectron microscopy: methods that increase the ultrastructural preservation of tissues, decrease their antigenicity, and thus a delicate balance between the quality of ultrastructure and the level of antigenicity has to be found. Similar to immunoelectron microscopy, multiplexed proteomic imaging requires good antigenicity, and the ability to resolve the protein location with great precision requires good ultrastructural preservation.
Table 1.
Comparison of antibody-based proteomic imaging methods.
| Imaging Technique | Resolution | Multiplexing capability | Live imaging compatible | References |
|---|---|---|---|---|
| Multiplex tissue immunoblotting | Multicellular | Up to 20 antibodies | No | [9–11] |
| 3D registration atlases (for organisms with high degree of stereotypy) | Cellular | Unlimited | Yes | [12] |
| Multi-epitope- ligand cartography / Toponomics | Subcellular (0.2 × 0.2 × 5 μm) | 100 antibodies demonstrated, theoretically more | No | [13,14] |
| Array tomography | Synaptic (200 × 200 × 70 nm) | 24 antibodies demonstrated, theoretically more | No | [15,16] |
| STED | Synaptic- Subsynaptic (80 × 80 × 80 nm in fixed tissue) (65 × 65 × 500 nm for live imaging) | 3 simultaneous colors. Potential for sequential multiplexing | Yes | [18, 19, 46] |
| STORM | Subsynaptic (14 × 14 × 35 nm in fixed tissue) (30 × 30 × 50 nm for live imaging) | 3 antibodies simultaneously; 13 demonstrated with registration using a synaptic coordinate system, theoretically more | Yes | [21, 29] |
| ATEM | Macromolecular (0.5 × 0.5 × 40 nm) | 11 antibodies demonstrated, theoretically more | No | [26–28] |
On one end of the spectrum of proteomic imaging methods are those that can resolve groups of cells, individual cells or even subcellular structures, without reaching the level of individual synapses. Multiplex tissue immunoblotting is based on protein transfer from a tissue onto a stack of membranes, which can then be labeled with multiple antibodies and protein distribution can be determined at multicellular level [9–11]. While this method has only been used with up to 20 antibodies, it can potentially allow labeling with a much larger number of antibodies, by using multiple immunofluorescence on each membrane and then eluting and restaining the membranes. Another approach to proteomic imaging takes advantage of the stereotypy of organisms like Drosophila and uses sophisticated 3D registration atlases that can align an unlimited number of similar tissues immunostained or labeled by a different method, thus basically achieving unlimited multiplexing capabilities with a cellular resolution [12]. On the other hand, multi-epitope-ligand cartography, or toponomics, exploits the bleaching property of fluorophores which can allow multiple fluorescently labeled antibodies to be applied sequentially to the same tissue, thus visualizing the distribution of up to a hundred of proteins within the same tissue with subcellular resolution [13,14]. The high multiplexing potential of all of these methods has fostered a new appreciation for the complexity of the nervous tissue [12,14] and has already hinted at the enormous diversity at synaptic level, even without the capabilities of individual synapse resolution [14].
On the other end of the spectrum of proteomic imaging methods are those that can resolve individual synapses and subsynaptic structures. Array tomography [15,16] has allowed the imaging of up to 24 different antibodies at individual synapses within brain tissue, and the potential for multiplexing is greater. The high spatial resolution of this wide-field fluorescence microscopy based method is enabled by the use of ultrathin serial sections (70 nm) of resin-embedded tissue, and its multiplexing capabilities are due to the possibility for multiple rounds of immunofluorescent labeling, imaging and antibody elution on such sections. This approach is currently limited to a lateral resolution of ~200 nm, the diffraction limit of conventional light microscopy.
Recent fluorescent microscopy methods have broken the diffraction limit, allowing imaging at resolutions intermediate between conventional light microscopy and electron microscopy. Two approaches in particular, STED and STORM, have shown significant potential in the analysis of neurons and synaptic structures. STED (stimulated emission depletion microscopy) is a confocal scanning method that achieves diffraction-unlimited resolution by spatially confining the emitting fluorophores with a second overlapped “depletion” beam that forces excited molecules back to the ground state [17]. As a scanning method, this approach has the speed of conventional confocal microscopy, but a resolution typically <50–80 nm in the lateral dimension, though preserving conventional axial resolution. To improve the axial resolution, STED was integrated with ultrathin sectioning and image alignment to achieve a reconstructed image of fixed hippocampal neurons stained with two different antibodies with <80 nm resolution in all dimensions [18]. While these experiments were carried out with sections stained prior to embedding, this approach seems well suited to integration with array tomography sections to increase its multiplexing capabilities. Implementing multicolor imaging with STED though is not as straightforward as with wide-field fluorescence, because it involves multiple wavelengths (excitation, emission and depletion of each fluorophore). A simultaneous three-color STED was demonstrated only recently in cultured cells immunostained with antibodies [19].
STORM [20] (stochastic optical reconstruction microscopy) is a superresolution fluorescence microscopy method that achieves resolution beyond the diffraction limit (typically ~30 nm lateral resolution) by single-molecule imaging of photoswitchable fluorescent probes. STORM has enabled the mapping of the immunofluorescence of 13 antibodies onto a common synaptic coordinate system [21], albeit across multiple distinct sections. Multiplexable STORM probes have been demonstrated using distinct pairs of activator and reporter dyes, which would allow for at least 3 color, and potentially up to 9 color simultaneous superresolution labeling [22]. Even though the full potential of STORM for antibody multiplexing has not yet been explored, it is easy to see how STORM could be a great asset in proteomic imaging, for example by using it with array tomography sections.
With STORM, localization works best for objects near the focal plane, because under single photon switching molecules outside the focal plane are activated, contributing out-of-focus background and introducing photobleaching artifacts. Two-photon photoactivation, which confines the activated fluorophores within the focused optical section [23] substantially enhances the performance of single-molecule based methods like STORM in thicker tissue specimens, though at the cost of speed, since each cycle of photoactivation involves scanning the two-photon excitation, and is limited by the speed of the scan-head. An improvement that overcomes the logistical scanning limit of this approach was implemented using temporal focusing of the photoactivation beam [24]. In this approach, a high speed galvo-mirror scans the beam along one axis, while structuring of the excitation pulse results in a spatial displacement of the focused beam across the other axis. Hence, the speed of photoactivation is limited by only one physically scanning mirror, decreasing the scan time and maintaining a resolution of <50 nm lateral and <100 nm axial. Generally, these approaches have been used in cells, not yet in tissues. Recently, however, a four-color super-resolution imaging based on single molecule switching and using only a single laser for excitation was applied to imaging in tissues as well [25].
Finally, the highest resolution (0.5 nm lateral resolution) has been achieved by ATEM (automated transmission electron microscopy) [26–28]. Although originally not a strictly speaking proteomic, but metabolomic imaging method, ATEM is based on small molecule immunostaining of ultrathin sections imaged by light microscopy intercalated within a large series of ultrathin sections imaged by automated electron microscopy. Alignment of the immunostained sections with the EM sections allows mapping of the immunolabels onto the tissue ultrastructure. Up to 11 different antibody labels have been used in this manner and in recent studies those have included antibodies not only for small molecules but also for proteins [28]. Remarkably, ATEM, despite being an electron microscopy method, also has the capability of imaging large tissue volumes. For example, it has allowed the imaging and full reconstruction at a 2 nm resolution of a retinal circular segment with a diameter of 0.22 mm and an approximate thickness of 0.03 mm [27]. To better illustrate the scale of this effort, the 16.5 terabyte imaged volume comprised more than 350, 000 image tiles that were captured over five months at a rate of 3,000 images/day.
The high-resolution methods, such as array tomography, STED, STORM and ATEM, are finally beginning to enable researchers to directly address long-standing questions about the scope of synaptic diversity, the plasticity of the synaptic molecular architecture and the complexity of synaptic connections in mammalian brain and retina.
Antibody-based proteomic imaging in vivo: are we there yet?
Protein location in the live brain is anything but fixed. And while a lot can be inferred about protein function from a snapshot of its ultrastructural localization and its spatial relationship to other proteins, one ultimately needs to know the dynamics of proteins in the live organism. Meanwhile, the great majority of proteomic imaging methods require the use of chemically fixed tissues. In a recent study, STORM was effectively used for two-color 3D super-resolution imaging in live BS-C-1 cells [29]. Two-color live STED imaging was also achieved recently [30], albeit not with antibody labeling but using self-labeling protein tags like SNAP- and CLIP-tags. And while two-color imaging is still very far from multiplexing, these studies suggest the enticing possibility that a high-resolution method with multiplexing capabilities can be applied to live imaging. Single particle tracking studies with enhanced multiplexing capabilities can also potentially be used for live imaging as suggested by a recent study [31] in which the mobility of individual α7 nicotinic receptors labeled with α-bungarotoxin linked to quantum dots was imaged at immunocytochemically defined synapses in cultured hippocampal neurons. Another recent approach demonstrated simple labeling for single particle tracking and potential superresolution imaging of endogenous proteins in living cells by binding labeled antibodies to targets at low concentration [32], allowing dynamic imaging across hundreds to thousands of trajectories for each molecule class. In principle, such an approach is limited only by the number of fluorophores that can be simultaneously detected and the ability of an antibody to access the target protein. Generally, the challenge to multiplexed proteomic imaging in living cells remains significant. The dynamics of cellular proteins are sufficiently fast that substantial motion occurs in the time it takes to switch filters on a microscope, potentially losing correlations that could emerge with simultaneous imaging. Innovations in multiplexing that resulted in more cleanly distinguishable signals with similar emission brightness could move proteomic imaging toward living cells.
More and better antibodies: Will they get us there or should we look elsewhere?
Despite all the great advances in biology that were enabled by the use of antibodies, the list of complaints against them is quite impressive. Here are just a few of them, which are most relevant to proteomic imaging:
Target proteins must be known in advance, so there is little room for spontaneous discoveries;
Antibodies are seldom able to distinguish between multiple isoforms of the same protein;
Proteins expressed at low levels may remain undetected by antibodies, even though they have an important function at the site of their expression – e.g. myosin motor II MyH7b in dendritic spines [33];
The primary and secondary antibodies are large enough to result in a displacement of the immunolabel of up to 30 nm away from the antigen [34].
The expanding search for better antibodies (chromobodies [35], fluobodies [36]) and other high affinity protein-binding ligands (affibodies [37], synbodies, [38]) may soon provide new powerful tools for proteomic imaging (Table 2). These probes are smaller than conventional antibodies, can be expressed in bacteria or chemically synthesized, and can be easily site-specifically linked to fluorescent reporters, by genetic or chemical means. This serves to simplify the labeling protocol, reducing both the issues that arise with antibody heterogeneity and with localization spread described above.
Table 2.
Recent antibody advances.
| Antibody advances | Advantages for proteomic imaging |
|---|---|
| Heavy-chain antibodies: nanobodies [47], chromobodies [35] fluobodies [36] | Much smaller than conventional antibodies: better resolution. Can recognize epitopes that are not accessible to conventional antibodies. Allow live cell imaging regardless of the compartmentalization of the protein. |
| Systematic high- throughput generation and screening of antibodies: The Human Protein Atlas Project [40] | High quality well characterized antibodies specific for human proteins Expected to cover all human proteins by 2015 |
| Beyond antibodies: engineered protein scaffolds [37]: affibodies, adnectins, DARPins, synbodies [38] | Possibility to engineer their affinity and specificity. Much smaller than conventional antibodies: better resolution. Can be directly fused with fluorescent proteins/reporters. Lower cost, reduced production time (synbodies). |
Future directions
Many of the antibody imaging methods introduced in this article are mutually compatible, which can allow a combined complementary approach building on the respective strengths of each method. For example, the superresolution advantage of STORM and STED can be combined with the high multiplexing capabilities of array tomography. Further down the road, after optimization of antibodies and tissue preparation, array tomography and ATEM (or ATLUM [39]) of the same tissue samples can provide detailed proteomic information of ultrastructurally resolved synapses with known connectivity. And while live imaging approaches lag behind in their multiplexing capabilities, tissues imaged in vivo can be retrospectively analyzed using array tomography, STORM, STED and ATEM, or a combination of these methods.
The ultimate goal of proteomic imaging will be to obtain a proteomic map of the human brain with subsynaptic resolution. In addition to the need to improve the existing proteomic imaging methods described above, the human brain presents further challenges because of its large size, great variability between different regions and individuals, and the lack of suitably preserved tissue except for small regions excised during surgeries. However, all the pieces now exist to begin the proteomic imaging analysis of the human brain– one small snippet at a time. Array tomography, STED, STORM and ATEM can be all applied to the human brain. Many of the antibodies produced against proteins from rodents or other species recognize the human proteins just as well and, most importantly, the arsenal of good quality human specific antibodies keeps growing [40] and is expected to cover all non-redundant human proteins by 2015. Even though the convenient transgenic technologies that allow labeling of specific neuronal populations [41,42] are not an option for human tissue, intracellular injections of dyes, e.g. Lucifer Yellow [43] can be successfully used for fixed human tissue. As an added bonus Lucifer Yellow withstands the harsh treatments needed for good tissue preservation at the EM level and can be detected with antibodies to visualize the ultrastructure of the filled neurons and their input and output [44].
Conclusions
The perfect method for multiplexed proteomic imaging of tissue ultrastructure does not exist. Arguably, there is not yet even a method that can do a passable job at this formidable task. But the handful of emerging approaches are already offering a fascinating glimpse at the proteomic diversity of the nervous system and the reassurance that comprehensive neuroproteomics at the ultrastructural level is an attainable goal in the near future. Perhaps a slight modification of Floyd Bloom’s famous statement is in order here: “The gain in brain lies mainly in the stains.” [45].
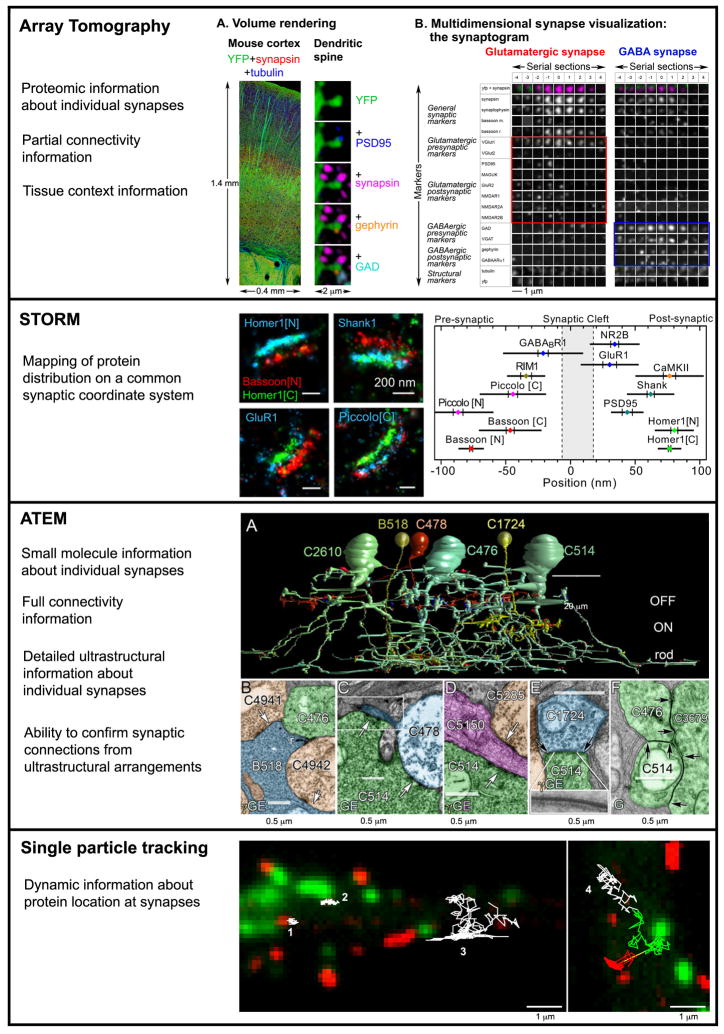
Figure 1. Different proteomic views of the synapse.
Array tomography, STORM and ATEM provide detailed snapshots of multitudes of individual synapses, while single particle tracking reveals the dynamic nature of proteins at smaller subsets of synapses. Array tomography: synapse visualization as volume renderings (A) or as “synaptograms” (B). Synaptograms display high dimensional proteomic data with columns representing individual serial sections through a synapse and rows representing each marker. The two synaptograms show examples of a glutamatergic synapse with glutamatergic markers boxed in red and a GABAergic synapse with the respective markers boxed in blue. Reproduced with permission from [16]. STORM: A synaptic coordinate system is defined by the positions of Bassoon (presynaptic) and Homer 1 (postsynaptic). Triple immunolabeling allows the localization of other synaptic proteins along these coordinates. Reproduced with permission from [21]. ATEM: A fragment of the mammalian AII amacrine cell network is visualized (A) with all synaptic and gap junction contacts mapped. Examples of synaptic connections between the reconstructed cells are shown in B. ATEM datasets contain both ultrastructural and molecular markers information. E, glutamate, G, glycine, γ, GABA. Reproduced with permission from [27]. Single particle tracking: The trajectories of single quantum dot-labeled α7-nicotinic acetylcholine receptors (white) are shown in relation to glutamatergic synapses (red, mCherry-Homer1c) and GABAergic synapses (green, EGFP-gephyrin) in cultured hippocampal interneurons. Reproduced from [31].
Highlights.
Recent advances in neuroproteomics have prompted the emergence of novel antibody-based multiplexed imaging methods
These methods are now beginning to offer a glimpse into the great proteomic diversity of synapses
A combination of these methods can allow a future comprehensive proteomic imaging at an ultrastructural level
Acknowledgments
This work was supported by NIH/NINDS RO1NS075252, R21NS063210, and Gatsby Charitable Foundation (KDM) and NIH 7U54RR022241 and NIH 5R01GM086237 (MPB). We thank Drs Nancy O’Rourke and Stephen J Smith (Stanford University) for their very helpful comments on the manuscript.
Footnotes
Conflict of interest
KDM is cofounder of Aratome, LLC, a company working on the commercial application of array tomography, and holds a patent about array tomography technology.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Kristina D. Micheva, Email: kmicheva@stanford.edu.
Marcel P. Bruchez, Email: bruchez@cmu.edu.
References
- 1.Masland RH. Neuronal diversity in the retina. Curr Opin Neurobiol. 2001;11 :431–436. doi: 10.1016/s0959-4388(00)00230-0. [DOI] [PubMed] [Google Scholar]
- 2.Klausberger T, Somogyi P. Neuronal diversity and temporal dynamics: the unity of hippocampal circuit operations. Science. 2008;321:53–57. doi: 10.1126/science.1149381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takamori S, Holt M, Stenius K, Lemke EA, Grønborg M, Riedel D, Urlaub H, Schenck S, Brügger B, Ringler P, Müller SA, Rammner B, Gräter F, Hub JS, De Groot BL, Mieskes G, Moriyama Y, Klingauf J, Grubmüller H, Heuser J, Wieland F, Jahn R. Molecular anatomy of a trafficking organelle. Cell. 2006;127:831–46. doi: 10.1016/j.cell.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 4.Cheng D, Hoogenraad CC, Rush J, Ramm E, Schlager MA, Duong DM, Xu P, Wijayawardana SR, Hanfelt J, Nakagawa T, Sheng M, Peng J. Relative and absolute quantification of postsynaptic density proteome isolated from rat forebrain and cerebellum. Mol Cell Proteomics. 2006;5:1158–70. doi: 10.1074/mcp.D500009-MCP200. [DOI] [PubMed] [Google Scholar]
- 5.Collins MO, Husi H, Yu L, Brandon JM, Anderson CN, Blackstock WP, Choudhary JS, Grant SG. Molecular characterization and comparison of the components and multiprotein complexes in the postsynaptic proteome. J Neurochem. 2006;97 (Suppl 1):16–23. doi: 10.1111/j.1471-4159.2005.03507.x. [DOI] [PubMed] [Google Scholar]
- 6.Dosemeci A, Makusky AJ, Jankowska-Stephens E, Yang X, Slotta DJ, Markey SP. Composition of the synaptic PSD-95 complex. Mol Cell Proteomics. 2007;6:1749–60. doi: 10.1074/mcp.M700040-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trinidad JC, Thalhammer A, Specht CG, Lynn AJ, Baker PR, Schoepfer R, Burlingame AL. Quantitative analysis of synaptic phosphorylation and protein expression. Mol Cell Proteomics. 2008;7:684–96. doi: 10.1074/mcp.M700170-MCP200. [DOI] [PubMed] [Google Scholar]
- 8.Coons AH, Creech HJ, Jones RN. Immunological properties of an antibody containing a fluorescent group. Proc Soc Expt Biol Med. 1941;47:200–202. [Google Scholar]
- 9.Chung J-Y, Braunschweig T, Baibakov G, Galperin M, Ramesh A, Skacel M, Gannot G, Knezevic V, Hewitt SM. Transfer and multiplex immunoblotting of a paraffin embedded tissue. Proteomics. 2006;6:767–774. doi: 10.1002/pmic.200401343. [DOI] [PubMed] [Google Scholar]
- 10.Gannot G, Tangrea MA, Erickson HS, Pinto PA, Hewitt SM, Chuaqui RF, Gillespie JW, Emmert-Buck MR. Layered peptide array for multiplex immunohistochemistry. J Mol Diagnostics. 2007;9:297–304. doi: 10.2353/jmoldx.2007.060143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chung J-Y, Hong SM, Choi BY, Cho H, Yu E, Hewitt SM. The expression of phospho-AKT, phospho-mTOR, and PTEN in extrahepatic cholangiocarcinoma. Clin Cancer Res. 2009;15:660–667. doi: 10.1158/1078-0432.CCR-08-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peng H, Chung P, Long F, Qu L, Jenett A, Seeds AM, Myers EW, Simpson JH. BrainAligner: 3D registration atlases of Drosophila brains. Nat Methods. 2011;8 :493–498. doi: 10.1038/nmeth.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schubert W, Bonnekoh B, Pommer AJ, Philipsen L, Bockelmann R, Malykh Y, Gollnick H, Friedenberger M, Bode M, Dress AWM. Analyzing proteome topology and function by automated multidimensional fluorescence microscopy. Nat Biotechnol. 2006;24:1270–1278. doi: 10.1038/nbt1250. [DOI] [PubMed] [Google Scholar]
- 14.Bode M, Irmler M, Friedenberger M, May C, Jung K, Stephan C, Meyer HE, Lach C, Hillert R, Krusche A, Beckers J, Marcus K, Schubert W. Interlocking transcriptomics, proteomics and toponomics technologies for brain tissue analysis in murine hippocampus. Proteomics. 2008;8:1170–1178. doi: 10.1002/pmic.200700742. [DOI] [PubMed] [Google Scholar]
- 15.Micheva KD, Smith SJ. Array tomography: a new tool for imaging the molecular architecture and ultrastructure of neural circuits. Neuron. 2007;55 :25–36. doi: 10.1016/j.neuron.2007.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **16.Micheva KD, Busse B, Weiler NC, O’Rourke N, Smith SJ. Single-synapse analysis of a diverse synapse population: proteomic imaging methods and markers. Neuron. 2010;68:639–653. doi: 10.1016/j.neuron.2010.09.024. High-throughput array tomography measures 18 molecular markers at individual synapses in mouse cortex with a resolution of approximately 200nm × 200nm × 70nm. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hell SW, Wichmann J. Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt Lett. 1994;19:780–782. doi: 10.1364/ol.19.000780. [DOI] [PubMed] [Google Scholar]
- **18.Punge A, Rizzoli SO, Jahn R, Wildanger JD, Meyer L, Schönle A, Kastrup L, Hell SW. 3D reconstruction of high-resolution STED microscope images. Microsc Res Tech. 2008;71:644–650. doi: 10.1002/jemt.20602. Thin serial sections were combined with 2D STED microscopy to achieve an 80 nm isotropic imaging resolution for a 3-d reconstruction of hippocampal neurons. [DOI] [PubMed] [Google Scholar]
- *19.Bückers J, Wildanger D, Vicidomini G, Kastrup L, Hell SW. Simultaneous multi-lifetime multi-color STED imaging for colocalization analyses. Opt Express. 2011;19:3130–43. doi: 10.1364/OE.19.003130. An interesting demonstration of a simplified approach for multiparamater STED imaging that relies on lifetime and color to decrease the number of optical constraints in the system (i.e. excitation, depletion, detection channels) [DOI] [PubMed] [Google Scholar]
- 20.Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **21.Dani A, Huang B, Bergan J, Dulac C, Zhuang X. Superresolution imaging of chemical synapses in the brain. Neuron. 2010;68:843–56. doi: 10.1016/j.neuron.2010.11.021. A powerful illustration of STORM imaging across the stereotypic synaptic reference frame to align 13 different proteins with the presynaptic or postsynaptic side of the terminal. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bates M, Huang B, Dempsey GT, Zhuang X. Multicolor super-resolution imaging with photo-switchable fluorescent probes. Science. 2007;317:1749–53. doi: 10.1126/science.1146598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Folling J, Belov V, Kunetsky R, Medda R, Schönle A, Egner A, Eggeling C, Bossi M, Hell SW. (2007) Photochromic rhodamines provide nanoscopy with optical sectioning. Angew Chem Int Ed. 2007;46:6266–6270. doi: 10.1002/anie.200702167. [DOI] [PubMed] [Google Scholar]
- 24.York AG, Ghitani A, Vaziri A, Davidson MW, Shroff H. (2011) Confined activation and subdiffractive localization enables whole-cell PALM with genetically expressed probes. Nat Methods. 2011;8:327–333. doi: 10.1038/nmeth.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baddeley D, Crossman D, Rossberger S, Cheyne JE, Montgomery JM, Jayasinghe ID, Cremer C, Cannell MB, Soeller C. 4D Super-Resolution Microscopy with Conventional Fluorophores and Single Wavelength Excitation in Optically Thick Cells and Tissues. PLoS One. 2011;6(5):e20645. doi: 10.1371/journal.pone.0020645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anderson JR, Jones BW, Yang J-H, Shaw MV, Watt CB, Koshevoy P, Spaltenstein J, Jurrus E, UVK, Whitaker RT, Mastronarde D, Tasdizen T, Marc RE. A computational framework for ultrastructural mapping of neural circuitry. PLoS Biol. 2009;7:e1000074. doi: 10.1371/journal.pbio.1000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **27.Anderson JR, Jones BW, Watt CB, Shaw MV, Yang JH, Demill D, Lauritzen JS, Lin Y, Rapp KD, Mastronarde D, Koshevoy P, Grimm B, Tasdizen T, Whitaker R, Marc RE. Exploring the retinal connectome. Mol Vis. 2011;17:355–79. A truly impressive 16.5 terabyte connectome data set from rabbit retina at 2 nm lateral resolution obtained with automated transmission electron microscope imaging. The data set represents a column of tissue 0.25 mm in diameter spanning the inner nuclear, inner plexiform and ganglion cell layers and containing 6 molecular markers, including an in vivo activity marker. [PMC free article] [PubMed] [Google Scholar]
- 28.Jones BW, Kondo M, Terasaki H, Watt CB, Rapp K, Anderson J, Lin Y, Shaw MV, Yang JH, Marc RE. Retinal remodeling in the TgP347L rabbit, a large-eye model of retinal degeneration. J Comp Neurol. 2011 Jun 16; doi: 10.1002/cne.22703. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- **29.Jones SA, Shim SH, He J, Zhuang X. Fast, three-dimensional super-resolution imaging of live cells. Nat Methods. 2011;8:499–505. doi: 10.1038/nmeth.1605. Using high excitation power and fast-switching probes, 25 nm resolved 2-d images can be obtained in 0.5 seconds and 3-d images with 30x30x50 nm can be obtained in 1-2 seconds for clatrhrin and transferrin in clathrin coated pits. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pellett PA, Sun X, Gould TJ, Rothman JE, Xu M-Q, Corrêa IR, Jr, Bewersdorf J. Two-color STED microscopy in living cells. Biomed Optics Express. 2011;2:2364–2371. doi: 10.1364/BOE.2.002364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *31.Bürli T, Baer K, Ewers H, Sidler C, Fuhrer C, Fritschy JM. Single particle tracking of alpha7 nicotinic AChR in hippocampal neurons reveals regulated confinement at glutamatergic and GABAergic perisynaptic sites. PLoS One. 2010;5:e11507. doi: 10.1371/journal.pone.0011507. QD labeled receptors were tracked in the context of post-synaptic site markers, and under a range of distinct stimulatory and inhibitory treatments, and the functional consequence of treatments on receptor motility was revealed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **32.Giannone G, Hosy E, Levet F, Constals A, Schulze K, Sobolevsky AI, Rosconi MP, Gouaux E, Tampé Choquet D, Cognet L. Dynamic superresolution imaging of endogenous proteins on living cells at ultra-high density. Biophys J. 2010;99:1303–1310. doi: 10.1016/j.bpj.2010.06.005. Low concentrations of labeled antibodies are used for dynamic labeling and tracking of single molecules in living cells. This approach is compatible with labeling endogenous proteins on natural cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rubio MD, Johnson R, Miller CA, Huganir RL, Rumbaugh G. Regulation of synapse structure and function by distinct myosin II motors. J Neurosci. 2011;31 :1448–1460. doi: 10.1523/JNEUROSCI.3294-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bergersen LH, Storm-Mathisen J, Gundersen V. Immunogold quantification of amino acids and proteins in complex subcellular compartments. Nat Protoc. 2008;3:144–152. doi: 10.1038/nprot.2007.525. [DOI] [PubMed] [Google Scholar]
- 35.Schmidthals K, Helma J, Zolghadr K, Rothbauer U, Leonhardt H. Novel antibody derivatives for proteome and high-content analysis. Anal Bioanal Chem. 2010;397:3203–3208. doi: 10.1007/s00216-010-3657-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olichon A, Surrey T. Selection of genetically encoded fluorescent single domain antibodies engineered for efficient expression in Escherichia coli. J Biol Chem. 2007;282:36314–36320. doi: 10.1074/jbc.M704908200. [DOI] [PubMed] [Google Scholar]
- 37.Gebauer M, Skerra A. Engineered protein scaffolds as next-generation antibody therapeurics. Curr Opin Chem Biol. 2009;13:245–255. doi: 10.1016/j.cbpa.2009.04.627. [DOI] [PubMed] [Google Scholar]
- 38.Diehnelt CW, Shah M, Gupta N, Belcher PE, Greving MP, Stafford P, Johnston SA. Discovery of high-affinity protein binding ligands – backwards. PLoS One. 2010;5(5):e10728. doi: 10.1371/journal.pone.0010728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kasthuri N, Lichtman JW. Neurocartography. Neuropsychopharm. 2010;35 :342–343. doi: 10.1038/npp.2009.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **40.Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S, Wernerus H, Björling L, Ponten F. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol. 2010;28:1248–1250. doi: 10.1038/nbt1210-1248. A highly ambitious project with the objective to generate at least two antibodies for all human protein-coding genes to allow knowledge-based annotated protein expression data for the complete human proteome. [DOI] [PubMed] [Google Scholar]
- 41.Feng G, Mellor RH, Bernstein M, Keller-Peck C, Nguyen QT, Wallace M, Nerbonne JM, Lichtman JW, Sanes JR. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28:41–51. doi: 10.1016/s0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- 42.Kuhlman SJ, Huang ZJ. High-resolution labeling and functional manipulation of specific neuron types in mouse brain by Cre-activated viral gene expression. PLoS One. 2008;3:e2005. doi: 10.1371/journal.pone.0002005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Benavides-Piccione R, Arellano JI, De Felipe J. Catecholaminergic innervation of pyramidal neurons in the human temporal cortex. Cereb Cortex. 2005;15 :1584–1591. doi: 10.1093/cercor/bhi036. [DOI] [PubMed] [Google Scholar]
- 44.Oberti D, Kirschmann MA, Hahnloser RH. Projection neuron circuits resolved using correlative array tomography. Front Neurosci. 2011;5:50. doi: 10.3389/fnins.2011.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Appel NM. Classical and contemporary histochemical approaches for evaluating central nervous system microanatomy. Ann N Y Acad Sci. 1997;820:14–28. doi: 10.1111/j.1749-6632.1997.tb46186.x. [DOI] [PubMed] [Google Scholar]
- 46.Lauterbach MA, Keller J, Schönle A, Kamin D, Westphal V, Rizzoli SO, Hell SW. Comparing video-rate STED nanoscopy and confocal microscopy of living neurons. J Biophotonics. 2010;3:417–424. doi: 10.1002/jbio.201000038. [DOI] [PubMed] [Google Scholar]
- 47.Muyldermans S. Single domain camel antibodies: current status. J Biotechnol. 2001;74:277–302. doi: 10.1016/s1389-0352(01)00021-6. [DOI] [PubMed] [Google Scholar]