Abstract
The influence of two binary vector systems, pGreen and pCAMBIA, on the Agrobacterium-mediated transformation ability of wheat and triticale was studied. Both vectors carried selection cassettes with bar or nptII driven by different promoters. Two cultivars of wheat, Kontesa and Torka, and one cultivar of triticale, Wanad, were tested. The transformation rates for the wheat cultivars ranged from 0.00 to 3.58% and from 0.00 to 6.79% for triticale. The best values for wheat were 3.58% for Kontesa and 3.14% for Torka, and these were obtained after transformation with the pGreen vector carrying the nptII selection gene under the control of 35S promoter. In the case of the bar selection system, the best transformation rates were, respectively, 1.46 and 1.79%. Such rates were obtained when the 35S::bar cassette was carried by the pCAMBIA vector; they were significantly lower with the pGreen vector. The triticale cultivar Wanad had its highest transformation rate after transformation with nptII driven by 35S in pCAMBIA. The bar selection system for the same triticale cultivar was better when the gene was driven by nos and the selection cassette was carried by pGreen. The integration of the transgenes was confirmed with at least three pairs of specific starters amplifying the fragments of nptII, bar, or gus. The expression of selection genes, measured by reverse transcriptase polymerase chain reaction (RT-PCR) in relation to the actin gene, was low, ranging from 0.00 to 0.63 for nptII and from 0.00 to 0.33 for bar. The highest relative transcript accumulation was observed for nptII driven by 35S and expressed in Kontesa that had been transformed with pGreen.
Keywords: Agrobacterium, Wheat, Triticale, Binary vectors, pGreen, pCAMBIA
Introduction
Transgenesis is a powerful technology that can be used to improve the traits of cultivated plants. It is also an excellent research tool for the functional analysis of genes. These two tasks can be approached by introducing expression or silencing gene cassettes into the plant genome via genetic transformation. The growing amount of information in public databases about the sequences of genes and their regulatory elements or EST is increasing the importance of the method.
The development of Agrobacterium-mediated transformation methods for wheat began in 1997 (Cheng et al. 1997) and for triticale eight years later (Nadolska-Orczyk et al. 2005). The main reason for applying this technique is its natural process of foreign DNA introduction into the plant genome, minimizing any rearrangements and multi-copy insertions (for a review, see Nadolska-Orczyk et al. 2000). This was recently approved for wheat (Wu et al. 2006) and barley (Travella et al. 2006; Bartlett et al. 2009). Since that time, several factors affecting the efficiency of transformation and the ability to transform new genotypes of wheat have been documented (reviewed by Jones et al. 2005; Bhalla 2006; Shrawat and Lörz 2006). These factors include the Agrobacterium strains, the binary vectors, the selectable genes and promoters, the inoculation method, and the in vitro culture conditions. Despite the moderate success in developing the method, the low transformation efficiency and low transgene expression level remain the main limitations. There are few reports on cereals comparing the direct effects of the promoters driving selectable genes (Prakash et al. 2008) or the same selection cassettes carried by different binary vector systems.
A method for the Agrobacterium-mediated transformation of the Polish cultivars of common wheat, Kontesa, Torka, and Eta, was developed by Przetakiewicz et al. (2004). The effects of three combinations of the bacterial strain and vectors containing different selection cassettes were compared. The highest selection rate, 12.6%, was obtained with EHA101(pGAH) on a kanamycin-containing medium, while it was much lower with AGL1(pDM805) on phosphinothricin and lower again on hygromycin with two different combinations. Since that time, new and versatile binary vector systems for plant transformation have been developed. The first one, the pGreen system, allowed any arrangements of the selectable and reporter cassettes (Hellens et al. 2000). The pGreen vector is small because its replication functions in trans were moved into another vector, allowing for replication in Agrobacterium. The system is supplemented with different selectable and reporter gene cassettes (http://www.pgreen.ac.uk/). The second system uses pCAMBIA vectors from the CAMBIA Institute (http://www.cambia.org/). These vectors are modified from pPZP vectors (Hajdukiewicz et al. 1994). They are small, stable in Agrobacterium, and have restriction sites and poly-linkers for the introduction of any DNA. The additional features of these two vector systems are: high copy numbers in Escherichia coli, high cloning capacity, improved compatibility with any given strains, and a high frequency of plant transformation (Komori et al. 2007). We tested the effectiveness of the two binary vector systems, which contain different selection cassettes for the transformation of polish cultivars of wheat and triticale. The low expression of the two selectable genes, bar and nptII, was proved through the analysis of the transcripts. This is the first report comparing the transformation ability for wheat and triticale of the two popular vector systems, pGreen and pCAMBIA.
Materials and methods
Plant material
Two cultivars of allohexaploid, spring wheat, Kontesa and Torka, and one allohexaploid cultivar of spring triticale, Wanad, were used for the experiments. The plants were grown in a growth chamber at 18/16°C (day/night), with a 16-h photoperiod, under 350 μE m−2 s−1 light. The kernels were isolated from 9- to 12-week-old plants and sterilized in 70% ethanol, followed by 0.1% HgCl2 with 0.1% Tween 20 for 3 to 4 min, and three washes in sterilized water. The immature 1- to 1.2-mm-long embryos were isolated from the kernels and placed on the medium.
In vitro culture of wheat and triticale
Immature embryos of wheat and triticale were precultured on MSB3 medium containing MS salts (Murashige and Skoog 1962), B5 vitamins (Gamborg et al. 1968), and 3 mg l−1 Dicamba, as previously described (Przetakiewicz et al. 2003). After 2 to 3 days, the embryos were inoculated with a drop of A. tumefaciens suspension and co-cultured for the next 2–3 days, depending on the growth rate of the bacteria. The subsequent culture was on MSB6 medium containing 2 mg l −1 Picloram, 1 mg l −1 2,4-D, 500 mg l−1 casein hydrolysate, 150 mg l −1 Timentin, and one of two selection agents, 50 mg l −1 kanamycin or 2 mg l −1 phosphinothricin. After a further 4–5 weeks, the embryonic calli were transferred onto plant regeneration medium R2-MSB and rooted on 0.5 MS medium (Przetakiewicz et al. 2003). The media contained the same selection agent. The culture conditions were as follows: 22°C day/night, with a 16-h photoperiod, under 65–70 μE m−2 s−1 light.
Bacterial strains and vectors
Two vector systems were used: pGreen II 0000 from the John Innes Centre, Norwich (http://www.pgreen.ac.uk/) and pCAMBIA 1305.2 from the Commonwealth Scientific and Industrial Research Organisation (CSIRO), Australia (http://www.cambia.org/). Four selection cassettes were cloned into the HpaI cloning site of the T-DNA of pGreen, namely, nos::nptII, 35S::nptII, nos::bar, and 35S::bar, according to the procedure described on the website http://www.pgreen.ac.uk/. The 35S::gus cassette was cloned into the StuI cloning site of the T-DNA of the vector (Fig. 1a). Two selection cassettes, 35S::nptII and 35S::bar, were cloned in place of the 35S::hpt cassette originally existing in the T-DNA of pCAMBIA (Fig. 1b). The 35S::GUSPlus cassette was already a part of the T-DNA of the original pCAMBIA 1305.2 vector. The lengths of the cloned cassettes, the cloning sites, and the lengths of the cloned vectors are listed in Table 1.
Fig. 1.
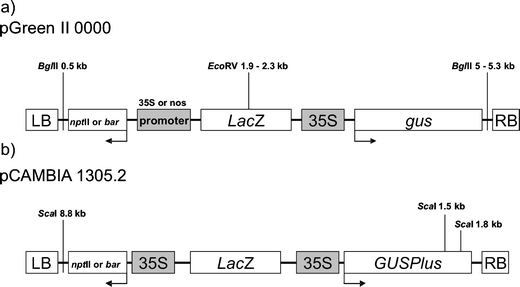
A schematic map of the T-DNA regions of the pGreen II 0000 (a) and pCAMBIA 1305.2 (b) vectors
Table 1.
The lengths of the cassettes used for the cloning of the whole cloned vectors and of the fragments of DNA after the restriction analysis
| Vector:cloned cassette | Cloning site | Length of cassette/cloned vector (bp) | Restriction analysis (Kb) | |
|---|---|---|---|---|
| pGreen II 0000: | EcoRV | BglII | ||
| nos::nptII | HpaI | 1,406/7,217 | 7.2 | 4.7 + 2.5 |
| 35S::nptII | 1,510/7,321 | 7.3 | 4.8 + 2.5 | |
| nos::bar | 1,148/6,961 | 6.9 | 4.5 + 2.4 | |
| 35S::bar | 1,220/7,031 | 7.0 | 4.5 + 2.5 | |
| 35S::gus | StuI | 2,507/3,304 | 5.8 | 3.3 + 2.5 |
| pCAMBIA: | ScaI | |||
| 35S::nptII | XhoI | 1,685/11,706 | 6.2 + 4.5 + 0.7 + 0.3 | |
| 35S::bar | 1,371/11,391 | 6.2 + 4.1 + 0.7 + 0.3 | ||
PCR analysis
Genomic DNA was isolated from the young leaves of plants according to the modified CTAB method (Murray and Thompson 1980). The polymerase chain reaction (PCR) assay was carried out in a 25-μl reaction mixture containing 200 ng of genomic DNA, 0.2 mM of each dNTP, 0.2–0.4 μM of each primer and 0.5–1 U Taq DNA polymerase, 1.6 mM MgCl2, and 1 × Taq DNA polymerase buffer. Four pairs of primers were designated to amplify fragments of the reporter and selection genes. The sequences of the primers and the length of the products were:
gus 7 5′-GGAGTATTGCCAACGAACC-3′ and
gus 8 5′-CGCCAGGAGAGTTGTTCATTC-3′ (606 bp);
npt 1 5′-GAGGCTATTCGGCTATGACTG-3′ and
npt 2 5′-ATCGGGAGCGGCGATACCGTA-3′ (700 bp);
npt 5 5′-GAAGGCGATAGAAGGCGATGCG-3′ and
npt 6 5′-TGAATGAACTCCAGGACGAGGCAG-3′ (609 bp); and
bar 7 5′-TCTGCACCATCGTCAACCACTACATC-3′ and
bar 8 5′-CAGAAACCCACGTCATGCCAGTTC-3’ (430 bp).
The PCR amplification conditions were: initialization of denaturation at 94°C for 5 min, 94°C for 30 s, then 57°C (for gus 7 and 8), 62°C (for npt 1 and 2), 66°C (for npt 5 and 6), or 68°C (for bar 7 and 8) for 30 s, then 72°C for 1 min, and, finally, 72°C for 10 min. The numbers of cycles were 36 to 40. The conditions were optimized for each pair of primers separately.
Semi-quantitative RT-PCR
RNA was isolated from the young leaves of six T1 seedlings per sample using TRI Reagent® Solution (Applied Biosystems Inc., USA), according to the manufacturer’s protocol. Isolated RNA was treated using DNase I Recombinant, RNase-free (Roche) to remove traces of genomic DNA. The first strand of cDNA was synthesized using a RevertAid™ First Strand cDNA Synthesis Kit (Fermentas), according to manufacturer’s protocol. The sequences of primers for the 153-bp cDNA of the nptII amplification were:
qRTnpt 5 5′-TAAAGCACGAGGAAGCGGTCAG-3′ and
qRTnpt 6 5′-GCCGCTTTTCTGGATTCATCG3′;
and for the 140-bp cDNA of the bar amplification, they were:
qRTbar 3 5′-GTCCACTCCTGCGGTTCCTGCG-3′ and
qRTbar 4 5′-ATGAGCCCAGAACGACGCCCG-3′.
The sequences of the primers for the 133-bp cDNA of the reference actin gene (accession numbers U60508 and AC209737.2) were:
qAct 1 5′-AGCAACTGGGATGACATGGAG-3′ and
qAct 2 5′ GGGTCATCTTCTCTCTGTTGGC 3′.
The RT-PCR amplification conditions were the same as for the PCR. The annealing temperatures for the nptII, bar, and actin amplifications were 58°C, 62°C, and 61°C, respectively.
Standard deviations were calculated based on the free repeats using the Microsoft Excel program.
Histochemical GUS assay
β-Glucuronidase activity was determined in the young leaves and coleoptiles collected from 5-day-old plants using a histochemical GUS assay (Jefferson et al. 1987). Fragments of the tissues were incubated overnight at 37°C in a buffer containing 2 mM X-Glu in 50 mM sodium phosphate buffer, pH 7.0.
Results
The transformation efficiency with pGreen and pCAMBIA vectors
Two wheat cultivars, Kontesa and Torka, and one triticale cultivar, Wanad, were successively transformed with two vector systems carrying phosphinothricin or kanamycin selection cassettes (Table 2). The range of the transformation rate for Kontesa was from 0.00 to 3.58% and was highest after transformation with pGreen carrying the 35S::nptII cassette. A similar result was obtained for Torka. The transformation rate ranged from 0.00 to 3.14% and was best with the same combination as for Kontesa. There was no selection of transgenic plants in the two wheat cultivars after transformation with pGreen carrying the 35S::bar cassette. However, the transformation efficiency with pCAMBIA carrying the same selection cassette was 1.46% for Kontesa and 1.79% for Torka. Similar rates of 1.46% and 1.96%, respectively, were obtained after transformation with the 35S::nptII cassette carried by pCAMBIA. The transformation efficiency was several times higher in the two cultivars of wheat after transformation with nptII driven by the 35S promoter than with the nos promoter. The pCAMBIA vector carrying the 35S::bar cassette was much more successful for the transformation of the two wheat cultivars than the pGreen vector with the same cassette. The transformation efficiency was 1.46% for Kontesa and 1.79% for Torka in the first case, compared with 0.00% in the second. By contrast, the pCAMBIA vector carrying 35S::nptII cassette was about two times less efficient for the two wheat cultivars transformation than pGreen.
Table 2.
The efficiency of transformation of two cultivars of wheat (Kontesa and Torka) and one cultivar of triticale (Wanad) transformed with the pGreen and pCAMBIA vector systems containing the bar and nptII selection genes under the control of the nos or 35S promoters
| Vector/selection cassette | Wheat | Triticale | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Kontesa | Torka | Wanad | ||||||||||
| Ne | Np | PCR+ | Tr (%) | Ne | Np | PCR+ | Tr (%) | Ne | Np | PCR+ | Tr (%) | |
| pGreen | ||||||||||||
| nos::bar | 495 | 3 | 1 | 0.20 | 609 | 3 | 3 | 0.49 | 358 | 9 | 4 | 1.12 |
| 35S::bar | 393 | 1 | 0 | 0.00 | 407 | 3 | 0 | 0.00 | 408 | 5 | 3 | 0.74 |
| nos::bar* | 203 | 1 | 1 | 0.49 | 101 | 6 | 0 | 0.00 | 200 | 0 | 0 | 0.00 |
| pCAMBIA | ||||||||||||
| 35S::bar | 205 | 3 | 3 | 1.46 | 167 | 5 | 3 | 1.79 | 187 | 8 | 2 | 1.07 |
| Total | 1,093 | 8 | 5 | 0.46 | 1,183 | 11 | 6 | 0.51 | 953 | 22 | 9 | 0.94 |
| pGreen | ||||||||||||
| nos::nptII | 469 | 11 | 3 | 0.64 | 373 | 4 | 1 | 0.27 | 318 | 16 | 6 | 1.89 |
| 35S::nptII | 642 | 33 | 23 | 3.58 | 510 | 22 | 16 | 3.14 | 391 | 7 | 7 | 1.79 |
| pCAMBIA | ||||||||||||
| 35S::nptII | 274 | 4 | 4 | 1.46 | 204 | 4 | 4 | 1.96 | 206 | 14 | 14 | 6.79 |
| Total | 1,385 | 48 | 30 | 2.16 | 1,087 | 30 | 21 | 1.93 | 915 | 37 | 27 | 2.95 |
Ne – number of explants
Np – number of plants selected
PCR+ − number of PCR-positive plants
Tr (%) – transformation rate
* – in the EHA105 strain
The transformation rates for Wanad ranged from 0.00 to 6.79%. The highest efficiency, 6.79%, was obtained after transformation with pCAMBIA carrying the 35S::nptII cassette. Significantly lower rates, 1.79% and 1.89%, were obtained after transformation with the pGreen vector carrying the 35S::nptII or nos::nptII cassettes, respectively. The transformation of Wanad with bar-containing cassettes was successful, but the rates were lower (0.00 to 1.12%). The highest transformation efficiency was obtained with nos::bar carried by pGreen.
Confirmation of the integration and expression of the selection genes
The integration of the transgenes into the genome of putative transgenic plants was proved by PCR amplification with three different pairs of specific primers amplifying fragments of the nptII (Fig. 2a, b), bar (Fig. 2c), or gus genes (Fig. 2d). Additionally, the expression of the selection genes nptII and bar was tested in bulk samples of T1 seedlings of selected lines via semi-quantitative RT-PCR using actin as a reference gene (Fig. 3a, b). The selection genes were expressed in most of the tested lines.
Fig. 2.
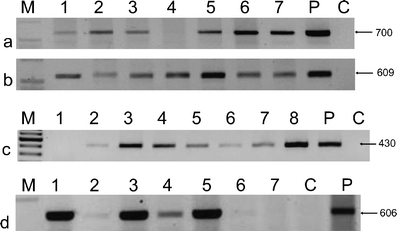
The results of a polymerase chain reaction (PCR) analysis of transformed T0 and T1 plants. The DNA of putative transgenic plants was amplified using specific primers: (a) npt 1 and 2 (700 bp), (b) npt 5 and 6 (609 bp), (c) bar 7 and 8 (430 bp), (d) gus 7 and 8 (606 bp). M – molecular weight marker; lines 1 to 8 – DNA from transformed plants; P – plasmid DNA; C – DNA from non-transgenic, control plants
Fig. 3.
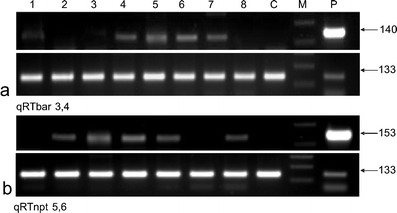
Semi-quantitative expression analysis of the bar (140-bp) and nptII (153-bp) genes. The amplification of the cDNA of the reference actin gene (133-bp) is shown at the bottom of the gels. M – molecular weight marker; lines 1 to 8 – cDNA from bulked samples of T1 plants; P – plasmid DNA; C – cDNA from non-transgenic, control plants
The relative levels of the nptII and bar transcript accumulation in the bulk samples of transgenic T1 plants of Kontesa, Torka, and Wanad transformed with different vectors and cassettes was measured in relation to the actin gene (Fig. 4a–d). The amounts of nptII transcript in the Kontesa T1 lines ranged from 0.00 to 0.63 (Fig. 4a). The highest levels of transcript, 0.63 and 0.43, were obtained in the two lines transformed with pGreen carrying the 35S::nptII cassette. It was several times higher than the most frequently measured for nptII relative value of 0.12. The transcript levels for nptII in Torka were only measured in the T1 lines transformed with pGreen carrying the 35S::nptII cassette (Fig. 4b). The relative amounts ranged from 0.10 to 0.32. Similar levels of the nptII transcript, ranging from 0.13 to 0.22, were obtained for the T1 lines of the triticale Wanad transformed with pGreen carrying the 35S::nptII cassette (Fig. 4c). The levels of the bar transcript were also low, from 0.00 to 0.33, with the highest level in the Wanad (0.33) and Torka (0.32) lines transformed with pCAMBIA carrying the 35S::bar cassette.
Fig. 4.
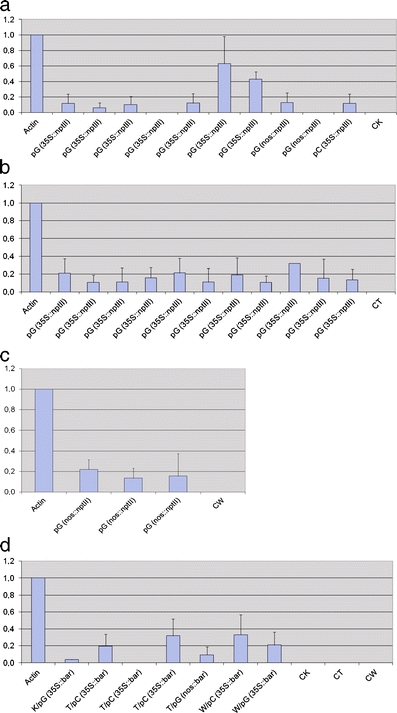
The relative transcript levels of nptII in Kontesa (a), Torka (b), and Wanad (c) lines transformed with different combinations of vectors and nptII constructs, and bar (d) in transgenic lines of Kontesa (K), Torka (T), and Wanad (W). CK, CT, CW – non-transgenic, control lines of Kontesa, Torka, and Wanad, respectively. The results are the means of three independent measurements
The expression of the reporter gus gene carried by the pGreen vector or GUSPlus carried by the pCAMBIA vector was proved by histochemical analysis of the GUS activity (Fig. 5). The analysis was also used to test the segregation of GUS-positive progeny in T1 lines (Table 3). There were no GUS-positive plants among 20 of the 83 lines tested. Most of the progeny from the 63 T1 lines were GUS-negative and did not show a Mendelian type of segregation.
Fig. 5.
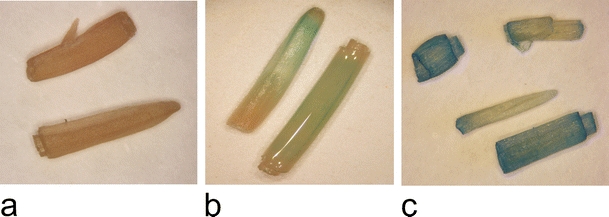
Histochemical analysis of GUS activity in the leaves of T1 plants: control, non-transgenic (a) and GUS-positive, transgenic plants (b–c)
Table 3.
The segregation analysis of GUS-positive plants in the T1 progeny from lines transformed with pGreen (35S::gus) and pCAMBIA (35S::GUSPlus)
| Vector (GUS cassette)/selection cassette | Wheat | Triticale | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Kontesa | Torka | Wanad | |||||||
| Nl Np GUS+ | Nl Np GUS+ | Nl Np GUS+ | |||||||
| pGreen (35S::gus) | |||||||||
| nos::nptII | 3 | 5 | 0 | 0 | 0 | 0 | 6 | 69 | 0 |
| 35S::nptII | 14 | 182 | 54 | 16 | 88 | 4 | 5 | 146 | 21 |
| nos::bar | 1 | 16 | 0 | 3 | 11 | 5 | 3 | 40 | 3 |
| 35S::bar | 1 | 0 | 0 | 0 | 0 | 0 | 3 | 5 | 0 |
| Total | 20 | 208 | 56 | 20 | 99 | 9 | 17 | 263 | 24 |
| pCAMBIA (35S::GUSPlus) | |||||||||
| 35S::nptII | 3 | 73 | 14 | 3 | 51 | 25 | 8 | 170 | 21 |
| 35S::bar | 2 | 32 | 0 | 5 | 32 | 2 | 2 | 22 | 1 |
| Total | 5 | 105 | 14 | 11 | 115 | 33 | 10 | 192 | 22 |
Nl – number of tested lines
Np – number of tested plants
GUS+ − number of GUS-positive plants
Discussion
The same selection cassettes carrying the nptII or bar selection genes driven by nos or 35S were cloned into two popular binary vector systems, pGreen and pCAMBIA. These combinations allowed us to test the effect of the chosen promoters, selection genes, and vectors on wheat and triticale transformation. The transformation events were proved by detailed PCR analysis and transcript analysis of the selection genes.
The selection system is one of the most important factors influencing transformation efficiency. We compared two commonly used selection genes, nptII and bar, under the control of different promoters. Based on the analysis of the transformation efficiency, we documented that, of the two selection genes used, nptII and bar, the former gave better results for wheat and triticale transformation. Similar data was already shown with the same cultivars (Przetakiewicz et al. 2004; Nadolska-Orczyk et al. 2005). However, this is the first direct test of these two selection genes, driven by the same promoters and carried by the same binary vector. A high transformation efficiency up to 4.3%, and even higher after the desiccation of post-infected tissue, reaching 19%, was also obtained in wheat transformed with the nptII selection genes under the control of 35S (Cheng et al. 1997, 2003). Lower transformation efficiency, albeit at the similar level to our results, was reported by Weir et al. (2001) after using the bar selection gene under the control of 35S.
Both selection genes were driven by constitutive promoters commonly used for dicotyledonous plant transformation: nos characterized as moderate and CaMV 35S (35S) described as a strong promoter. In cereals, these promoters are considered to confer comparatively weak expression (Jones et al. 2005; Hensel et al. 2011). More frequently used are constitutive promoters of cereal origin, like maize Ubi1 and rice Act1. On the other hand, it was already proved that both the 35S and nos promoters could be successfully used for the Agrobacterium-mediated transformation of cereals (Cheng et al. 1997, 2003; Przetakiewicz et al. 2004; Nadolska-Orczyk et al. 2005; Weir et al. 2001). However, as emphasized above, there was no research directly testing the two promoters driving the same selection genes carried by the same vectors in wheat and triticale. The only research on the effect of the 35S and Actin1 promoter driving the nptII gene in corn transformation was done by Prakash et al. (2008). They showed no appreciable difference in the frequency of corn plant transformation. The direct effect of the nos or 35S promoters driving the nptII and bar selection genes on the transformation efficiency was documented here and in earlier reports as positive. As expected, several-fold higher transformation efficiencies were obtained in combinations transformed with nptII, which was driven by the strong 35S, independently of the binary vector used. An unexpected result was the lack of any selection in the two cultivars of wheat transformed with 35S::bar compared with the low efficiency after transformation with nos::bar in the pGreen vector. However, the transformation efficiency of wheat cultivars transformed with the same 35S::bar selection cassette in the pCAMBIA vector was high. The final result of the transformation efficiency in the two cultivars of wheat was dependent on the transformation vector used for the experiments. The bar selection system was more effective when carried by pCAMBIA, and the nptII-based selection was more effective in pGreen. This dependence of the transformation efficiency and the vector used for transformation was also observed in the triticale cultivar, Wanad. Both the bar and the nptII selection systems were more effective in pCAMBIA.
The high efficiency of transformation obtained in the selected combinations indirectly indicates that the expression of the selection cassette occurring at the tissue culture level was also high. We detected nptII and bar transcripts in calli transformed with both of the tested selection genes (data not shown). However, the levels of transcripts were very low, probably because the integration of transgenes and selection usually only occurs in some cells of transformed tissue.
The expression of nptII and bar measured in the leaves of developing T1 seedlings was low. There are several aspects which might be taken into consideration. The transcript level was measured for bulk samples of five seedlings of each line, because T1 is the segregating generation. It means that the values showed us the mean level of transcript for each line. Additionally, it was determined in relation to the actin gene, showing a stable and high level of the transcript in the leaf tissues of wheat and triticale, and representing the gene family. We might also presume that the nos or 35S promoters confer comparatively weak expression in the leaves, as suggested by other researchers. This low expression of both genes in the leaves was indirectly proved by the kanamycin and phosphinothricin resistance tests, done on the leaves of young plants (data not shown). The selection of resistant progeny was rare. On the other hand, RNAi studies performed on cereals proved the advantageous effect of having the 35S promoter drive the silencing cassette. The silencing of native genes of barley (Zalewski et al. 2010) and wheat (Gasparis et al. 2011) detected in different tissues of transgenic lines was distinct.
Histochemical β-glucuronidase activity of reporter gus gene in the leaves of transgenic wheat plants is very rarely reported. We also obtained a low number or complete lack in some lines of GUS-positive plants. The detailed study of this reporter gene activity in transgenic wheat leaves and roots showed mRNA and GUS protein detected only by immunoblot (Bahieldin et al. 2005). Fluorometric or histochemical assays of the same plants indicated that wheat leaf and root tissues contain strong, non-proteinaceous inhibitor of β-glucuronidase activity. The activity of this/these endogenous inhibitor(s) in vegetative tissues of wheat could explain the difficulties with histochemical GUS detection reported in our study and other papers.
Acknowledgments
This research was supported by the Polish Ministry of Science and Higher Education, grant PBZ-MNiSW-2/3/2006/31.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Bahieldin A, Eissa HF, Mahfouz HT, Dyer WE, Madkour MA, Qu R. Evidence for non-proteinaceous inhibitor(s) of β-glucuronidase in wheat (Triticum aestivum L.) leaf and root tissues. Plant Cell Tissue Org Cult. 2005;82:11–17. doi: 10.1007/s11240-004-5890-8. [DOI] [Google Scholar]
- Bartlett JG, Snape JW, Harwood WA. Intron-mediated enhancement as a method for increasing transgene expression levels in barley. Plant Biotechnol J. 2009;7:856–866. doi: 10.1111/j.1467-7652.2009.00448.x. [DOI] [PubMed] [Google Scholar]
- Bhalla PL. Genetic engineering of wheat—current challenges and opportunities. Trends Biotechnol. 2006;24:305–311. doi: 10.1016/j.tibtech.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Cheng M, Fry JE, Pang S, Zhou H, Hironaka CM, Duncan DR, Conner TW, Wan Y. Genetic transformation of wheat mediated by Agrobacterium tumefaciens. Plant Physiol. 1997;115:971–980. doi: 10.1104/pp.115.3.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng M, Hu TC, Layton J, Liu C-N, Fry JE. Desiccation of plant tissues post-Agrobacterium infection enhances T-DNA delivery and increases stable transformation efficiency in wheat. In Vitro Cell Dev Biol Plant. 2003;39:595–604. doi: 10.1079/IVP2003471. [DOI] [Google Scholar]
- Gamborg OL, Miller RA, Ojima K. Nutrient requirements of suspension cultures of soybean root cells. Exp Cell Res. 1968;50:151–158. doi: 10.1016/0014-4827(68)90403-5. [DOI] [PubMed] [Google Scholar]
- Gasparis S, Orczyk W, Zalewski W, Nadolska-Orczyk A. The RNA-mediated silencing of one of the Pin genes in allohexaploid wheat simultaneously decreases the expression of the other, and increases grain hardness. J Exp Bot. 2011;62:4025–4036. doi: 10.1093/jxb/err103. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol. 1994;25:989–994. doi: 10.1007/BF00014672. [DOI] [PubMed] [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM. pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol. 2000;42:819–832. doi: 10.1023/A:1006496308160. [DOI] [PubMed] [Google Scholar]
- Hensel G, Himmelbach A, Chen W, Douchkov DK, Kumlehn J. Transgene expression systems in the Triticeae cereals. J Plant Physiol. 2011;168:30–44. doi: 10.1016/j.jplph.2010.07.007. [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones HD, Doherty A, Wu H. Review of methodologies and a protocol for the Agrobacterium-mediated transformation of wheat. Plant Methods. 2005;1:5. doi: 10.1186/1746-4811-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori T, Imayama T, Kato N, Ishida Y, Ueki J, Komari T. Current status of binary vectors and superbinary vectors. Plant Physiol. 2007;145:1155–1160. doi: 10.1104/pp.107.105734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
- Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acid Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadolska-Orczyk A, Orczyk W, Przetakiewicz A. Agrobacterium-mediated transformation of cereals—from technique development to its application. Acta Physiol Plant. 2000;22:77–88. doi: 10.1007/s11738-000-0011-8. [DOI] [Google Scholar]
- Nadolska-Orczyk A, Przetakiewicz A, Kopera K, Binka A, Orczyk W. Efficient method of Agrobacterium-mediated transformation for triticale (x Triticosecale Wittmack) J Plant Growth Regul. 2005;24:2–10. doi: 10.1007/s00344-004-0046-y. [DOI] [Google Scholar]
- Prakash NS, Prasad V, Chidambram TP, Cherian S, Jayaprakash TL, Dasgupta S, Wang Q, Mann MT, Spencer TM, Boddupalli RS. Effect of promoter driving selectable marker on corn transformation. Transgenic Res. 2008;17:695–704. doi: 10.1007/s11248-007-9149-0. [DOI] [PubMed] [Google Scholar]
- Przetakiewicz A, Orczyk W, Nadolska-Orczyk A. The effect of auxin on plant regeneration of wheat, barley and triticale. Plant Cell Tissue Org Cult. 2003;73:245–256. doi: 10.1023/A:1023030511800. [DOI] [Google Scholar]
- Przetakiewicz A, Karas A, Orczyk W, Nadolska-Orczyk A. Agrobacterium-mediated transformation of polyploid cereals. The efficiency of selection and transgene expression in wheat. Cell Mol Biol Lett. 2004;9:903–917. [PubMed] [Google Scholar]
- Shrawat AK, Lörz H. Agrobacterium-mediated transformation of cereals: a promising approach crossing barriers. Plant Biotechnol J. 2006;4:575–603. doi: 10.1111/j.1467-7652.2006.00209.x. [DOI] [PubMed] [Google Scholar]
- Travella S, Klimm TE, Keller B. RNA interference-based gene silencing as an efficient tool for functional genomics in hexaploid bread wheat. Plant Physiol. 2006;142:6–20. doi: 10.1104/pp.106.084517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir B, Gu X, Wang MB, Upadhyaya N, Elliott AR, Brettell RIS. Agrobacterium tumefaciens-mediated transformation of wheat using suspension cells as a model system and green fluorescent protein as a visual marker. Aust J Plant Physiol. 2001;28:807–818. [Google Scholar]
- Wu H, Sparks CA, Jones HD. Characterisation of T-DNA loci and vector backbone sequences in transgenic wheat produced by Agrobacterium-mediated transformation. Mol Breed. 2006;18:195–208. doi: 10.1007/s11032-006-9027-0. [DOI] [Google Scholar]
- Zalewski W, Galuszka P, Gasparis S, Orczyk W, Nadolska-Orczyk A. Silencing of the HvCKX1 gene decreases the cytokinin oxidase/dehydrogenase level in barley and leads to higher plant productivity. J Exp Bot. 2010;61:1839–1851. doi: 10.1093/jxb/erq052. [DOI] [PubMed] [Google Scholar]


