Abstract
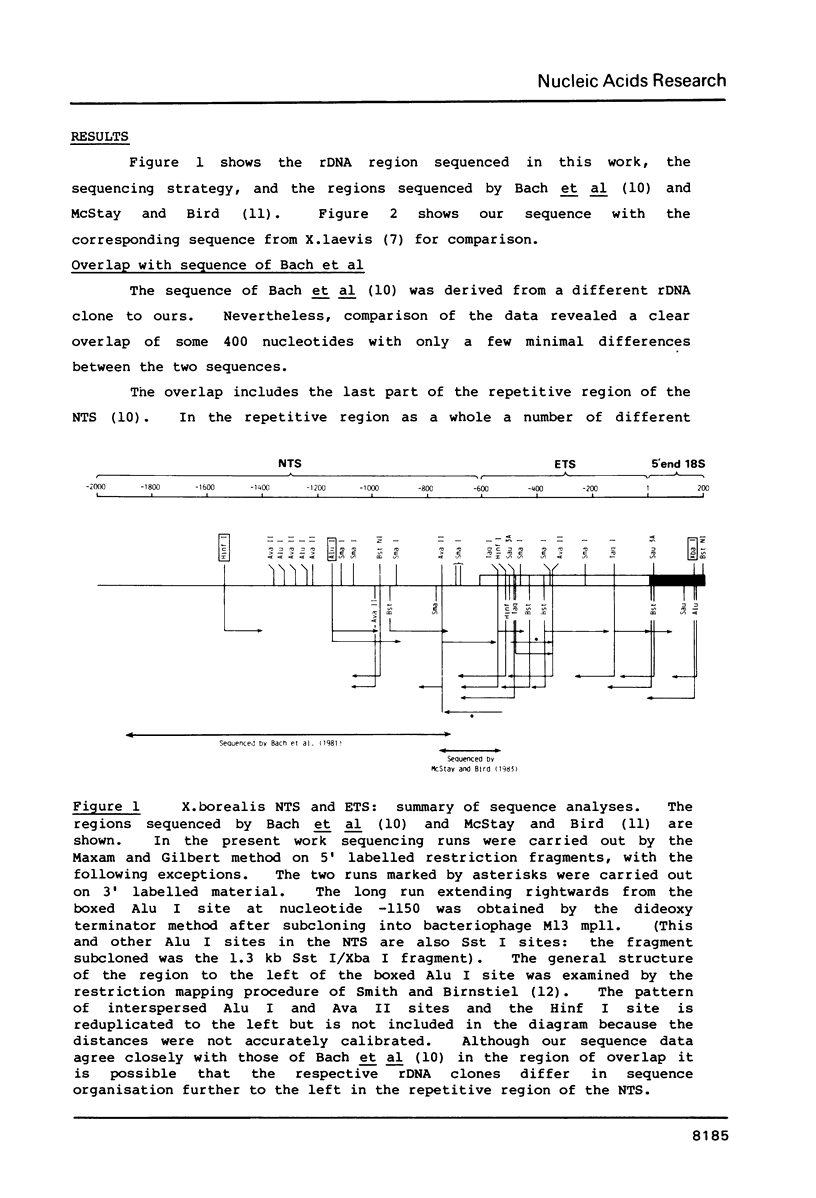
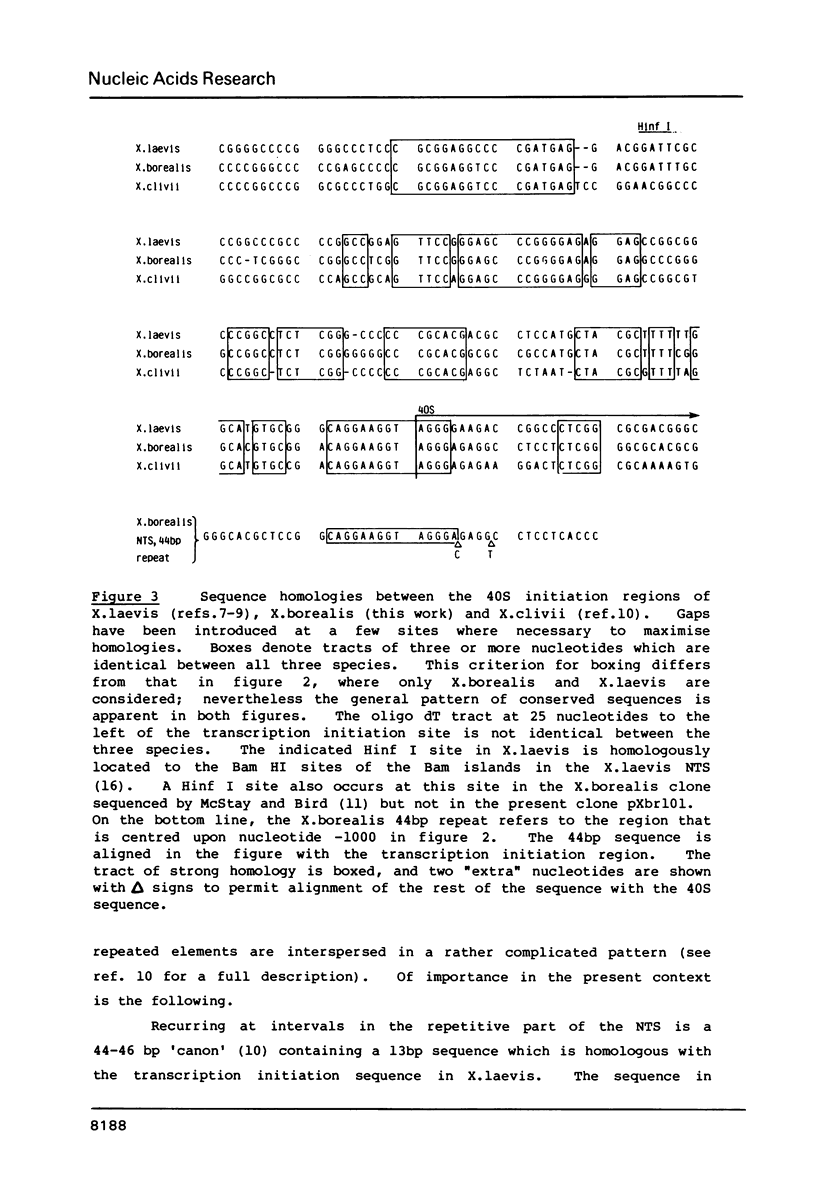
We report sequence data from a cloned rDNA unit from Xenopus borealis, extending leftwards from the 18S gene to overlap a region previously sequenced by R. Bach, B. Allet and M. Crippa (Nucleic Acids Research 9, 5311-5330). Comparison with data from other species of Xenopus leads to the inference that the transcription initiation site in X.borealis is in the newly sequenced region and not, as was previously thought, in the region sequenced earlier. The X.borealis external transcribed spacer thus defined is some 612 nucleotides long, about 100 nucleotides shorter than in X.laevis. The X.borealis and X.laevis external transcribed spacers show a pattern of extensive but interrupted sequence divergence, with a large conserved tract starting about 100 nucleotides downstream from the transcription initiation site and shorter conserved tracts elsewhere. The regions in between the conserved tracts differ in length between the respective external transcribed spacers indicating that insertions and deletions have contributed to their divergence, as previously inferred for the internal transcribed spacers. Much of the overall length difference is in the region flanking the 18S gene, where there are also length microheterogeneities in X.laevis rDNA. As in X.laevis, the transcribed spacer sequences flanking the 18S gene in X.borealis contain no major tracts of mutual complementarity. The accumulated data on transcribed spacers in Xenopus render it unlikely that processing of ribosomal precursor RNA involves interaction between the regions flanking 18S RNA.
Full text
PDF











Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bach R., Allet B., Crippa M. Sequence organization of the spacer in the ribosomal genes of Xenopus clivii and Xenopus borealis. Nucleic Acids Res. 1981 Oct 24;9(20):5311–5330. doi: 10.1093/nar/9.20.5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bram R. J., Young R. A., Steitz J. A. The ribonuclease III site flanking 23S sequences in the 30S ribosomal precursor RNA of E. coli. Cell. 1980 Feb;19(2):393–401. doi: 10.1016/0092-8674(80)90513-9. [DOI] [PubMed] [Google Scholar]
- Coen E., Strachan T., Dover G. Dynamics of concerted evolution of ribosomal DNA and histone gene families in the melanogaster species subgroup of Drosophila. J Mol Biol. 1982 Jun 15;158(1):17–35. doi: 10.1016/0022-2836(82)90448-x. [DOI] [PubMed] [Google Scholar]
- Furlong J. C., Maden B. E. Patterns of major divergence between the internal transcribed spacers of ribosomal DNA in Xenopus borealis and Xenopus laevis, and of minimal divergence within ribosomal coding regions. EMBO J. 1983;2(3):443–448. doi: 10.1002/j.1460-2075.1983.tb01442.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall L. M., Maden B. E. Nucleotide sequence through the 18S-28S intergene region of a vertebrate ribosomal transcription unit. Nucleic Acids Res. 1980 Dec 20;8(24):5993–6005. doi: 10.1093/nar/8.24.5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maden B. E., Moss M., Salim M. Nucleotide sequence of an external transcribed spacer in Xenopus laevis rDNA: sequences flanking the 5' and 3' ends of 18S rRNA are non-complementary. Nucleic Acids Res. 1982 Apr 10;10(7):2387–2398. doi: 10.1093/nar/10.7.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Michot B., Bachellerie J. P., Raynal F. Structure of mouse rRNA precursors. Complete sequence and potential folding of the spacer regions between 18S and 28S rRNA. Nucleic Acids Res. 1983 May 25;11(10):3375–3391. doi: 10.1093/nar/11.10.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss T. A transcriptional function for the repetitive ribosomal spacer in Xenopus laevis. Nature. 1983 Mar 17;302(5905):223–228. doi: 10.1038/302223a0. [DOI] [PubMed] [Google Scholar]
- Moss T., Boseley P. G., Birnstiel M. L. More ribosomal spacer sequences from Xenopus laevis. Nucleic Acids Res. 1980 Feb 11;8(3):467–485. doi: 10.1093/nar/8.3.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss T. Transcription of cloned Xenopus laevis ribosomal DNA microinjected into Xenopus oocytes, and the identification of an RNA polymerase I promoter. Cell. 1982 Oct;30(3):835–842. doi: 10.1016/0092-8674(82)90288-4. [DOI] [PubMed] [Google Scholar]
- Schibler U., Wyler T., Hagenbüchle O. Changes in size and secondary structure of the ribosomal transcription unit during vertebrate evolution. J Mol Biol. 1975 May 25;94(3):503–517. doi: 10.1016/0022-2836(75)90217-x. [DOI] [PubMed] [Google Scholar]
- Smith H. O., Birnstiel M. L. A simple method for DNA restriction site mapping. Nucleic Acids Res. 1976 Sep;3(9):2387–2398. doi: 10.1093/nar/3.9.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner-Webb B., Reeder R. H. The nucleotide sequence of the initiation and termination sites for ribosomal RNA transcription in X. laevis. Cell. 1979 Oct;18(2):485–499. doi: 10.1016/0092-8674(79)90066-7. [DOI] [PubMed] [Google Scholar]
- Stewart M. A., Hall L. M., Maden B. E. Multiple heterogeneities in the transcribed spacers of ribosomal DNA from Xenopus laevis. Nucleic Acids Res. 1983 Feb 11;11(3):629–646. doi: 10.1093/nar/11.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldman G. M., Brand R. C., Klootwijk J., Planta R. Some characteristics of processing sites in ribosomal precursor RNA of yeast. Nucleic Acids Res. 1980 Jul 11;8(13):2907–2920. doi: 10.1093/nar/8.13.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg R. A., Penman S. Processing of 45 s nucleolar RNA. J Mol Biol. 1970 Jan 28;47(2):169–178. doi: 10.1016/0022-2836(70)90337-2. [DOI] [PubMed] [Google Scholar]
- Wellauer P. K., Dawid I. B., Kelley D. E., Perry R. P. Secondary structure maps of ribosomal RNA. II. Processing of mouse L-cell ribosomal RNA and variations in the processing pathway. J Mol Biol. 1974 Oct 25;89(2):397–407. doi: 10.1016/0022-2836(74)90527-0. [DOI] [PubMed] [Google Scholar]
- Young R. A., Steitz J. A. Complementary sequences 1700 nucleotides apart form a ribonuclease III cleavage site in Escherichia coli ribosomal precursor RNA. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3593–3597. doi: 10.1073/pnas.75.8.3593. [DOI] [PMC free article] [PubMed] [Google Scholar]



