Abstract
Bacterial pathogens detect and integrate multiple environmental signals to coordinate appropriate changes in gene expression including the selective expression of virulence factors, changes to metabolism and the activation of stress response systems. Mutations that abolish the ability of the pathogen to respond to external cues are typically attenuating. Here we discuss our recent discovery of a novel post-transcriptional regulatory pathway critical for Salmonella virulence and stress resistance. The enzymes PoxA and YjeK coordinately attach a unique β-amino acid onto a highly conserved lysine residue in the translation factor elongation factor P (EF-P). Strains in which EF-P is unmodified due to the absence of PoxA or YjeK are attenuated for virulence and display highly pleiotropic phenotypes, including hypersusceptibility to a wide range of unrelated antimicrobial compounds. Work from our laboratory and others now suggests that EF-P, previously thought to be essential, instead plays an ancillary role in translation by regulating the synthesis of a relatively limited subset of proteins. Other observations suggest that the eukaryotic homolog of EF-P, eIF5A, may illicit similar changes in the translation machinery during stress adaptation, indicating that the role of these factors in physiology may be broadly conserved.
Key words: Salmonella, elongation factor P, lysyl-tRNA synthetase, beta-lysine, antibiotic resistance
Introduction
Stress adaptation pathways are necessarily multi-layered and complex in order to accurately integrate information from multiple environmental inputs into appropriate responses. Studies of virulence regulation have largely focused on transcriptional regulation including the roles of alternative sigma factors1 and two-component regulatory systems.2,3 Post-transcriptional regulation of stress adaptation and virulence pathways can be mediated by small RNAs,4 riboswitches,5,6 and nucleotide-derived second messengers like (p)ppGpp, c-diGMP and cAMP.7,8 Additional levels of post-transcriptional regulation are provided by toxin-antitoxin systems that can cleave mRNA under conditions of stress and nutrient deprivation.9 A common assumption is that, aside from the stringent response, the prokaryotic ribosome is a relatively passive player in stress adaptation and that most post-transcriptional regulation occurs at the level of mRNA structure rather than through alterations of the ribosome itself. Only limited reports exist where the prokaryotic translational apparatus appears to play a direct role in selectively directing the expression of specific transcripts.10,11 This is despite an extensive body of work indicating that components of the eukaryotic translation apparatus, like eIF2 and eIF4, are modified under certain conditions to block global protein synthesis and promote translation of stress-response genes.12
Our laboratories, in collaboration Dr. Ferric Fang at the University of Washington, recently identified a unique post-translational modification of a poorly understood bacterial elongation factor, EF-P, that is essential for Salmonella virulence and resistance to a variety of unrelated stressors including acid, detergents and antibiotics.13 As shown in Figure 1, the modification of EF-P is carried out by two enzymes, PoxA (also called GenX or YjeA) and YjeK. Our findings regarding post-translational modification of EF-P in Salmonella were corroborated by an independent report regarding EF-P from E. coli by the laboratory of Shigeyuki Yokoyama at RIKEN.14 As described below, these two recent reports support the hypothesis that EF-P interacts with the translational machinery to reprogram synthesis of a subset of proteins necessary for bacterial virulence and survival against multiple forms of stress.
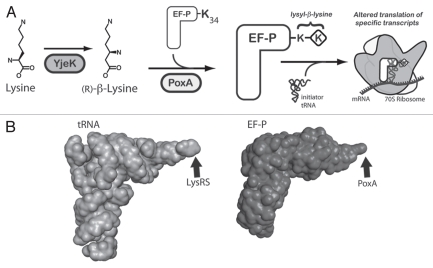
Figure 1.
The role of YjeK and PoxA in the modification of EF-P. (A) A model for the post-translational modification of the conserved lysyl 34 residue of EF-P by the PoxA and YjeK enzymes. As shown the YjeK enzyme converts L-lysine to (R)-β-lysine, which is subsequently ligated onto EF-P to generate the lysyl-β-lysine modification, although the order of these two enzymes in this pathway could be reversed. The modified elongation factor subsequently interacts with the ribosome and the initiator tRNA to facilitate the formation of the first peptide bond of specific transcripts. The mechanism underlying why only certain transcripts are regulated by EF-P remains unclear. (B) The three dimensional structures of tRNA (left, PDB 1EHZ) and EF-P (right, PDB 1UEB)24 reveal their similar overall structure. The sites of their modification by LysRS or by the aaRS paralog, PoxA, are indicated with arrows.
PoxA and YjeK are Critical for Salmonella Virulence and Stress Resistance
We first identified PoxA and YjeK during a screen for Salmonella mutants that were able to resist the cytostatic effects of S-nitroso-glutathione (GSNO), a compound that, in some aspects, mimics the nitrosative stress encountered by the bacteria within an activated macrophage. However, Salmonella poxA and yjeK mutants did not display enhanced resistance to other nitric oxide-generating compounds, indicating that they are not resistant to nitric oxide per se. Furthermore, rather than being hypervirulent as their GSNO-resistance might predict, the Salmonella poxA and yjeK mutants displayed profound virulence defects in a mouse infection model.5,13
PoxA was initially identified in 1982 in a screen for E. coli mutants that displayed reduced pyruvate oxidase activity.16 The pyruvate oxidase enzyme was later found to be encoded by an unlinked gene, poxB.17 PoxA appears to be critical for optimal expression of PoxB but its role appears to be post-translational given that mRNA levels in poxA mutants are similar to wild-type. PoxA is an “orphan” member of the lysyl-tRNA synthetases (LysRS) family that shares a high degree of sequence and structural similarity with the catalytic domain of LysRS but lacks the canonical anticodon recognition domain.15 A number of residues critical for PoxA function in vivo map to residues that are known to be critical for aminoacyl-tRNA synthetase (aaRS) activity.13 YjeK is a lysyl-2-3-aminomutase enzyme that catalyzes the conversion of L-lysine to R-β-lysine.18
The enzymatic properties of the PoxA and YjeK proteins gave little clue as to their biological function. To address the role of these proteins in bacterial physiology, we performed a large-scale phenotypic analysis of poxA and yjeK mutants using a Biolog phenotype microarray,19 which measures bacterial viability under several different growth media and against several antimicrobial compounds by quantitating the turnover of a colorimetric tetrazolium redox dye. This assay revealed that poxA and yjeK mutant phenotypes are highly pleiotropic and almost identical for each one of the nearly 2000 conditions tested. Under nutrient-limiting conditions, the mutant strains exhibited enhanced respiration compared to wild-type Salmonella. This enhanced respiratory activity was particularly evident for the utilization of amino acids and dipeptides as nitrogen sources (in particular, those containing methionine or branched-chain amino acids) as well as for a number of inorganic and organic sources of phosphate or sulfur. In striking contrast, the poxA and yjeK mutant strains are hypersusceptible to a broad range of external stressors, which is consistent with a more limited study on poxA mutants by Van Dyk and colleagues.20 Both mutants displayed enhanced susceptibility to antibiotics that fall into several distinct pharmacological and structural classes including antimicrobial peptides, detergents, lipophilic chelators, heavy metals and various inhibitors of cell wall synthesis, protein synthesis, RNA synthesis and DNA gyrase. Importantly, the large number of phenotypes shared by the yjeK and poxA mutants provided strong support that they function in a common pathway. This was further supported by our epistasis analysis that demonstrated that the phenotypes of the Salmonella poxA yjeK double mutant were no more or less severe than the poxA single mutant.
PoxA and YjeK Modify EF-P
The underlying cause of the pleiotropic phenotypes displayed by our mutants came to light through bioinformatic analysis indicating that both genes were closely linked with the efp gene, encoding EF-P.13,21 The poxA, yjeK and efp genes are linked, sometimes in the same operon, in a variety of Gram-negative bacteria. Genes that remain closely associated with one another despite large evolutionary distances often participate in a common pathway. EF-P was first identified in 1975 but neither its role in protein synthesis nor its function in cellular physiology are well understood.22 In an in vitro model of peptide bond synthesis, EF-P was found to stimulate the formation of a peptide bond between N-formyl-Met-tRNA and puromycin. Furthermore, the degree of stimulation was dependent on the amino acid acceptor, such that the stimulatory effect was the greatest for small, nonpolar amino acid acceptors such as glycine and leucine.23 The EF-P protein mimics a tRNA in both shape and size24 and a recent structure of EF-P bound to the 70S ribosome demonstrates that EF-P binds at a unique site between the peptidyl- and exit-tRNA sites where it interacts with ribosomal protein L1.25 This supported the general conclusion that EF-P plays a central role in protein translation, perhaps in the formation of the first peptide bond, despite the fact that the basic reactions of protein synthesis could be reconstituted in vitro in the absence of EF-P.
Earlier studies on EF-P purified from E. coli revealed that it carried a post-translational modification that had a mass consistent with that of lysine.26 We pursued the hypothesis that PoxA and YjeK may be responsible for modification of EF-P, perhaps with YjeK generating β-lysine that is subsequently ligated to EF-P by PoxA. In a reconstituted in vitro modification assay, we showed that EF-P becomes labeled with radioactive lysine in a PoxA-dependent manner.13 We also showed that this modification occurs at a specific lysine residue (Lys34) such that mutation of Lys34 to an alanine inhibits PoxA-mediated modification of EF-P and abolishes EF-P function in vivo, a finding corroborated by Yanagisawa et al.14 The recent crystal structure of EF-P in complex with PoxA showed that their interaction mimics that of an aaRS and its cognate tRNA, lending further support for the idea that the modification of EF-P by PoxA is analogous to the aminoacylation of tRNA by its cognate aaRS.14 YjeK enhanced the efficiency of the EF-P modification reaction, likely by generating in situ the preferred substrate for PoxA, β-lysine, from lysine.
On the Role of EF-P in Bacterial Physiology
The efp gene was previously reported to be essential for bacterial viability, consistent with the hypothesis that EF-P is critical for global protein synthesis.27,28 Systematic high-throughput deletion studies, however, have established that efp is not essential in either E. coli or Salmonella typhi.29–31 An earlier paper also demonstrated that EF-P is not essential in Agrobacterium tumefaciens.32 The Agrobacterium homolog of efp, chvH, was identified in a screen for mutants defective in plant virulence.33 chvH mutants exhibited reduced virulence gene expression and hypersusceptibility to detergents such as SDS and sodium deoxycholate, similar to what we observed for Salmonella poxA and yjeK mutants. Furthermore, the chvH mutant phenotypes could be rescued by complementation with the E. coli efp gene, supporting the hypothesis that chvH and efp are functional homologs. Together these findings support the hypothesis that EF-P does not play a role in global protein synthesis and, further, that its role in stress resistance and virulence is not limited to E. coli and Salmonella but may be universal in all bacterial species.
To assess the changes that might occur in response to EF-P modification, we assessed the protein expression patterns of wild-type, poxA and yjeK mutant strains. Our 2D-gel analysis of the mutant strains revealed that the changes in their soluble proteomes were relatively modest. Fewer than 90 spots differed in intensity, more than half of which appeared to be more abundant in the mutant strains. The majority of the downregulated proteins appear to have roles related to metabolism, such as nutrient acquisition and ATP generation. Interestingly a number of proteins belonging to the Salmonella pathogenicity island 1 (SPI-1) were upregulated in the poxA mutant strain. The upregulation of these proteins may be due to indirect effects in the poxA mutant strain. Given that the expression of relatively few proteins is affected by loss of PoxA or YjeK, it is highly plausible that modified EF-P does not act as a global translation factor, but rather as a posttranscriptional regulator of a specific subset of mRNAs. Our finding that most of the differentially expressed proteins are upregulated in the mutant strains provides important insight into the nature of EF-P regulation.
Given the pleiotropic nature of the Salmonella poxA and yjeK mutants, it is hard to tell at this point if there is a specific defect that directly attenuates virulence or if the virulence defect is caused by a combination of several factors. The detergent sensitivity observed in our Salmonella mutants as well as in the Agrobacterium chvH (efp) mutant, together with the hypersusceptibility to unrelated antimicrobial compounds, suggests that the underlying cause may involve changes in membrane biogenesis or permeability.
Is There a Common Physiological Link with eIF5A?
The functions of the eukaryotic homolog of EF-P, eIF5A and its archaeal counterpart, aIF5A, are equally enigmatic. Interestingly, both eIF5A and aIF5A are modified at a highly conserved lysine residue to generate the unique amino acid hypusine (Fig. 2).34 This residue maps to the same position that is modified in EF-P. However, despite the similarities between the eIF5A and EF-P modifications, the enzymes involved in the generation of hypusine bear no similarity to PoxA or YjeK.
Figure 2.
The structural similarity of EF-P and its modification to eIF5A. The structure of EF-P (left) is simliar to that of eIF5A of S. cerevisiae (PDB 3ER0) except that EF-P contains an additional C-terminal domain that is absent from eIF5A. Both factors contain a highly conserved loop, containing the modified lysine (circled). The structures of the modified amino acids are shown in boxes below each structure. The modification on EF-P is proposed to be lysyl-β-lysine although its structure has not been formally determined in the context of the native protein.
Like EF-P, the cellular function of eIF5A remains poorly understood but the factor is likely to have an ancillary role in protein synthesis. Many eukaryotes encode multiple isoforms of eIF5A. For example, human and yeast have two isoforms whereas Arabidopsis has three.35–37 Intriguingly many of the phenotypes associated with eIF5A are involved with stress adaptation, cell cycle control and apoptosis.34,38–40 Following the induction of oxidative stress, eIF5A has also been shown to be essential for polysome disassembly and stress granule formation, most likely through its role in enhancing translation elongation.41 Recently several eIF5A paralogs were shown to play a role in tolerance to heat, osmotic and oxidative stress in Arabidopsis thaliana.42,43 Overexpression of eIF5A under any of these stress conditions elicited a stronger response from existing stress response pathways, thereby providing enhanced stress protection.
Another similarity between EF-P and eIF5A centers on their possible role in the biogenesis of the cell envelope. A synthetic lethal screen in yeast identified Ypt1, a GTPase essential for vesicular trafficking and secretion, as a novel genetic interactor of eIF5A.36 These data point towards a specialized role for eIF5A in regulating protein translation and stress response in eukaryotes under adverse conditions, perhaps via changes on the cell surface, similar to what we observe for EF-P in Salmonella.
Summary
The recent work on EF-P indicates that this factor, previously presumed to be essential for protein synthesis, instead plays an auxiliary role that is required for virulence and resistance to multiple forms of stress. PoxA and YjeK function to modify EF-P and may play a role in translational reprogramming during infection or under conditions of stress. Beta-lysylation, the novel modification reaction carried out by PoxA and YjeK represents a striking example of molecular mimicry where a tRNA synthetase paralog modifies a tRNA-shaped protein instead of a tRNA.
Future work will focus on identifying the targets of EF-P mediated regulation and the mechanism by which EF-P specifically recognizes those targets. Given that eIF5A has been shown to bind RNA in a sequence-dependent manner and EF-P itself contains an RNA-binding domain, the existence of a consensus EF-P binding motif seems likely.44 However, the role of secondary mRNA structure in conferring specificity cannot be ruled out as many RNA-binding proteins are known to target specific structural elements. The recently solved structure of EF-P on the ribosome also revealed direct interactions between the modified region of EF-P and the 3′-end of initiator tRNA in the P-site, suggesting that EF-P may have rather direct effects on translation initiation.25 In addition to possible roles for EF-P in modulating ribosomal mRNA binding and initiation, a further effect to optimize the efficiency of translation elongation is also possible given that such a role has recently been demonstrated for eIF5a.45,46 Work is now needed to understand exactly how EF-P functions in translation, and how modification regulates its various possible activities.
The underlying reason for the virulence attenuation observed for both Salmonella and Agrobacterium will also be a central area of future inquiry. The apparent paradox that Salmonella poxA mutants are avirulent despite increased SPI-1 expression is likely explained by the stress resistance defects displayed by these mutants. Another possibility is that SPI-1 expression, which is rapidly shut down upon entry into macrophages, is not properly controlled in these mutants.47 We also do not currently know whether the lysyl-β-lysine modification of EF-P occurs constitutively or is only triggered under specific conditions. Given that EF-P appears to play a role in stress tolerance and virulence, it will be important to determine whether the modification system itself is regulated by those conditions. Alternatively, if EF-P is constitutively modified, there must exist additional inputs that determine the substrate specificity of EF-P-mediated regulation.
All bacterial genomes sequenced to date encode at least one EF-P homolog, with many genomes containing additional paralogs.21 While many of the secondary EF-P paralogs lack the conserved lysine residue, there is a strong correlation between the presence of the lysine residue and whether or not the bacterium has PoxA and YjeK homologs. Ninety-seven percent of EF-P homologs from species with a PoxA and YjeK homolog have Lys34 compared to 70% of EF-P homologs from species lacking PoxA and YjeK.21 This raises the question of whether EF-P is modified in those species without a PoxA or YjeK enzyme, such as Pseudomonas. Since the modified lysine is a conserved feature in both the EF-P and eIF5A proteins, it will be of interest to determine if and how lysyl modification is carried out in other bacterial species. The fact that many bacterial species have more than one EF-P paralog is reminiscent of the varied distribution of eIF5A among eukaryotes. Indeed, the two human isoforms of eIF5A appear to be differentially expressed and fulfill unique roles within the cell.35,48 Whether this is the case for bacterial EF-P paralogs remains to be determined and will be an important area for future research.
Acknowledgements
The Navarre laboratory is supported by an Operating Grant and New Investigator Award from the Canada Institutes for Health Research (MOP-86683 and MSH-87729) and a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada (RGPIN 386286-10). S. Betty Zou is supported by a Vanier Canada Graduate Scholarship from the Natural Sciences and Engineering Research Council of Canada. Work on EF-P in the Ibba lab described above was supported by the National Institutes of Health (GM65183).
References
- 1.Sharma UK, Chatterji D. Transcriptional switching in Escherichia coli during stress and starvation by modulation of sigma activity. FEMS Microbiol Rev. 2010;34:646–657. doi: 10.1111/j.1574-6976.2010.00223.x. [DOI] [PubMed] [Google Scholar]
- 2.Raghavan V, Groisman EA. Orphan and hybrid two-component system proteins in health and disease. Curr Opin Microbiol. 2010;13:226–231. doi: 10.1016/j.mib.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gooderham WJ, Hancock REW. Regulation of virulence and antibiotic resistance by two-component regulatory systems in Pseudomonas aeruginosa. FEMS Microbiol Rev. 2009;33:279–294. doi: 10.1111/j.1574-6976.2008.00135.x. [DOI] [PubMed] [Google Scholar]
- 4.Vogel J. A rough guide to the non-coding RNA world of Salmonella. Mol Microbiol. 2009;71:1–11. doi: 10.1111/j.1365-2958.2008.06505.x. [DOI] [PubMed] [Google Scholar]
- 5.Block KF, Hammond MC, Breaker RR. Evidence for widespread gene control function by the ydaO riboswitch candidate. J Bacteriol. 2010;192:3983–3989. doi: 10.1128/JB.00450-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park S, Cromie MJ, Lee E, Groisman EA. A bacterial mRNA leader that employs different mechanisms to sense disparate intracellular signals. Cell. 2010;142:737–748. doi: 10.1016/j.cell.2010.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dalebroux ZD, Svensson SL, Gaynor EC, Swanson MS. ppGpp conjures bacterial virulence. Microbiol Mol Biol Rev. 2010;74:171–199. doi: 10.1128/MMBR.00046-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pesavento C, Hengge R. Bacterial nucleotide-based second messengers. Curr Opin Microbiol. 2009;12:170–176. doi: 10.1016/j.mib.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 9.Gerdes K, Christensen SK, Løbner-Olesen A. Prokaryotic toxin-antitoxin stress response loci. Nat Rev Microbiol. 2005;3:371–382. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- 10.Milon P, Tischenko E, Tomšic J, Caserta E, Folkers G, La Teana A, et al. The nucleotide-binding site of bacterial translation initiation factor 2 (IF2) as a metabolic sensor. Proc Nat Acad Sci USA. 2006;103:13962–13967. doi: 10.1073/pnas.0606384103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giuliodori AM, Brandi A, Giangrossi M, Gualerzi CO, Pon CL. Cold-stress-induced de novo expression of infC and role of IF3 in cold-shock translational bias. RNA. 2007;13:1355–1365. doi: 10.1261/rna.455607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spriggs KA, Bushell M, Willis AE. Translational regulation of gene expression during conditions of cell stress. Mol Cell. 2010;40:228–237. doi: 10.1016/j.molcel.2010.09.028. [DOI] [PubMed] [Google Scholar]
- 13.Navarre WW, Zou SB, Roy H, Xie JL, Savchenko A, Singer A, et al. PoxA, YjeK and elongation factor P coordinately modulate virulence and drug resistance in Salmonella enterica. Mol Cell. 2010;39:209–221. doi: 10.1016/j.molcel.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yanagisawa T, Sumida T, Ishii R, Takemoto C, Yokoyama S. A paralog of lysyl-tRNA synthetase aminoacylates a conserved lysine residue in translation elongation factor P. Nat Struct Mol Biol. 2010;17:1136–1143. doi: 10.1038/nsmb.1889. [DOI] [PubMed] [Google Scholar]
- 15.Kaniga K, Compton M, Curtiss R, Sundaram P. Molecular and functional characterization of Salmonella enterica serovar typhimurium poxA gene: effect on attenuation of virulence and protection. Infect Immun. 1998;66:5599–5606. doi: 10.1128/iai.66.12.5599-5606.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang Y, Cronan J. Mapping nonselectable genes of Escherichia coli by using transposon Tn10: location of a gene affecting pyruvate oxidase. J Bacteriol. 1982;151:1279–1289. doi: 10.1128/jb.151.3.1279-1289.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang Y, Cronan J. Genetic and biochemical analyses of Escherichia coli strains having a mutation in the structural gene (poxB) for pyruvate oxidase. J Bacteriol. 1983;154:756–762. doi: 10.1128/jb.154.2.756-762.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Behshad E, Ruzicka F, Mansoorabadi S, Chen D, Reed GH, Frey PA. Enantiomeric free radicals and enzymatic control of stereochemistry in a radical mechanism: the case of lysine 2,3-aminomutases. Biochem. 2006;45:12639–12646. doi: 10.1021/bi061328t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bochner B, Gadzinski P, Panomitros E. Phenotype microarrays for high-throughput phenotypic testing and assay of gene function. Genome Res. 2001;11:1246–1255. doi: 10.1101/gr.186501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Dyk T, Smulski D, Chang Y. Pleiotropic effects of poxA regulatory mutations of Escherichia coli and Salmonella typhimurium, mutations conferring sulfometuron methyl and alpha-ketobutyrate hypersensitivity. J Bacteriol. 1987;169:4540–4546. doi: 10.1128/jb.169.10.4540-4546.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bailly M, de Crécy-Lagard V. Predicting the pathway involved in post-translational modification of elongation factor P in a subset of bacterial species. Biol Direct. 2010;5:3. doi: 10.1186/1745-6150-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glick B, Ganoza M. Identification of a soluble protein that stimulates peptide bond synthesis. Proc Natl Acad Sci USA. 1975;72:4257–4260. doi: 10.1073/pnas.72.11.4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glick B, Chládek S, Ganoza M. Peptide bond formation stimulated by protein synthesis factor EF-P depends on the aminoacyl moiety of the acceptor. Eur J Biochem. 1979;97:23–28. doi: 10.1111/j.1432-1033.1979.tb13081.x. [DOI] [PubMed] [Google Scholar]
- 24.Hanawa-Suetsugu K, Sekine S, Sakai H, Hori-Takemoto C, Terada T, Unzai S, et al. Crystal structure of elongation factor P from Thermus thermophilus HB8. Proc Natl Acad Sci USA. 2004;101:9595–9600. doi: 10.1073/pnas.0308667101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blaha G, Stanley R, Steitz T. Formation of the first peptide bond: the structure of EF-P bound to the 70S ribosome. Science. 2009;325:966–970. doi: 10.1126/science.1175800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aoki H, Xu J, Emili A, Chosay JG, Golshani A, Ganoza MC. Interactions of elongation factor EF-P with the Escherichia coli ribosome. FEBS J. 2008;275:671–681. doi: 10.1111/j.1742-4658.2007.06228.x. [DOI] [PubMed] [Google Scholar]
- 27.Aoki H, Dekany K, Adams S, Ganoza M. The gene encoding the elongation factor P protein is essential for viability and is required for protein synthesis. J Biol Chem. 1997;272:32254–32259. doi: 10.1074/jbc.272.51.32254. [DOI] [PubMed] [Google Scholar]
- 28.Gerdes SY, Scholle MD, Campbell JW, et al. Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J Bacteriol. 2003;185:5673–5684. doi: 10.1128/JB.185.19.5673-5684.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamamoto N, Nakahigashi K, Nakamichi T, Yoshino M, Takai Y, Touda Y, et al. Update on the Keio collection of Escherichia coli single-gene deletion mutants. Mol Syst Biol. 2009;5:335. doi: 10.1038/msb.2009.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006–2008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langridge GC, Phan M, Turner DJ, Perkins TT, Parts L, Haase J, et al. Simultaneous assay of every Salmonella typhi gene using one million transposon mutants. Genome Res. 2009;19:2308–2316. doi: 10.1101/gr.097097.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peng W, Banta L, Charles T, Nester E. The chvH locus of Agrobacterium encodes a homologue of an elongation factor involved in protein synthesis. J Bacteriol. 2001;183:36–45. doi: 10.1128/JB.183.1.36-45.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Charles TC, Nester EW. A chromosomally encoded two-component sensory transduction system is required for virulence of Agrobacterium tumefaciens. J Bacteriol. 1993;175:6614–6625. doi: 10.1128/jb.175.20.6614-6625.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park M, Nishimura K, Zanelli C, Valentini S. Functional significance of eIF5A and its hypusine modification in eukaryotes. Amino Acids. 2010;38:491–500. doi: 10.1007/s00726-009-0408-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clement P, Johansson H, Wolff E, Park M. Differential expression of eIF5A-1 and eIF5A-2 in human cancer cells. FEBS J. 2006;273:1102–1114. doi: 10.1111/j.1742-4658.2006.05135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frigieri M, Thompson G, Pandolfi J, Zanelli C, Valentini S. Use of a synthetic lethal screen to identify genes related to TIF51A in Saccharomyces cerevisiae. Genet Mol Res. 2007;6:152–165. [PubMed] [Google Scholar]
- 37.Liu Z, Duguay J, Ma F, Wang TW, Tshin R, Hopkins MT, et al. Modulation of eIF5A1 expression alters xylem abundance in Arabidopsis thaliana. J Exp Bot. 2008;59:939–950. doi: 10.1093/jxb/ern017. [DOI] [PubMed] [Google Scholar]
- 38.Li A, Li H, Jin B, Ye QN, Zhou T, Yu XD, et al. A novel eIF5A complex functions as a regulator of p53 and p53-dependent apoptosis. J Biol Chem. 2004;279:49251–49258. doi: 10.1074/jbc.M407165200. [DOI] [PubMed] [Google Scholar]
- 39.Sun Z, Cheng Z, Taylor C, Mcconkey B, Thompson J. Apoptosis induction by eIF5A1 involves activation of the intrinsic mitochondrial pathway. J Cell Physiol. 2010;223:798–809. doi: 10.1002/jcp.22100. [DOI] [PubMed] [Google Scholar]
- 40.Rahman-Roblick R, Roblick UJ, Hellman U, Conrotto P, Liu T, Becker S, et al. p53 targets identified by protein expression profiling. Proc Natl Acad Sci USA. 2007;104:5401–5406. doi: 10.1073/pnas.0700794104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li C, Ohn T, Ivanov P, Tisdale S, Anderson P. eIF5A promotes translation elongation, polysome disassembly and stress granule assembly. PLoS ONE. 2010;5:9942. doi: 10.1371/journal.pone.0009942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma F, Liu Z, Wang T, Hopkins MT, Peterson CA, Thompson JE. Arabidopsis eIF5A3 influences growth and the response to osmotic and nutrient stress. Plant Cell Environ. 2010;33:1682–1696. doi: 10.1111/j.1365-3040.2010.02173.x. [DOI] [PubMed] [Google Scholar]
- 43.Xu J, Zhang B, Jiang C, Ming F. RceIF5A, encoding an eukaryotic translation initiation factor 5A in Rosa chinensis, can enhance thermotolerance, oxidative and osmotic stress resistance of Arabidopsis thaliana. Plant Mol Biol. 2010;75:167–178. doi: 10.1007/s11103-010-9716-2. [DOI] [PubMed] [Google Scholar]
- 44.Xu A, Jao DL, Chen KY. Identification of mRNA that binds to eukaryotic initiation factor 5A by affinity co-purification and differential display. Biochem J. 2004;384:585–590. doi: 10.1042/BJ20041232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saini P, Eyler D, Green R, Dever T. Hypusine-containing protein eIF5A promotes translation elongation. Nature. 2009;459:118–121. doi: 10.1038/nature08034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gregio A, Cano V, Avaca J, Valentini S, Zanelli C. eIF5A has a function in the elongation step of translation in yeast. Biochem Biophys Res Commun. 2009;380:785–790. doi: 10.1016/j.bbrc.2009.01.148. [DOI] [PubMed] [Google Scholar]
- 47.Bustamante V, Martínez L, Santana F, Knodler LA, Steele-Mortimer O, Puente JL. HilD-mediated transcriptional cross-talk between SPI-1 and SPI-2. Proc Natl Acad Sci USA. 2008;105:14591–14596. doi: 10.1073/pnas.0801205105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clement PMJ, Henderson CA, Jenkins ZA, Smit-McBride Z, Wolff EC, Hershey JW, et al. Identification and characterization of eukaryotic initiation factor 5A-2. Eur J Bioche. 2003;270:4254–4263. doi: 10.1046/j.1432-1033.2003.03806.x. [DOI] [PubMed] [Google Scholar]
- 49.Shi H, Moore PB. The crystal structure of yeast phenylalanine tRNA at 1.93 Å resolution: a classic structure revisited. RNA. 2000;6:1091–1105. doi: 10.1017/s1355838200000364. [DOI] [PMC free article] [PubMed] [Google Scholar]