Abstract
The proto-oncogene MDM2 inhibits p53 and plays a key role in cell growth control and apoptosis. Identification of two antagonizing MDM2 polymorphisms, SNP285 and SNP309, affecting cancer risk through modulation of Sp1 transcription factor binding, shed new light on the biological activity and phylogeny of this gene.
Key words: MDM2, polymorphism, breast cancer, ovarian cancer, endometrial cancer
Introduction
MDM2 (mouse double minute 2 homolog) is a major regulator of p53, popularly coined the “guardian of the genome.” The levels of these two important proteins are tightly regulated and linked in a feedback loop where p53 acts as a transcription factor inducing MDM2 transcription, while MDM2 binds, inhibits and degrades the p53 protein through E3 ligase activity.1–3 The functional importance of this balanced interaction is illustrated by the findings that knock-out of the MDM2 gene leads to embryonic lethality in mice, an effect rescued by concomitant p53 deletion.4,5 In addition to the tight link to p53, MDM2 is also known to interact with several other major players involved in cellular growth control. Among others, MDM2 binds to and inhibits the function of the retinoblastoma protein (pRB),6 leading to increased levels of free E2F1, promoting cell cycle progression. Enhancing this effect, MDM2 also bind directly to E2F1 and stimulates its activity as a transcription factor.7 Given this role as a negative regulator of p53 and pRb functional pathways, one may regard MDM2 as a “master regulator gene,” with a key position governing cellular homeostasis. Thus, it is clear that MDM2 is a classical protooncogene that, if hyperactive, contributes to malignant cell proliferation. Elevated MDM2 activity is found in many tumors and is considered as an alternative way of p53 inactivation.
The two classical underlying mechanisms of MDM2 hyperactivity in cells are gene amplification (increased gene copy number)8,9 and enhanced translation,10–12 both of which are observed in many human cancer forms harboring wild-type TP53 (the gene coding for the p53 protein). Recently, a third mechanism of MDM2 hyperactivation was suggested: Inuzuka and co-workers demonstrated MDM2 to be phosphorylated by Casein Kinase I (CKI), subsequently enhancing MDM2 degradation by the SCFβ-TRCP ubiquitin ligase.13 In cases where CKI and/or β-TRCP are inactivated, this would lead to abnormally high levels of MDM2 and, thus, tumor promoting MDM2 hyperactivity.14 This hypothesis is indirectly supported by the findings of β-TRCP deletions in several tumor forms.14–16
In this review, we will focus on a fourth mechanism influencing MDM2 activity: promoter single nucleotide polymorphisms (SNPs) affecting MDM2 transcription. The promoter SNPs (and haplotypes of combined SNPs) are unique in the sense that they are the only described cis-acting mechanisms affecting MDM2 through altering transcription levels. Also, while the three above mentioned mechanisms are all tumor specific, promoter SNPs are the only described germ line MDM2 alterations. Thus, ethnic distribution of these variants may provide important information about evolutionary mechanisms selecting for different MDM2 variants, subsequently extending our biological knowledge about the multiple biological functions of this important gene.
MDM2 SNP309 as a Cancer Risk Factor
In 2004, the group headed by Arnold Levine made the first discovery of a MDM2 functional polymorphism located in the intronic promoter of MDM2 (promoter P2).17 The polymorphism was a T to G transversion and was termed SNP309 (SNP309T>G; rs2279744) due to its position 309 bps downstream of MDM2 exon 1. In their original paper, Bond et al.17 showed that the G-variant of this polymorphism (SNP309G) extended a binding site for the transcription factor Sp1 and, thus, lead to increased transcription of MDM2, as compared to the SNP309T allele. Further, the tumorigenic effect of the SNP309G allele was supported by the findings of an association between this allele and early age at cancer diagnosis among individuals with Li-Fraumeni syndrome (carrying germ-line TP53 mutations), as well as an early age at diagnosis of soft tissue sarcomas, large B-cell lymphomas and colorectal cancers in women without any concomitant cancer predisposing germline mutations.17,18 In addition, an association between SNP309G and an early diagnosis of estrogen receptor rich breast cancer was recorded.19 Recently, novel data from mouse models strongly supported the initial findings: Post and co-workers generated mice carrying the human versions of promoter P2. In this model, mice carrying the SNP309GG genotype were observed to be more tumor prone than those carrying the SNP309TT genotype.20
However, while numerous subsequent case-control studies assessing the role of SNP309 as a cancer risk factor or a predictor of early cancer development have been performed, the data generated have been somewhat conflicting. So far, there is no clear pattern with respect to a potential difference in the effect of SNP309 on different cancer forms. However, with respect to ethnicity, most of the studies linking the SNP309G variant allele to increased cancer risk or a young age at diagnosis have been performed on Asian or Ashkenazi Jewish populations. In contrast, the majority of studies performed in Caucasians have revealed negative results.21,22
MDM2 SNP285 Reduces Sp1 Transcription Factor Binding
While studying SNP309 status in Norwegian breast cancer patients, we discovered a second MDM2 polymorphism (SNP285G>C; rs117039649) located only 24 bps upstream of SNP309 in the MDM2 intronic promoter (P2).23 SNP285C was located on the SNP309G allele forming a distinct SNP285C/309G haplotype. As was the case for SNP309, SNP285 was located within a predicted binding site for the Sp1 transcription factor. Notably, the two SNPs were located at two adjacent Sp1 binding sites. Preliminary in silico analyses indicated that the presence of the SNP285C-allele would reduce the binding strength between Sp1 and the MDM2 promoter, as compared to the SNP285G-allele. Thus, we hypothesized that SNP285C may counteract the effect of SNP309G.
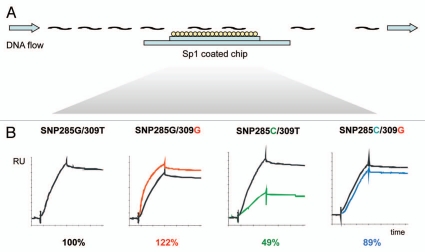
Applying a highly sensitive surface plasmon resonance (SPR) assay (Fig. 1) using the SNP309T allele as reference standard (100%), we demonstrated that the SNP309G-allele enhances Sp1 protein binding to the MDM2 promoter to 122%. In contrast, SNP285C reduced Sp1 binding to only 48.5% of that of the wild-type SNP285G. Interestingly, the double polymorphic SNP285C/309G haplotype bound Sp1 with an affinity of 89.4% of that of the wild-type SNP285G/309T haplotype, indicating that the presence of SNP285C not only neutralizes, but also overcompensates for, the effect of SNP309G. Thus, a “double polymorphic allele” (SNP285C/309G) should have a moderately reduced transcriptional activity as compared to the wild-type (SNP285G/309T) allele (Fig. 2).23 Further, the in vitro binding curves did not fit a 1:1 binding model but rather a heterogeneous ligand model, in line with a model where two or more Sp1 molecules bind each MDM2 promoter. Thus, the binding curves support the results from in silico prediction of SNP285 and 309 to reside within two separate but adjacent Sp1 binding sites.
Figure 1.
Surface plasmon resonance (SPR) assay determining the effect of SNP285 and SNP309 on Sp1 binding to the MDM2 promoter P2. DNA fragments amplified from the MDM2 promoter were injected into a flow over an Sp1 coated chip (A), while changes in the SPR were recorded (B). The wild-type SNP285G/309T-allele was used as reference (100%), to which the other variant alleles were compared. Illustration modified from reference 23.
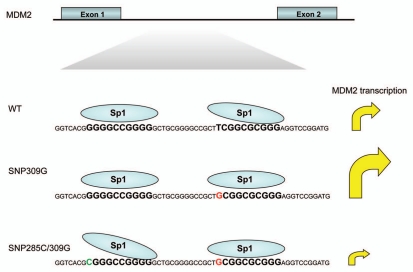
Figure 2.
Effects of MDM2 SNP285 and SNP309 on transcription. The enhanced Sp1 binding to the MDM2 promoter caused by SNP309G leads to increased transcription, while the reduced Sp1 binding to an adjacent binding site, caused by SNP285C reduces transcription.
SNP285C Reduces the Risk of Female Cancers
Subsequent to the in vitro findings we have analyzed the distribution of SNP285 and SNP309 in several female cancer forms. Based on the finding of reduced binding strength between the MDM2 promoter and the Sp1 transcription factor in the presence of SNP285C, one would assume that this polymorphism was associated with reduced risk of cancer. Data so far on three major female cancers (breast, ovarian and endometrial) establish that this is in fact the case. In our initial study,23 we compared the frequencies of SNP285 in 1,973 breast cancer and 1,993 ovarian cancer patients with 2,465 healthy controls. We found SNP285C to reduce the risk of the two malignancies among carriers of the SNP309G-allele (SNP309TG heterozygotes and SNP309GG homozygotes combined) with 21 and 26%, respectively. Interestingly, there was a clear difference with respect to the effect on individuals heterozygous or homozygous for the SNP309G variant between the two cancer forms. While the effect of SNP285C on breast cancer was most profound in SNP309GG homozygotes (risk reduction of 45 versus 9% only in SNP309TG heterozygotes), the opposite was the case for ovarian cancer: we found a risk reduction of 37% for SNP285C carriers among SNP309TG heterozygotes, while no effect was observed among SNP309GG homozygotes.
Recently, we have completed a similar screening among patients suffering from endometrial cancer.24 The results here mirrors the data obtained for ovarian cancer: SNP285C reduce the risk of endometrial cancer by 39% among SNP309TG heterozygotes. No effect was observed on SNO309GG homozygotes.
The results from the screenings of ovarian and endometrial cancers is very much in line with the in vitro data, indicating that one SNP285C may neutralize the effect of one SNP309G allele but not two. The breast cancer data does not fit this model. So far we lack an explanation for this observation, which should be confirmed in other trials. However, it is well known from genes other than MDM2 that heterozygote carriers of a genetic alteration may be at risk of different diseases from the homozygotes.25
Hypothesizing an Influence of MDM2 SNPs on Diseases Other than Cancer
In addition to the case-control part of our initial screening, we also assessed the distribution of SNP285C in different ethnic populations.23 While the frequency of the C allele was similar in Norway, The Netherlands and the UK (approximately 8%), it was absent in an Asian population (Chinese). Further, in a Caucasian population with a slightly different migratory pattern than the rest of western Europe (Finns) the frequency of SNP285C was <2%. These data indicate the C variant of SNP285 to be a relatively young polymorphism, which seems to have spread rapidly among western Caucasians.26 A positive selection for this variant, making such a rapid spread possible, most likely must be due to survival advantages other than a reduction in cancer risk, since cancers, with a few exception, appear after the age of childbirth. Given the role of the p53 pathway in processes like inflammation, one may speculate if the MDM2 promoter polymorphism may affect resistance towards infectious diseases. Future studies are warranted to address this topic.
SNP285C May Have Confounded Contemporary SNP309 Studies
Considering the potential effects of the SNP309G variant, as mentioned above, most studies performed on Asian populations are positive for a cancer promoting effect of this polymorphism, while the data from Caucasian populations are more conflicting.21,22 The presence of the SNP309G counteracting SNP285C variant among ∼8% of western Caucasians (i.e., ∼12% of the SNP309G alleles) may have affected the conclusions of case-control studies performed on Caucasian populations so far. It may well be that the studies performed on the SNP285C-free Asian cohorts represent the “true” effect of SNP309G, while studies in western Europe need to be “corrected” for SNP285 status, in order to reveal the impact of SNP309G on cancer risk in these populations.
Conclusion
In conclusion, contrasting the previously described mechanisms of MDM2 hyperactivation, the SNPs in promoter P2 link transcriptional regulation of this gene to tumorigenesis. SNP285C significantly reduces the risk of three major female cancers and may have confounded case-control studies of SNP309 in Caucasians.
Abbreviations
- MDM2
mouse double-minute 2 homologue
- SNP
single nucleotide polymorphism
- Sp1
specificity protein 1
References
- 1.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 2.Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 3.Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 4.Jones SN, Roe AE, Donehower LA, Bradley A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature. 1995;378:206–208. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- 5.Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378:203–206. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 6.Xiao ZX, Chen J, Levine AJ, Modjtahedi N, Xing J, Sellers WR, et al. Interaction between the retinoblastoma protein and the oncoprotein MDM2. Nature. 1995;375:694–698. doi: 10.1038/375694a0. [DOI] [PubMed] [Google Scholar]
- 7.Martin K, Trouche D, Hagemeier C, Sorensen TS, La Thangue NB, Kouzarides T. Stimulation of E2F1/DP1 transcriptional activity by MDM2 oncoprotein. Nature. 1995;375:691–694. doi: 10.1038/375691a0. [DOI] [PubMed] [Google Scholar]
- 8.Oliner JD, Kinzler KW, Meltzer PS, George DL, Vogelstein B. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature. 1992;358:80–83. doi: 10.1038/358080a0. [DOI] [PubMed] [Google Scholar]
- 9.Momand J, Jung D, Wilczynski S, Niland J. The MDM2 gene amplification database. Nucleic Acids Res. 1998;26:3453–3459. doi: 10.1093/nar/26.15.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Landers JE, Cassel SL, George DL. Translational enhancement of mdm2 oncogene expression in human tumor cells containing a stabilized wild-type p53 protein. Cancer Res. 1997;57:3562–3568. [PubMed] [Google Scholar]
- 11.Sheikh MS, Shao ZM, Hussain A, Fontana JA. The p53-binding protein MDM2 gene is differentially expressed in human breast carcinoma. Cancer Res. 1993;53:3226–3228. [PubMed] [Google Scholar]
- 12.Trotta R, Vignudelli T, Candini O, Intine RV, Pecorari L, Guerzoni C, et al. BCR/ABL activates mdm2 mRNA translation via the La antigen. Cancer Cell. 2003;3:145–160. doi: 10.1016/s1535-6108(03)00020-5. [DOI] [PubMed] [Google Scholar]
- 13.Inuzuka H, Tseng A, Gao D, Zhai B, Zhang Q, Shaik S, et al. Phosphorylation by casein kinase I promotes the turnover of the Mdm2 oncoprotein via the SCF(beta-TRCP) ubiquitin ligase. Cancer Cell. 2010;18:147–159. doi: 10.1016/j.ccr.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inuzuka H, Fukushima H, Shaik S, Wei W. Novel insights into the molecular mechanisms governing mdm2 ubiquitination and destruction. Oncotarget. 2010;1:685–690. doi: 10.18632/oncotarget.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Momand J, Wu HH, Dasgupta G. MDM2—master regulator of the p53 tumor suppressor protein. Gene. 2000;242:15–29. doi: 10.1016/s0378-1119(99)00487-4. [DOI] [PubMed] [Google Scholar]
- 16.Rayburn E, Zhang R, He J, Wang H. MDM2 and human malignancies: expression, clinical pathology, prognostic markers and implications for chemotherapy. Curr Cancer Drug Targets. 2005;5:27–41. doi: 10.2174/1568009053332636. [DOI] [PubMed] [Google Scholar]
- 17.Bond GL, Hu W, Bond EE, Robins H, Lutzker SG, Arva NC, et al. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell. 2004;119:591–602. doi: 10.1016/j.cell.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 18.Bond GL, Menin C, Bertorelle R, Alhorpuro P, Aaltonen LA, Levine AJ. MDM2 SNP309 Accelerates colorectal tumour formation in women. J Med Genet. 2006;43(12):950–952. doi: 10.1136/jmg.2006.043539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bond GL, Hirshfield KM, Kirchhoff T, Alexe G, Bond EE, Robins H, et al. MDM2 SNP309 accelerates tumor formation in a gender-specific and hormone-dependent manner. Cancer Res. 2006;66:5104–5110. doi: 10.1158/0008-5472.CAN-06-0180. [DOI] [PubMed] [Google Scholar]
- 20.Post SM, Quintas-Cardama A, Pant V, Iwakuma T, Hamir A, Jackson JG, et al. A high-frequency regulatory polymorphism in the p53 pathway accelerates tumor development. Cancer Cell. 2010;18:220–230. doi: 10.1016/j.ccr.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Economopoulos KP, Sergentanis TN. Differential effects of MDM2 SNP309 polymorphism on breast cancer risk along with race: a meta-analysis. Breast Cancer Res Treat. 2010;120:211–216. doi: 10.1007/s10549-009-0467-1. [DOI] [PubMed] [Google Scholar]
- 22.Hu Z, Jin G, Wang L, Chen F, Wang X, Shen H. MDM2 promoter polymorphism SNP309 contributes to tumor susceptibility: evidence from 21 case-control studies. Cancer Epidemiol Biomarkers Prev. 2007;16:2717–2723. doi: 10.1158/1055-9965.EPI-07-0634. [DOI] [PubMed] [Google Scholar]
- 23.Knappskog S, Bjornslett M, Myklebust LM, Huijts PE, Vreeswijk MP, Edvardsen H, et al. The MDM2 promoter SNP285C/309G haplotype diminishes Sp1 transcription factor binding and reduces risk for breast and ovarian cancer in Caucasians. Cancer Cell. 2011;19:273–282. doi: 10.1016/j.ccr.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 24.Knappskog S, Trovik J, Marcickiewicz J, Tingulstad S, Staff AC, Hveem K, et al. Impact of the MDM2 promoter polymorphsims SNP285 and SNP309 on endometrial cancer risk among Caucasians. ASCO. 2011:5092. [Google Scholar]
- 25.Smirnov DA, Cheung VG. ATM gene mutations result in both recessive and dominant expression phenotypes of genes and microRNAs. Am J Hum Genet. 2008;83:243–253. doi: 10.1016/j.ajhg.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knappskog S, Lonning PE. MDM2 promoter SNP285 and SNP309; phylogeny and impact on cancer risk. Oncotarget. 2011;2:251–258. doi: 10.18632/oncotarget.243. [DOI] [PMC free article] [PubMed] [Google Scholar]