Abstract
Splicing can occur co-transcriptionally. What happens when the splicing reaction lags after the completed transcriptional process? We found that elongation rates are independent of ongoing splicing on the examined genes and suggest that when transcription has completed but splicing has not, the splicing machinery is retained at the site of transcription, independently of the polymerase.
Key words: co-transcriptional splicing, transcription kinetics, live-cell imaging, RNA polymerase
The Connection Between Transcription Rates and Co-transcriptional Splicing
The unraveling of unique nuclear compartments termed nuclear bodies has led to the quest for the spatial detection of unique sites of DNA replication, mRNA transcription and other gene expression related processes. Among these, the actual site of pre-mRNA splicing in the cell nucleus has been examined. While it seemed reasonable that splicing would occur sequentially to transcription, and perhaps only after the release of the mRNA from the gene, immunocytological findings of the distribution of transcribing genes and splicing factors actually pointed to the fact that these processes are occurring in close proximity.1 Currently, it is agreed upon that much of the splicing events occur co-transcriptionally. This means that the spliceosomal machinery can assemble on the nascent transcript while still tethered to the actively transcribing polymerase.2,3 Co-transcriptional splicing also implies that some introns are removed prior to the completion of mRNA synthesis.4–11
The significance of a cotranscriptional processing mechanism is a highly discussed issue,12 especially as it becomes evident that this is a widespread process occurring on many genes and in different organisms. The almost immediate detection of splice sites on the emerging nascent transcript in a cotranscriptional model would replace the need for a scanning mechanism that would be required to run through the completed pre-mRNA sequence and search for exons, which tend to be rather small sized and lying in between the comparably huge intronic sequences. The understanding that the spliceosomal machinery can piggy-back on the pre-mRNA while still attached to the polymerase has raised some interesting ideas. For instance, it was appealing to suggest that the long C-terminal tail (CTD) of the polymerase, which one could imagine swaying behind the enzyme in proximity with the emerging pre-mRNA, could serve as a landing pad for splicing factors traveling with the polymerase and as a launching pad for targeting them onto the pre-mRNA once splice sites emerged.13–16 Indeed, some studies pointed in this direction.17–21 However, conclusive data on this subject has not accumulated, particularly since the CTD itself was not found to have a stimulating effect on splicing in yeast or in mammalian cells.22,23 Yet, the involvement of the CTD in the regulation of alternative splicing has been demonstrated through the SRp20 SR protein.24 This suggests that the CTD does have a role in the recruitment of splicing factors to the pre-mRNA, possibly in an indirect manner. Therefore, at the moment it remains unclear whether indeed such direct interactions between the splicing machinery and the polymerase actually occur and, if they do, to what purpose.
Recent studies now point to the role of histone modifications in the marking of exon and intron boundaries, in the recruitment of splicing factors, and in the outcome of alternative splicing12,25,26 and, since the connection between transcription and alternative splicing is well established, they therefore support the cotranscriptional splicing concept.27,28 These studies have shown that RNAP II elongation rates can influence the choice of alterative exons and thus the generation of alternatively spliced products. By reducing RNAP II elongation rates either via mutations in the enzyme or by use of drug treatments, alternative exon inclusion was enhanced, similar to experiments that have placed a pause site downstream of weak alternative exons thereby promoting exon inclusion.29 Recent evidence points to a crucial role of polymerase pausing in the outcome of co-transcriptional splicing.12 Several pausing mechanisms have been described, such as pausing at the 3′ splice site30 or pausing at the terminal exon.31 Together with the emerging information on favored nucleosome positioning on exons versus reduced positioning on exon-intron junctions, and RNAP II enrichment on exons, it would be tempting to suggest that transcriptional rates are reduced on exons thereby providing the sufficient time for the recognition of the 5′ and 3′ splice sites and for co-transcriptional splicing to occur. This would also mean that most of the introns situated at the 5′-end of genes are co-transcriptionally removed. However, the question of how much splicing is actually completed during RNA synthesis remains open, since only a small selection of mammalian genes have been analyzed.
Following Co-Transcriptional Splicing in Living Cells
David Bentley suggested three ways by which the coupling of RNAP II transcription with pre-mRNA processing can influence processing reactions.6 Localization: by virtue of simple proximity between the different machineries, an increase in the local concentration is obtained thereby increasing the probability and efficiency of the biochemical reactions. Allostery: the close contact obtained between various mRNA processing factors and subunits of the RNAP II elongation machinery can allosterically activate or inhibit mRNA processing factors.16,32 Kinetic coupling: elongation rates influence mRNA folding and the assembly of RNA-protein complexes, and as described above modulate alternative splicing decisions.
If transcription rates can regulate alternative splicing outcomes, and the splicing machinery is co-transcriptionally assembled on the pre-mRNA, could it be possible that reciprocal coupling occurs as well, and that the assembled spliceosome can modulate transcription rates? We set out to examine this relationship using live-cell approaches for measuring transcriptional kinetics.33 Transcriptional elongation rates on specific genes in living mammalian cells are obtained by tagging the mRNA34 followed by photobleaching experiments (fluorescence recovery after photobleaching, FRAP) for measuring the rate of synthesis as it occurs in real-time.35–37 These approaches have measured RNAP II transcription rates that can reach 4.5 kb/min.35,38 This measurement is three-fold higher than previously measured polymerase transcription speeds. Recently, additional approaches that do not use live-cell analysis have measured ∼4 kb/min elongation rates as well.39,40
A crucial aspect of the live-cell measurements of transcription is that they are conducted on a specific intron-containing gene harboring the RNA tagging sequence, and that the gene is stably integrated and forms a tandem gene array consisting of many copies of the gene. In this manner, a “transcription factory”41 recruiting a high concentration of polymerases is generated and easily visualized.42 With respect to co-transcriptional splicing, using such a tandem array should help in defining if splicing is actually occurring as the polymerase is transcribing i.e., splicing factors should be detected at the site of transcription. Indeed, RNA FISH has shown that intron sequences can be detected at the transcription site only, and not throughout the nucleoplasm suggesting that splicing is occurring at the gene locus itself.35,43,44 We also showed that all U snRNAs and associated snRNP proteins, as well as a variety of SR proteins and RNA-binding proteins were recruited and enriched at the site of transcription.44 Spliceosome recruitment is transcription-dependent since it is observed only when the gene is in a transcribing state. Because intron sequences are detected by RNA FISH on the gene only and not elsewhere in the nucleus, we concluded that splicing on the studied gene was occurring co-transcriptionally. In addition, quantification of spliceosome accumulation on the transcripts was proportional to the number of introns in each gene, suggesting independent processing for each intron.
The Strange Case of U1 snRNP
As a comparison to spliced genes we examined a similar but intronless gene containing one exon only. Recruitment of splicing factors to this gene is not expected since no introns or splice sites are found in the sequence. This was the general finding; however, U1 snRNA and the U1A protein were enriched on the actively transcribing intronless gene. Immunostaining for the unique 2,2,7-trimethylguanosine cap of snRNAs confirmed the presence of snRNA on the actively transcribing intronless gene.44 Previously, a proteomic analysis of the human RNAP II protein complex identified all the components of the U1 snRNP and SR proteins, but no other snRNPs or splicing factors, indicating the possible association of U1 snRNP with the polymerase.17 In another recent study, U1 snRNA co-immuoprecipitated with RNAP II from mitotic extracts in which RNAP II is transcriptionally inactive. Further investigation using tandem arrays containing integrated intron-containing genes showed similar results. The expected battery of splicing factors were recruited to the spliced genes only, whereas identical genes containing site-specific mutations in the splice sites that rendered the genes splicing deficient, recruited U1 snRNA, U1-70K and SP2/ASF only.45 It should be noted that U1-70K was not found associated with the intronless heat-shock genes as examined by chromatin immunoprecipitation techniques.8 Also, U1 snRNP was found to protect pre-mRNAs from premature cleavage and polyadenylation in a splicing-independent manner.46 Altogether, it is possible to suggest a mechanism by which U1 snRNP is associated with RNAP II even in the absence of splicing in order to scan for the presence of a 5′ splice site. When a 5′ splice site is identified by U1 this could then trigger the step-wise recruitment of the spliceosomal machinery and the induction of co-transcriptional splicing.
RNAP II Elongation Rates are Independent of Ongoing Splicing
Returning to our assay for measuring transcription rates on specific genes in living cells, we monitored RNAP II elongation rates on a series of genes containing the β-globin exons and introns.44 The genes differed in the number of introns: 2 introns, 3 introns, 5 introns and an intronless gene. The mRNA tag served as a real-time fluorescent output for transcriptional activity, as explained below. The tagged region was located immediately downstream to the splicing events, and therefore we could measure the rate at which the polymerase was transcribing while engaged in co-transcriptional splicing, and compare the different genes.
The technique used to measure transcription rates in vivo is the FRAP method.47 The 3′UTR of the gene, which is downstream to the splice sites, contains a series of MS2 sequence repeats. The latter are transcribed and form stem-loop structures in the pre-mRNA that are immediately coated by a specific RNA binding protein called YFP-MS2. Therefore, an actively transcribing gene array is visualized as a unique yellow dot in the nucleus (see image in Fig. 1). In this live-cell assay, the YFP-MS2 signal is photobleached and the recovery of fluorescence is monitored over time. The rate at which the YFP-MS2 fluorescence recovers on the active gene is a reflection of the rate at which RNAP II is transcribing new MS2 stem-loops. The rates of transcriptional elongation are retrieved from the FRAP recovery curves by either modeling of the data with differential equations or with computational simulations.35,38,48
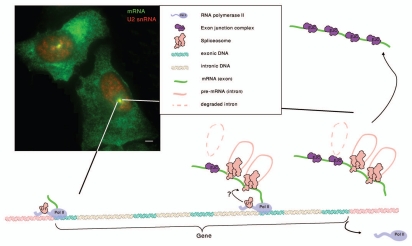
Figure 1.
When transcription has completed but splicing has not, the splicing machinery is retained at the site of transcription, independently of the polymerase. Image from an RNA FISH experiment on the U2OS E3 (3 exons, 2 introns) cell line. Co-transcriptional accumulation of the spliceosomal U2 snRNA (red) on the actively transcribing gene site (mRNA in green). Bar, 5 µM. The scheme depicts RNAP II transcription on an intron-containing gene. U1 snRNP might accompany the polymerase by associating with the CTD prior to transcription of the splice sites, and could thereby be in close proximity to the emerging transcript. Splicoeosomes are recruited to each intron and after splicing an exon-junction complex (EJC) is deposited on the exon-exon junctions. If the polymerase has completed transcription but splicing has not, the polymerase leaves the site of transcription while the transcript is delayed until splicing has completed. Only then can the transcript diffuse out into the nucleoplasm.
In this manner we could measure the rates at which RNAP II transcribed the portion of the gene starting from the MS2 region until the end of the gene, hence measuring polymerase elongation rates after the generation of the splice sites. We found that elongation rates were similar on the genes with 2 or 3 introns as well as on the intronless gene. This indicated that the co-transcriptional recruitment of the spliceosomal machinery did not increase or reduce the rate at which the polymerase was moving along this part of the genes. Interestingly, in support of our data, a recent study using quantitative RT-PCR found that polymerase rates were similar on all examined genes and did not differ in exonic or intronic regions.40
Unprocessed mRNA is Retained at the Site of Transcription
In contrast to the above three genes, significantly slower kinetics were measured on the gene containing 5 introns and 6 exons (termed E6 gene), even though the gene sequences were identical. This finding could suggest that RNAP II elongation rates on this gene were somehow reduced. However, live-cell experiments monitoring GFP-RNAP II kinetics on the E6 gene, in conjunction with ChIP experiments examining the distribution of RNAP II along the gene, indicated that this was not the case. Rather, the data pointed to a link between splicing and the slowed kinetics. When splicing was inhibited with specific inhibitors (Spliceostatin A or Meayamycin) the observed reduction in kinetics was relieved and returned to normal, implying that the splicing reaction was leading to the measured slow kinetics. Indeed, inhibition of transcription (actinomycin D) while monitoring the release of the remaining transcripts from the different genes demonstrated that the E6 mRNA clearance from the gene was significantly slower.
What could be the reason for the prolonged association of the transcripts with the E6 gene? We performed several quantifications in order to examine this issue. One possibility is that 3′ processing and polyadenylation were incomplete thereby causing an accumulation of the unprocessed mRNAs. But when the ratio of polyA signal to the last exon signal was compared by quantitative RNA FISH, it was found to be similar on all genes, or in other words—there was no out of the ordinary accumulation of non-polyadenylated mRNAs on the E6 gene. Another possibility is that the polymerase is somehow stalled at the end of the gene. Here we quantified the ratio of the first exon to the polymerase, assuming that normally there should be a 1:1 correlation between them. Surprisingly, when the polymerase signal on the gene was quantified and compared to the signal of the transcripts, an unusually high ratio of the E6 transcript to the polymerase was measured. This accumulation of the transcript was relieved and returned to a 1:1 mRNA:polymerase ratio when splicing was inhibited. Putting it simply, this analysis showing the accumulation of E6 transcripts on the gene means that the polymerase did not “hang around” at the end of the E6 gene when transcription had terminated and pre-mRNA processing had not.
Computer simulation of the data showed that the E6 transcripts were being retained for up to 10 min in comparison to 50 seconds for the other genes. We used the computer simulation and these kinetic parameters to predict and analyze the experimental results. When we simulated the E6 transcript:polymerase quantitative FISH experiment, the simulation showed a 3-fold increase in the ratio of transcript per polymerase, exactly as found in the experimental results. We also simulated and obtained a ten minute difference between the times for mRNA clearance from the gene during actinomycin D treatment, as in the experimental results. Moreover, computational simulation not only provides kinetic parameters but can also better explain the experimental results. For instance, the simulation explained the results of quantitative FISH measuring the removal of introns from the pre-mRNA at the site, and showed that the kinetics of intron removal was not influenced by the retained mRNA, pointing to the fact that the splicing process might be the limiting factor for mRNA retention. Therefore, we propose that the E6 gene serves as a model for co-transcriptional splicing that has not been completed before the polymerase has completed its job. In such a case the polymerase disengages from the gene while the pre-mRNA remains in the gene vicinity until splicing has completed.
“Ready to Go” Transcripts are Released from the Gene
These data point to the preferential release of completed and “ready to go” transcripts. Although mRNPs remodel between the nucleus and the cytoplasm, it is unclear whether remodeling occurs during the nucleoplasmic pathway from the gene to the nuclear pore.49 Completion of processing at the gene itself prior to release would make sense especially for constitutive splicing, since all the splicing factors have already assembled. On the other hand alternative splicing might be controlled through the choice between co- or post-transcriptional processing, since the stoichiometry of splicing factors can lead to different alternative outcomes, and thereby changing of the splicing environment (gene vs. nucleoplasm/speckle) would help in controlling the specific splicing factors associated with the transcript. Still, the phenomena of co-transcriptional splicing has yet to be quantified for a large set of mammalian genes. It remains to be seen what fraction of the genome is co-transcriptionally or post-transcriptionally spliced, what are the rules that govern these choices, and whether they are spatially compartmentalized.
Acknowledgments
Y.S.T. is supported by the European Research Council (ERC), the Israel Science Foundation (ISF) (250/06), ISF-Bikura, Israel Cancer Research Fund (ICRF), German-Israeli Foundation for Scientific Research and Development (GIF), USA-Israel Binational Science Foundation (BSF), German-Israeli Project Cooperation (DIP) and is the Jane Stern Lebell Family Fellow in Life Sciences at Bar-Ilan University. Y.B. is grateful to the Azrieli Foundation for the award of an Azrieli fellowship.
Abbreviations
- RNAP
RNA polymerase
- CTD
C-terminal domain
- ChIP
chromatin immunoprecipitation
References
- 1.Huang S, Spector DL. Will the real splicing sites please light up? Curr Biol. 1992;2:188–190. doi: 10.1016/0960-9822(92)90516-d. [DOI] [PubMed] [Google Scholar]
- 2.Osheim YN, Miller OL, Jr, Beyer AL. RNP particles at splice junction sequences on Drosophila chorion transcripts. Cell. 1985;43:143–151. doi: 10.1016/0092-8674(85)90019-4. [DOI] [PubMed] [Google Scholar]
- 3.Beyer AL, Osheim YN. Splice site selection, rate of splicing and alternative splicing on nascent transcripts. Genes Dev. 1988;2:754–765. doi: 10.1101/gad.2.6.754. [DOI] [PubMed] [Google Scholar]
- 4.Bauren G, Wieslander L. Splicing of Balbiani ring 1 gene pre-mRNA occurs simultaneously with transcription. Cell. 1994;76:183–192. doi: 10.1016/0092-8674(94)90182-1. [DOI] [PubMed] [Google Scholar]
- 5.Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- 6.Bentley DL. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol. 2005;17:251–256. doi: 10.1016/j.ceb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 7.Kornblihtt AR, de la Mata M, Fededa JP, Munoz MJ, Nogues G. Multiple links between transcription and splicing. RNA. 2004;10:1489–1498. doi: 10.1261/rna.7100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Listerman I, Sapra AK, Neugebauer KM. Cotranscriptional coupling of splicing factor recruitment and precursor messenger RNA splicing in mammalian cells. Nat Struct Mol Biol. 2006;13:815–822. doi: 10.1038/nsmb1135. [DOI] [PubMed] [Google Scholar]
- 9.Pandya-Jones A, Black DL. Co-transcriptional splicing of constitutive and alternative exons. RNA. 2009;15:1896–1908. doi: 10.1261/rna.1714509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li X, Manley JL. Cotranscriptional processes and their influence on genome stability. Genes Dev. 2006;20:1838–1847. doi: 10.1101/gad.1438306. [DOI] [PubMed] [Google Scholar]
- 11.Proudfoot NJ, Furger A, Dye MJ. Integrating mRNA processing with transcription. Cell. 2002;108:501–512. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- 12.Oesterreich FC, Bieberstein N, Neugebauer KM. Pause locally, splice globally. Trends Cell Biol. 2011;21:328–335. doi: 10.1016/j.tcb.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 13.Greenleaf AL. Positive patches and negative noodles: linking RNA processing to transcription? Trends Biochem Sci. 1993;18:117–119. doi: 10.1016/0968-0004(93)90016-g. [DOI] [PubMed] [Google Scholar]
- 14.de Almeida SF, Carmo-Fonseca M. The CTD role in cotranscriptional RNA processing and surveillance. FEBS Lett. 2008;582:1971–1976. doi: 10.1016/j.febslet.2008.04.019. [DOI] [PubMed] [Google Scholar]
- 15.Lewis JD, Tollervey D. Like attracts like: getting RNA processing together in the nucleus. Science. 2000;288:1385–1389. doi: 10.1126/science.288.5470.1385. [DOI] [PubMed] [Google Scholar]
- 16.Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- 17.Das R, Yu J, Zhang Z, Gygi MP, Krainer AR, Gygi SP, et al. SR proteins function in coupling RNAP II transcription to pre-mRNA splicing. Mol Cell. 2007;26:867–881. doi: 10.1016/j.molcel.2007.05.036. [DOI] [PubMed] [Google Scholar]
- 18.McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, et al. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 19.Fong N, Bird G, Vigneron M, Bentley DL. A 10 residue motif at the C-terminus of the RNA pol II CTD is required for transcription, splicing and 3′ end processing. EMBO J. 2003;22:4274–4282. doi: 10.1093/emboj/cdg396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosonina E, Blencowe BJ. Analysis of the requirement for RNA polymerase II CTD heptapeptide repeats in pre-mRNA splicing and 3′-end cleavage. RNA. 2004;10:581–589. doi: 10.1261/rna.5207204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Emili A, Shales M, McCracken S, Xie W, Tucker PW, Kobayashi R, et al. Splicing and transcription-associated proteins PSF and p54nrb/nonO bind to the RNA polymerase II CTD. RNA. 2002;8:1102–1111. doi: 10.1017/s1355838202025037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Licatalosi DD, Geiger G, Minet M, Schroeder S, Cilli K, McNeil JB, et al. Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol Cell. 2002;9:1101–1111. doi: 10.1016/s1097-2765(02)00518-x. [DOI] [PubMed] [Google Scholar]
- 23.Natalizio BJ, Robson-Dixon ND, Garcia-Blanco MA. The carboxyl-terminal domain of RNA polymerase II is not sufficient to enhance the efficiency of pre-mRNA capping or splicing in the context of a different polymerase. J Biol Chem. 2009;284:8692–8702. doi: 10.1074/jbc.M806919200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de la Mata M, Kornblihtt AR. RNA polymerase II C-terminal domain mediates regulation of alternative splicing by SRp20. Nat Struct Mol Biol. 2006;13:973–980. doi: 10.1038/nsmb1155. [DOI] [PubMed] [Google Scholar]
- 25.Luco RF, Misteli T. More than a splicing code: integrating the role of RNA, chromatin and non-coding RNA in alternative splicing regulation. Curr Opin Genet Dev. 2011;21:366–372. doi: 10.1016/j.gde.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwartz S, Ast G. Chromatin density and splicing destiny: on the cross-talk between chromatin structure and splicing. EMBO J. 2010;29:1629–1636. doi: 10.1038/emboj.2010.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, et al. A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell. 2003;12:525–532. doi: 10.1016/j.molcel.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 28.Howe KJ, Kane CM, Ares M., Jr Perturbation of transcription elongation influences the fidelity of internal exon inclusion in Saccharomyces cerevisiae. RNA. 2003;9:993–1006. doi: 10.1261/rna.5390803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roberts GC, Gooding C, Mak HY, Proudfoot NJ, Smith CW. Co-transcriptional commitment to alternative splice site selection. Nucleic Acids Res. 1998;26:5568–5572. doi: 10.1093/nar/26.24.5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alexander RD, Innocente SA, Barrass JD, Beggs JD. Splicing-dependent RNA polymerase pausing in yeast. Mol Cell. 2010;40:582–593. doi: 10.1016/j.molcel.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carrillo Oesterreich F, Preibisch S, Neugebauer KM. Global analysis of nascent RNA reveals transcriptional pausing in terminal exons. Mol Cell. 2010;40:571–581. doi: 10.1016/j.molcel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 32.Ho CK, Shuman S. Distinct roles for CTD Ser-2 and Ser-5 phosphorylation in the recruitment and allosteric activation of mammalian mRNA capping enzyme. Mol Cell. 1999;3:405–411. doi: 10.1016/s1097-2765(00)80468-2. [DOI] [PubMed] [Google Scholar]
- 33.Lamond AI, Swedlow JR. RNA polymerase II transcription in living color. Nat Struct Mol Biol. 2007;14:788–790. doi: 10.1038/nsmb0907-788. [DOI] [PubMed] [Google Scholar]
- 34.Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. Localization of ASH1 mRNA particles in living yeast. Mol Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 35.Darzacq X, Shav-Tal Y, de Turris V, Brody Y, Shenoy SM, Phair RD, et al. In vivo dynamics of RNA polymerase II transcription. Nat Struct Mol Biol. 2007;14:796–806. doi: 10.1038/nsmb1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boireau S, Maiuri P, Basyuk E, de la Mata M, Knezevich A, Pradet-Balade B, et al. The transcriptional cycle of HIV-1 in real-time and live cells. J Cell Biol. 2007;179:291–304. doi: 10.1083/jcb.200706018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brody Y, Shav-Tal Y. Visualizing transcription in real-time. Cent Eur J Biol. 2008;3:11–18. [Google Scholar]
- 38.Ben-Ari Y, Brody Y, Kinor N, Mor A, Tsukamoto T, Spector DL, et al. The life of an mRNA in space and time. J Cell Sci. 2010;123:1761–1774. doi: 10.1242/jcs.062638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wada Y, Ohta Y, Xu M, Tsutsumi S, Minami T, Inoue K, et al. A wave of nascent transcription on activated human genes. Proc Natl Acad Sci USA. 2009;106:18357–18361. doi: 10.1073/pnas.0902573106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singh J, Padgett RA. Rates of in situ transcription and splicing in large human genes. Nat Struct Mol Biol. 2009;16:1128–1133. doi: 10.1038/nsmb.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iborra FJ, Pombo A, Jackson DA, Cook PR. Active RNA polymerases are localized within discrete transcription “factories” in human nuclei. J Cell Sci. 1996;109:1427–1436. doi: 10.1242/jcs.109.6.1427. [DOI] [PubMed] [Google Scholar]
- 42.Darzacq X, Singer RH, Shav-Tal Y. Dynamics of transcription and mRNA export. Curr Opin Cell Biol. 2005;17:332–339. doi: 10.1016/j.ceb.2005.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang G, Taneja KL, Singer RH, Green MR. Localization of pre-mRNA splicing in mammalian nuclei. Nature. 1994;372:809–812. doi: 10.1038/372809a0. [DOI] [PubMed] [Google Scholar]
- 44.Brody Y, Neufeld N, Bieberstein N, Causse SZ, Bohnlein EM, Neugebauer KM, et al. The in vivo kinetics of RNA polymerase II elongation during co-transcriptional splicing. PLoS Biol. 2011;9:1000573. doi: 10.1371/journal.pbio.1000573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spiluttini B, Gu B, Belagal P, Smirnova AS, Nguyen VT, Hebert C, et al. Splicing-independent recruitment of U1 snRNP to a transcription unit in living cells. J Cell Sci. 2010;123:2085–2093. doi: 10.1242/jcs.061358. [DOI] [PubMed] [Google Scholar]
- 46.Kaida D, Berg MG, Younis I, Kasim M, Singh LN, Wan L, et al. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature. 2010;468:664–668. doi: 10.1038/nature09479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shav-Tal Y. The living test-tube: Imaging of real-time gene expression. Soft Matter. 2006;2:361–370. doi: 10.1039/b600234j. [DOI] [PubMed] [Google Scholar]
- 48.Brody Y, Shav-Tal Y. Measuring the kinetics of mRNA transcription in single living cells. J Vis Exp. 2011;(54):e2898. doi: 10.3791/2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mor A, Suliman S, Ben-Yishay R, Yunger S, Brody Y, Shav-Tal Y. Dynamics of single mRNP nucleocytoplasmic transport and export through the nuclear pore in living cells. Nat Cell Biol. 2010;12:543–552. doi: 10.1038/ncb2056. [DOI] [PubMed] [Google Scholar]