Abstract
Addition of straw is common practice in rice agriculture, but its effect on the path of microbial CH4 production and the microbial community involved is not well known. Since straw from rice (C3 plant) and maize plants (C4 plant) exhibit different δ13C values, we compared the effect of these straw types using anoxic rice field soils from Italy and China, and also a soil from Thailand that had previously not been flooded. The temporal patterns of production of CH4 and its major substrates H2 and acetate, were slightly different between rice straw and maize straw. Addition of methyl fluoride, an inhibitor of acetoclastic methanogenesis, resulted in partial inhibition of acetate consumption and CH4 production. The δ13C of the accumulated CH4 and acetate reflected the different δ13C values of rice straw versus maize straw. However, the relative contribution of hydrogenotrophic methanogenesis to total CH4 production exhibited a similar temporal change when scaled to CH4 production irrespectively of whether rice straw or maize straw was applied. The composition of the methanogenic archaeal communities was characterized by terminal restriction fragment length polymorphism (T-RFLP) analysis and was quantified by quantitative PCR targeting archaeal 16S rRNA genes or methanogenic mcrA genes. The size of the methanogenic communities generally increased during incubation with straw, but the straw type had little effect. Instead, differences were found between the soils, with Methanosarcinaceae and Methanobacteriales dominating straw decomposition in Italian soil, Methanosarcinaceae, Methanocellales, and Methanobacteriale in China soil, and Methanosarcinaceae and Methanocellales in Thailand soil. The experiments showed that methanogenic degradation in different soils involved different methanogenic population dynamics. However, the path of CH4 production was hardly different between degradation of rice straw versus maize straw and was also similar for the different soil types.
Keywords: methanogenesis, rice field soil, straw, archaea, pathway
Introduction
Methane production in rice fields is driven by the anaerobic degradation of organic matter. The organic matter that is converted into CH4 is basically derived from three different sources, i.e., straw, soil organic matter (SOM), and plant photosynthesis (Watanabe et al., 1999). Addition of straw, which is common practice in rice agriculture to improve soil fertility, enhances CH4 production in flooded rice fields and results in increased CH4 emission (Schütz et al., 1989; Denier van der Gon and Neue, 1995; Watanabe et al., 1995; Yan et al., 2005). Under such conditions, CH4 production derived from SOM can be negligible in comparison to that derived from straw (Chidthaisong and Watanabe, 1997; Watanabe et al., 1998). The methanogenic degradation of straw is achieved by fermentation producing acetate and H2/CO2 as major intermediates from which CH4 is formed (Glissmann and Conrad, 2002). Besides rice straw also the addition of straw from other plants, e.g., vetch, rape seed, wheat, was studied (Cai et al., 2001; Ma et al., 2008). However, maize straw has not received much attention, despite the fact that crop rotation between maize and rice has recently increased and is now already covering about 3.5 million hectare in Asia (Timsina et al., 2010).
Both rice and maize straw consist mainly of cellulose (30–40%), hemicellulose (25–35%), and lignin (4–8%; Tian et al., 1992; Singh et al., 2011). However, maize is a C4 plant while rice is a C3 plant resulting in distinct δ13C values of the plants and their degradation products. While plants with a C3 photosynthesis have δ13C values on the average of about −27‰, plants with a C4 photosynthesis have much higher δ13C values, about −13‰ (Farquhar et al., 1989). This difference in natural 13C abundance has been exploited to study carbon turnover in soil (Amelung et al., 2008), for example, to determine the partitioning between CO2 derived from root organic matter versus SOM (Balesdent and Balabane, 1992; Hanson et al., 2000; Kuzyakov, 2006). Such determination of partitioning is possible by measuring the δ13C of CO2 produced in a field that undergoes vegetation change from C3 to C4 plant or vice versa (e.g., Rochette et al., 1999; Flessa et al., 2000; Kuzyakov and Cheng, 2001).
The partitioning of the CH4 emission should in principle also be possible and would be of great interest in order to allow better management of this important greenhouse gas. However, partitioning studies have so far only been conducted by pulse labeling experiments with 13CO2. These studies indicate that about 50% of the CH4 emitted may originate from the rice plants, either by root exudation or root decay (Minoda and Kimura, 1994; Watanabe et al., 1999). However, pulse labeling experiments are cumbersome and invasive. Experiments using the natural abundance of δ13C have so far not been conducted for partitioning the CH4 production. One problem is the potentially large isotopic fractionation factors that are involved in the conversion of organic matter to CH4 (Conrad, 2005). These fractionation factors are on the order of 10–70‰ and thus, more than an order of magnitude larger than the fractionation factors involved in conversion of organic matter to CO2, which are only about 1–3‰ (Werth and Kuzyakov, 2010). The fractionation factors involved in CH4 production strongly depend on the path of CH4 formation. Thus, it is much larger for CH4 formation from H2/CO2 (typically 50–70‰) than for CH4 formation from acetate (typically 10–20‰; Conrad, 2005). The extent to which H2/CO2 and acetate contribute to CH4 formation thus crucially affects the stable carbon isotope fractionation during CH4 production from organic matter. Since the path of CH4 production may be dependent on type of organic matter degraded, we were interested whether methanogenic degradation of maize straw would be different from that of maize straw.
Methanogenic degradation of organic matter in flooded soils is achieved by a complex microbial community consisting of hydrolytic, fermenting, and methanogenic microorganisms (Conrad, 2007). The CH4 is usually produced by acetoclastic and hydrogenotrophic methanogenic archaea. Rice field soils frequently exhibit a rather large diversity of such methanogenic archaea, including acetoclastic Methanosarcinaceae and Methanosaetaceae as well as hydrogenotrophic Methanocellales, Methanomicrobiales, and Methanobacteriales. The abundance of methanogens in rice field soils is usually rather high (>106 cells g−1 dry soil) throughout the season, even when the soil is drained (Krüger et al., 2005; Ma et al., 2012). Upland soils, on the other hand, contain only very low numbers (<103) of methanogens. However, these numbers may dramatically increase upon flooding of the soil (Peters and Conrad, 1996; Angel et al., 2011, 2012).
We comparatively studied carbon isotopic fractionation during CH4 production in three different soils (two rice field soils and one upland soil) amended with either rice straw or maize straw, determined the paths of CH4 formation and the methanogenic archaea involved in order to address the following research questions: (1) Is the relative contribution of hydrogenotrophic and acetoclastic methanogenesis to CH4 production different during degradation of maize straw versus rice straw; (2) is there a difference when different soils, including rice field soil and upland soil, are amended; (3) how does straw affect the composition and abundance of the methanogenic archaeal community; and (4) can amendment with maize straw serve as a labeling technique to determine partitioning of C-flux to CH4?
Materials and Methods
Soil incubation
Soils were collected from Italy (Vercelli), China (Fuyang), and Thailand (Suwan). The Italian soil (Vercelli), a silt loam, was originally from a rice field (45.9°N, 171.6°E) at the Italian Rice Research Institute in Vercelli, Italy (Holzapfel-Pschorn et al., 1986), and has subsequently been used for cultivation of rice in our glasshouse. The China soil (Fuyang), a clay loam, was collected in 2007 from a rice field (30.1°N, 119.9°E) at the China National Rice Research Institute in Fuyang, Hangzhou (Peng et al., 2008). The Thailand soil (Suwan), a clay soil, was collected in 2010 from an upland field site (14.6°N, 101.3°E) at the National Corn and Sorghum Research Center of Kasetsart University (Suwanwajokkasikit Field Corps Research Station), Nakornratchasima province, Thailand (Thongsaga et al., 2010). The field had been cropped with maize for several years. The content of C (%) of the soils from Vercelli, Fuyang, and Suwan was 1.3 ± 0.01, 2.4 ± 0.1, and 2.0 ± 0.1, respectively. The N content (%) was 0.13 ± 0.01, 0.25 ± 0.01, and 0.19 ± 0.03, respectively. The soils were air-dried and stored in dry state at room temperature. This treatment ensures that methanogenic activity rapidly recovers after flooding (Mayer and Conrad, 1990). Rice straw originated from rice plants and maize straw from maize plants, both grown in our glasshouse using Italian rice field soil or commercial standard soil, respectively. The straw was air-dried and ground using a blender. The C and N contents of maize straw were 40.2% C and 2.4% N, of rice straw 41.7% C and 0.7% N. The δ13C of the maize straw was −12.97 ± 0.05‰, that of rice straw was −30.96 ± 0.10‰.
Soil slurries were prepared by mixing 600 g dry soil with 600 mL of deionized, sterile, anoxic water, and incubating the mixture in 2 L bottles under a headspace of N2 at 25°C for 4 weeks. The soil slurry was then amended with either rice straw or maize straw at a ratio of 10 mg dry ground straw per gram dry soil, and aliquots (10 g slurry) dispensed into 26 mL pressure tubes. The tubes were closed with black rubber stoppers, flushed with N2, pressurized to 0.5 bar overpressure, and incubated at 25°C. The tubes with soil slurry were prepared in numerous parallels, of which triplicates were sacrificed for chemical analyses. The gas phase of some incubations was amended with CH3F, an inhibitor of acetoclastic methanogenesis (Janssen and Frenzel, 1997; Conrad and Klose, 1999b), at a ratio of 1 or 2%. The initial pH values of the soil slurries were between pH 6.1 (Vercelli) and 7.3 (Suwan), but upon anoxic incubation narrowed to pH values between pH 6.7 and 7.0.
Chemical measurements
Methane was quantified in a gas chromatograph (GC) equipped with methanizer and flame ionization detector, H2 in a GC with a reduced gas detector (Conrad and Klose, 1999a). The partial pressures of CH4 measured in the headspace of the incubations were converted into molar quantities using the molar volumes of an ideal gas at the different temperatures. The isotopic composition (δ13C) of CH4 and CO2 were determined in a Finnigan gas chromatograph combustion isotope ratio mass spectrometer (GC–C–IRMS) system (Thermoquest, Bremen, Germany; Fey et al., 2004). Concentrations of acetate in the liquid phase of the soil incubations were analyzed by high pressure liquid chromatography (HPLC; Krumböck and Conrad, 1991), the δ13C of acetate by a HPLC–C–IRMS as described before (Conrad et al., 2007). The δ13C of the methyl group of acetate was determined by off-line pyrolysis followed by GC–C–IRMS analysis (Conrad et al., 2007). The C and N content of soil and straw were analyzed on a CHNS-elemental analyzer by the Analytical Chemical Laboratory of the University of Marburg. The δ13Corg of straw and the straw-amended soils were analyzed at the Centre for Stable Isotope Research and Analysis (KOSI) at the University of Göttingen using an elemental analyzer coupled to an IRMS. The δ13Corg were for Italian, China, and Thailand soil, respectively, −27.1, −27.6, and −22.3‰ for rice straw-amended and −24.6, −25.9, and −18.6‰ for maize straw-amended soil.
Molecular analyses
DNA was extracted from the soil using the Fast DNA Spin Kit for soil (MP, Heidelberg, Germany) following the manufacturer’s instructions as described in detail by Kolb et al. (2005). DNA extraction was optimized to obtain maximum copy numbers of archaeal 16S rRNA in quantitative PCR (qPCR; see below), which was by combining the DNA from 1, 2, and 3 extractions for soil from Vercelli, Suwan, and Fuyang, respectively. The analysis of terminal restriction fragment length polymorphism (T-RFLP) of archaeal 16S rRNA genes was done as described (Chin et al., 1999) using the primer combination Ar109f/Ar915r (Grosskopf et al., 1998), with the reverse primer labeled with FAM (6-caboxyfluorescein). The 16S rRNA gene amplicons were digested with TaqI (Fermentas, St. Leon Rot, Germany), and the products were size-separated in an ABI 3130 DNA sequencer (Applera, Darmstadt, Germany). The abundance of total archaeal 16S rRNA gene copies was determined by qPCR (Angel et al., 2011, 2012). The abundance of individual groups of archaea was calculated from the relative abundances of their T-RFs multiplied with the copy numbers of total archaeal 16S rRNA genes determined by qPCR (Conrad and Klose, 2006). The abundance of total copies of the mcrA gene, coding for a subunit of the methyl coenzyme M reductase of methanogens was also determined (only in Suwan soil) by qPCR (Angel et al., 2011). The abundance of individual groups of methanogens (only in Suwan soil) was determined by TaqMan qPCR targeting specific 16S rRNA gene sequences (Yu et al., 2005) as described by Angel et al. (2012).
Calculations
Values of δ13C are defined by
| (1) |
with R = 13C/12C of sample (sa) and standard (st) respectively, using VPDB carbonate as standard. Fractionation factors for a reaction A → B are defined after (Hayes, 1993; Angel et al., 2012):
| (2) |
sometimes expressed as isotopic enrichment factor ε ≡ 1 − α (in units of per mile). The fractionation factor for conversion of CO2 to CH4 is given by
| (3) |
where is the δ13C of the CH4 derived from H2 + CO2, i.e., the CH4 produced in the presence of CH3F, assuming that acetoclastic methanogenesis was then completely inhibited.
Relative contribution of H2 + CO2-derived CH4 to total CH4 was determined using the following mass balance equation (Conrad, 2005):
| (4) |
where is the fraction of CH4 formed from H2 + CO2, the δ13C of total produced methane, the δ13C of CH4 derived from H2 + CO2, and the δ13C of CH4 derived from acetate determined by:
| (5) |
where is the δ13C of the methyl group of acetate accumulated and is the isotopic enrichment factor (equivalent to ) during acetoclastic methanogenesis. It is assumed, that the methyl group of acetate was converted to CH4 with ‰, which is close to the values determined previously for straw-amended or acetate-amended rice field soil dominated by Methanosarcina species as acetoclastic methanogens (Penning and Conrad, 2007; Goevert and Conrad, 2009).
Results
Methane production
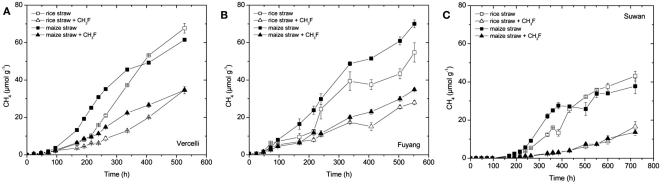
Methane was produced from each soil amended with either rice straw or maize straw (Figure 1). Vigorous CH4 production started after about 2 days in soil from Vercelli and Fuyang, and after about 7 days in soil from Suwan. The soil from Suwan was not an authentic rice field soil and had so far not been flooded. Note that the soil from Suwan was preincubated under anoxic conditions without straw amendment for 38 days before the experiment shown in Figure 1C was started. Furthermore, in contrast to soils from Vercelli and Fuyang, production of CH4 from Suwan soil was only observed when straw was added (data not shown). However, the maximum rates of CH4 production were in a similar range (140–200 nmol h−1 g−1 dry soil) in all three soils, irrespectively which type of straw was added. Addition of methyl fluoride (1–2%) partially inhibited CH4 production in all incubations (Figure 1). The pH values during methanogenesis were between pH 6.8 and 7.1.
Figure 1.
Accumulation of CH4 in soil from (A) Vercelli, (B) Fuyang, and (C) Suwan, amended with either rice straw or maize straw and incubated under anoxic conditions in the absence or presence of CH3F (1, 2, and 2% in Vercelli, Fuyang, and Suwan soil, respectively); mean ± SE, n = 3.
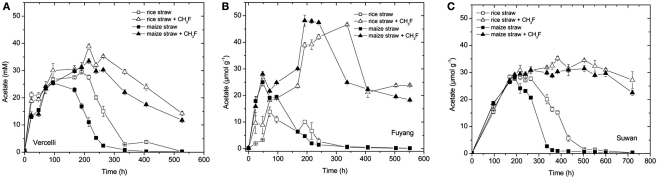
Acetate transiently accumulated in all incubations indicating that it was produced during degradation of organic matter (straw) and consumed by methanogenesis (Figure 2). The maximum amount of acetate accumulated accounted for about 30–50% of CH4 produced until the end of the incubation (a concentration of 1 mM acetate corresponds to an amount of 1 μmol g−1 dry soil). Addition of CH3F resulted in inhibition of acetate consumption so that acetate concentrations stayed high until the end of incubation (Figure 2). Only in Vercelli soil, acetate steadily decreased after 300 h of incubation, probably since Vercelli soil was only treated with 1% CH3F, whereas the other soils were treated with 2% CH3F. Since CH3F concentrations usually decrease slowly during incubation (Conrad and Klose, 1999b; Penning and Conrad, 2006), the CH3F concentrations in Vercelli soil probably fell below the critical inhibitory threshold of 1% so that acetate could again be consumed by methanogens (Penning et al., 2006b). Increase of acetate (Fuyang) and/or decrease of acetate (Vercelli, Suwan) was generally faster with maize straw than with rice straw (Figure 2), being probably a reason for why CH4 production was also somewhat faster with maize straw than with rice straw (Figure 1).
Figure 2.
Transient accumulation of acetate in soil from (A) Vercelli, (B) Fuyang, and (C) Suwan, amended with either rice straw or maize straw and incubated under anoxic conditions in the absence or presence of CH3F (1, 2, and 2% in Vercelli, Fuyang, and Suwan soil, respectively); mean ± SE, n = 3.
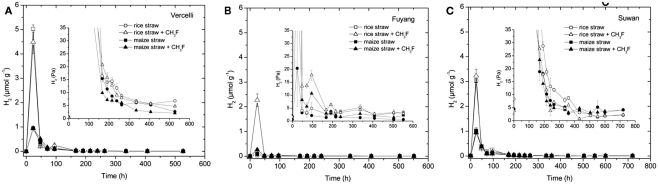
Hydrogen, which is next to acetate another important substrate for methanogenesis, also accumulated transiently (Figure 3). As previously reported (Conrad, 2002) the highest H2 concentrations were reached during the initial lag phase of CH4 production. During the main phase of CH4 production H2 partial pressures were generally lower than 50 Pa. In Vercelli and Fuyang soil, but not in Suwan soil, H2 partial pressures seemed to be generally about 2 Pa higher with rice straw than with maize straw (Figure 3).
Figure 3.
Transient accumulation of H2 in soil from (A) Vercelli, (B) Fuyang, and (C) Suwan, amended with either rice straw or maize straw and incubated under anoxic conditions in the absence or presence of CH3F (1, 2, and 2% in Vercelli, Fuyang, and Suwan soil, respectively); mean ± SE, n = 3. The insert shows the H2 accumulated in units of partial pressure (Pa).
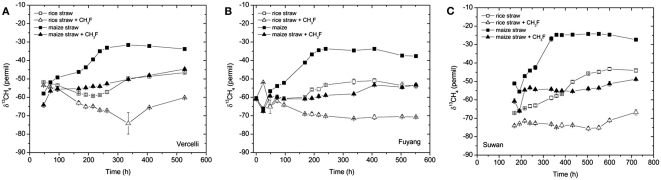
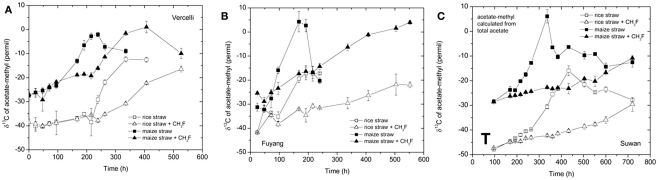
The δ13C of the produced CH4 was generally higher for the soils amended with maize straw than with rice straw and was generally lower in the treatments with CH3F than in the untreated controls (Figure 4). The δ13C of the produced acetate was also generally higher for the soils amended with maize straw than with rice straw, and increased faster with time in the absence than in the presence of CH3F (Figure 5). This behavior is reasonable, since δ13C of the residual acetate should increase because of carbon isotope fractionation during acetate consumption (cf. Penning and Conrad, 2007).
Figure 4.
Values of δ13C of the CH4 accumulated in soil from (A) Vercelli, (B) Fuyang, and (C) Suwan, amended with either rice straw or maize straw and incubated under anoxic conditions in the absence or presence of CH3F (1, 2, and 2% in Vercelli, Fuyang, and Suwan soil, respectively); mean ± SE, n = 3.
Figure 5.
Values of δ13C of the methyl group of acetate accumulated in soil from (A) Vercelli, (B) Fuyang, and (C) Suwan, amended with either rice straw or maize straw and incubated under anoxic conditions in the absence or presence of CH3F (1, 2, and 2% in Vercelli, Fuyang, and Suwan soil, respectively); mean ± SE, n = 3. The data in Vercelli and Fuyang soil were directly measured; those in Suwan soil were calculated from measured δ13C of total acetate using = δ13Cac − 10‰.
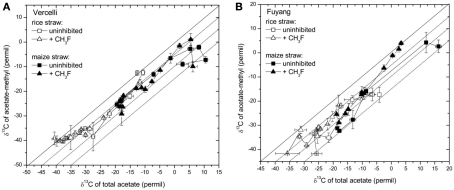
Note that δ13C of acetate-methyl is given in Figure 5. The data were directly measured in the incubations of Vercelli and Fuyang soil. The δ13C of acetate-methyl was about 10‰ lower than that of total acetate (Figure 6). For Suwan soil, we only analyzed the δ13C of total acetate. The corresponding values of acetate-methyl shown in Figure 5 were calculated by subtracting 10‰ from the δ13C of total acetate.
Figure 6.
Relation between the δ13C of acetate-methyl and the δ13C of total acetate measured in soil from (A) Vercelli, and (B) Fuyang, amended with either rice straw or maize straw and incubated under anoxic conditions in the absence or presence of CH3F (1 and 2% in Vercelli and Fuyang soil, respectively); mean ± SE, n = 3.
The fractionation factors () for hydrogenotrophic methanogenesis (calculated by Eq. 3) were in a range of 1.045–1.060, 1.050–1.060, and 1.040–1.055 for soils from Vercelli, Fuyang, and Suwan, respectively. In general, values were by about 0.005–0.010 units higher in soil amended with rice straw than with maize straw (data not shown).
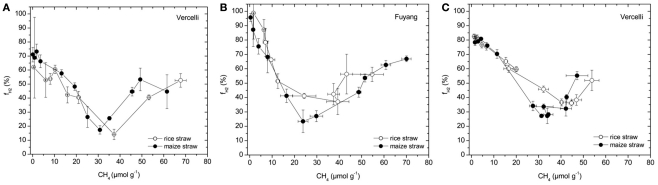
The percentage contribution of hydrogenotrophic methanogenesis to total CH4 production was calculated (using Eq. 4) by mass balance of δ13C values of CH4 (Figure 4) and acetate-methyl (Figure 5) and scaled against the amount of CH4 produced (Figure 1). The results show that this contribution was initially high, decreased during the time when the transiently accumulated acetate was consumed, and then increased again (Figure 7). Interestingly, the pattern was very similar for treatments with maize straw versus rice straw.
Figure 7.
Percentage of CH4 produced from H2/CO2 () in soil from (A) Vercelli, (B) Fuyang, and (C) Suwan, amended with either rice straw or maize straw, as calculated using Equation 4, and the data shown in Figures 4 and 5.
Methanogenic archaea
The abundance of methanogenic archaea in the authentic rice field soils from Vercelli and Fuyang was quantified before and after incubation with straw using qPCR and T-RFLP targeting archaeal 16S rRNA genes. The T-RFs detected could be affiliated to different methanogenic groups based on previous studies using T-RFLP and cloning/sequencing of archaeal genes (Chin et al., 1999; Lueders and Friedrich, 2000; Peng et al., 2008; Wu et al., 2009). The following T-RFs (in brackets) could be affiliated: Methanomicrobiales (84 bp), Methanobacteriales (92 bp), Methanosarcinaceae (186 bp), Methanosaetaceae (284 bp), and Methanocellales (393 bp). The T-RFs of 74, 381, >738 bp were only detected in low relative abundance (<2%), and thus were not further considered. In Vercelli and Fuyang soil the T-RF = 186 bp represents mainly Methanosarcinaceae. Although this T-RF can also represent non-methanogenic crenarchaeotes (Rice cluster VI or group I.1b), they generally constitute only a minor fraction of T-RF = 186 bp in rice field soil and can be neglected.
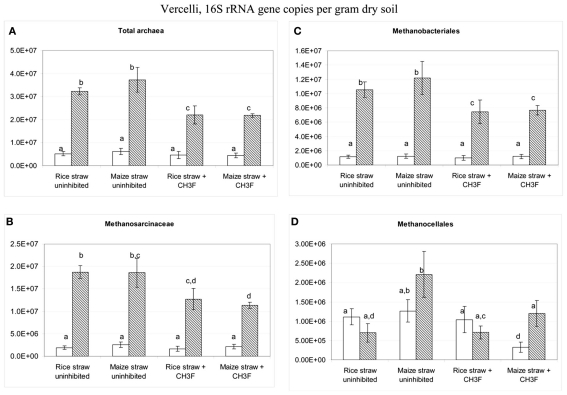
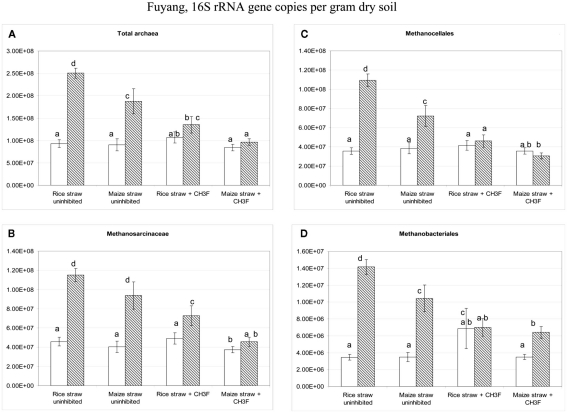
Terminal restriction fragment length polymorphism patterns of archaeal 16S rRNA genes slightly changed during incubation with rice and maize straw in both Vercelli and Fuyang soil with the relative abundance of the 186 bp peak increasing during the phase of active acetate consumption (data not shown). Similar patterns have been observed before and were interpreted as growth of acetoclastic Methanosarcina spp. (Conrad and Klose, 2006; Peng et al., 2008). In both Vercelli (Figure 8) and Fuyang (Figure 9) soil the numbers of total archaea significantly increased after incubation with straw, more in Vercelli than in Fuyang, and more with maize straw than with rice straw. The proliferating archaeal community in Vercelli soil consisted mainly of Methanosarcinaceae and Methanobacteriales, while Methanocellales were much less abundant (Figure 8). In Fuyang soil, on the other hand, Methanosarcinaceae and Methanocellales were the most abundant methanogens, but Methanobacteriales were also of relatively large importance (Figure 9). Addition of CH3F inhibited the proliferation of the methanogenic populations in Vercelli and Fuyang soil (Figures 8 and 9).
Figure 8.
Copy numbers in Vercelli soil of (A) total archaeal 16S rRNA genes, and of those archaeal 16S rRNA genes belonging to (B) Methanosarcinceae, (C) Methanobacteriales, and (D) Methanocellales at the beginning (left column) and the end (right column) of incubation of Vercelli soil with either rice straw or maize straw in the presence and absence of CH3F; mean ± SE, n = 3. Different letters indicate significant (p < 0.05) differences between columns. Beginning and end of incubation are as in Figure 1.
Figure 9.
Copy numbers in Fuyang soil of (A) total archaeal 16S rRNA genes, and of those archaeal 16S rRNA genes belonging to (B) Methanosarcinceae, (C) Methanocellales, and (D) Methanobacteriales at the beginning (left column) and the end (right column) of incubation of Fuyang soil with either rice straw or maize straw in the presence and absence of CH3F; mean ± SE, n = 3. Different letters indicate significant (p < 0.05) differences between columns. Beginning and end of incubation are as in Figure 1.
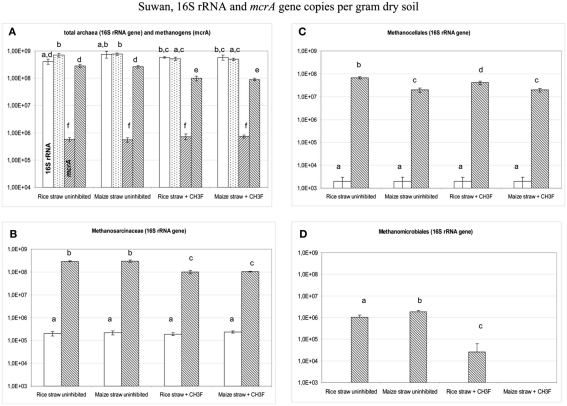
The soil from Suwan had previously not been used for cultivating rice. Although archaea were abundant, the T-RFLP pattern was almost exclusively dominated by the 186 bp T-RF. In contrast to Vercelli and Fuyang soil, the abundance of total archaea did not increase upon incubation with straw (Figure 10). Sequencing of six clones of archaeal 16S rRNA genes generally resulted in assignment to crenarchaeotes (accession numbers: JQ286402–JQ286407). We therefore assumed that the archaeal 16S rRNA in this soil was mainly represented by crenarchaeotes, whose abundance was not enhanced by addition of straw. We therefore decided to quantify methanogens directly by applying qPCR assays targeting the mcrA gene that is unique to methanogens. Using this assay, a significant proliferation of methanogens by two orders of magnitude was detected after incubation with straw reaching almost the abundance of total archaeal 16S rRNA gene copies (Figure 10). The methanogenic community consisted almost equally of Methanosarcinaceae and Methanocellales, with Methanomicrobiales as only minor additional component (Figure 10). Proliferation of Methanosarcinaceae and total methanogens was slightly inhibited by the presence of CH3F (Figure 10).
Figure 10.
Copy numbers in Suwan soil of (A) total archaeal 16S rRNA genes (the two left columns in each group) and total mcrA genes (the two right columns in each group), and of 16S rRNA genes belonging to (B) Methanosarcinceae, (C) Methanocellales, and (D) Methanomicrobiales at the beginning (left column) and the end (right column) of incubation of Suwan soil with either rice straw or maize straw in the presence and absence of CH3F; mean ± SE, n = 3. Different letters indicate significant (p < 0.05) differences between columns. Beginning and end of incubation are as in Figure 1.
Discussion
Our study showed differences in the temporal patterns and rates of CH4 production, and of the transient production of the methanogenic precursors acetate and H2 especially across the different soils but also between the treatments with maize straw versus rice straw. Similarly, the composition and dynamics of the methanogenic populations were quite different in the different soils. Finally, substantial differences were found in the isotopic signatures of CH4 and acetate-methyl between maize straw and rice straw treatments, basically since the organic matter serving as substrate for methanogenic degradation had different isotopic signatures (C4 versus C3 plant). However, despite all these differences, CH4 production from both maize straw versus rice straw followed similar pathways, which were also quite similar for the different soils.
Methanogenic pathway
Hence, the differences were all in details of the methanogenic degradation process and the active methanogenic populations, while the overall methanogenic path was very similar. This demonstrates an amazing constancy in system functioning despite all the differences in substrate quality (mainly based on straw type) and archaeal community composition and abundance (mainly based on soil type). We had previously observed a similar constancy in methanogenic paths in soils vegetated with cultivated rice versus wild rice, although differences in turnover rates existed (Conrad et al., 2009). However, these soils had a similar archaeal community composition. On the other hand, we have observed dramatic differences in CH4 production and emission when rice was grown on different soils, and roots were colonized with crossly different methanogenic populations (Conrad et al., 2008). Therefore, we do not generally exclude that the archaeal methanogenic community composition and abundance might have a substantial effect on methanogenic functioning. In the present study, however, the methanogenic functioning was generally similar.
In general, CH4 production was at first dominated by hydrogenotrophic methanogenesis which subsequently decreased as acetoclastic methanogenesis became progressively more important, and finally increased again. This temporal pattern was paralleled by the initial peak in H2 accumulation, followed by the slower transient accumulation of acetate, which was eventually depleted to rather low concentrations characteristic for steady state between fermentative production and methanogenic consumption of acetate. Similar temporal patterns of the relative contribution of hydrogenotrophic versus acetoclastic methanogenesis have been observed before (Conrad et al., 2002; Fey et al., 2004; Penning and Conrad, 2007). Such temporal pattern has been explained by a conceptual model based on the sequential degradation of organic matter by hydrolytic fermentative, and methanogenic microbes, by the competition of sulfate-reducing and iron-reducing bacteria for H2 and by the relatively late activation of acetoclastic methanogens (Conrad, 2002; Glissmann and Conrad, 2002).
Methanogenic archaea
Incubation of maize straw or rice straw under methanogenic conditions generally resulted in increase of total methanogenic archaea irrespectively of the soil used. In the Vercelli and Fuyang soils, the archaeal 16S rRNA genes almost exclusively belonged to methanogens and copy numbers were relatively abundant (106–108 g−1 dry soil) right from the beginning of incubation, as expected for authentic rice field soils (Krüger et al., 2005; Ma et al., 2010). In the Suwan soil by contrast, copy numbers were in the beginning rather low (103 g−1 dry soil), which is normal for upland soil (Angel et al., 2012). In all of the soils, however, numbers of methanogens eventually increased during methanogenic degradation of straw. The most abundant methanogens always belonged to the family of Methanosarcinaceae, presumably since acetoclastic Methanosarcina species proliferated by consuming the transiently accumulated acetate. Such proliferation during degradation of straw has been observed before and the final abundance was found to be consistent with theoretical calculations based on the energetic conditions in the soil for acetate conversion to CH4 (Conrad and Klose, 2006; Angel et al., 2012). The most abundant hydrogenotrophic methanogens were Methanobacteriales in Vercelli soil, and Methanocellales in the other two soils. The results on Vercelli (Weber et al., 2001; Conrad and Klose, 2006) and Fuyang (Peng et al., 2008) soil is consistent with previous observations. Suwan soil had not been flooded before and thus initially contained only a low abundance of methanogens. In the end, however, Methanosarcinaceae and Methanocellales dominated. In other upland soils these two methanogenic groups were also found to be the most prevalent ones, both before first flooding and after an extensive phase of methanogenesis (Angel et al., 2012). These two methanogenic groups are in particular known for potentially being able to deal with toxic O2 species and surviving oxic soil conditions (Erkel et al., 2006; Angel et al., 2011).
Addition of CH3F generally resulted in substantial inhibition of CH4 production, irrespectively of the type of soil or straw. As CH3F is known as specific inhibitor of acetoclastic methanogens (Janssen and Frenzel, 1997), one would expect that acetate consumption by this group of methanogens is inhibited. Thus, members of the Methanosarcinaceae should be impeded in growth unless they use an alternative energy source such as CH4 production from H2/CO2. A previous study using methanogenic rice roots has indeed shown that growth of acetoclastic Methanosarcinaceae was inhibited by CH3F (Penning and Conrad, 2006). In the present study using Vercelli soil addition of CH3F also significantly inhibited the growth of the potentially acetoclastic Methanosarcinaceae, but it also inhibited growth of non-acetoclastic Methanobacteriales. The same was observed in Fuyang soil amended with maize straw, while the methanogenic population in the rice straw treatments were not significantly inhibited at all. In Suwan soil, on the other hand, again only the growth of Methansarcinaceae but not of Methanocellales was inhibited by CH3F. Although we have no final explanation for the different behavior, we assume that it is due to the fact that Methanosarcina species are not restricted to acetoclastic methanogenic metabolism, and may have used for example H2/CO2 for growth in the presence of CH3F, and that growth of hydrogenotrophic methanogens was unspecifically inhibited (Conrad and Klose, 1999b).
Stable carbon isotope fractionation
Due to inhibition of acetoclastic methanogenesis addition of CH3F inhibited or delayed the consumption of the accumulated acetate and resulted in lower δ13C of the produced CH4. The δ13C of the CH4 produced in the presence of CH3F should represent the δ13C of hydrogenotrophically produced CH4 (). Since the fractionation factor for hydrogenotrophic methanogenesis is larger than for acetoclastic methanogenesis (Conrad, 2005), is expected of being lower than the δ13C of total CH4 or of acetoclastically produced CH4 (), which was actually observed. The fractionation factors of hydrogenotrophic methanogenesis that were calculated from the values of (Eq. 3) were in a range of which have quite commonly been observed in rice field soils (Sugimoto and Wada, 1993; Chidthaisong et al., 2002; Penning and Conrad, 2007; Conrad et al., 2009). The same range of αCO2, CH4 has been used for calculation of () of total CH4 produced from CO2 reduction (Eq. 4) during studies on rice fields in TX, USA (Bilek et al., 1999) and Italy (Krüger et al., 2002). If acetoclastic methanogenesis is not completely inhibited (which might have been the case in Vercelli and Fuyang soil, but not in Suwan soil), would be less negative than expected and values of would be overestimated (sensitivity analysis see Conrad et al., 2002). However, the range of values was similar in soil from Suwan (25–80%) as in soils from Vercelli (15–75%) and Fuyang (25–100%). Nevertheless, it should be noted that the values that were finally reached were relatively high (50–70%) compared to a value of 33% that is expected when carbohydrates are methanogenically decomposed under steady state conditions (Conrad, 1999).
Computation of is also affected by the value of , which was based on measured values of δ13C of acetate-methyl and the assumption of a fractionation factor for acetoclastic methanogenesis of (equivalent to ‰). Such value is reasonable because acetoclastic methanogenesis was apparently dominated by Methanosarcina. In soil, Methanosarcina species exhibit values that are about 5–10‰ less negative than in pure culture which is probably due to limitation by acetate (Penning and Conrad, 2007; Goevert and Conrad, 2009). In systems dominated by Methanosaeta species, values of are even less negative, i.e., on the order of > −10‰ (Valentine et al., 2004; Penning et al., 2006a), and fractionation may become actually zero in systems where acetate supply is so low that it is quantitatively utilized as it is produced. Calculation of results in increasing values with becoming smaller (equivalent to becoming less negative). However, we may assume that this was not the case in our experiments, which generally showed acetate concentrations well above 100 μM.
Taking all these uncertainties into account it is striking that amendment with either maize straw or rice straw had a similar effect on the overall value of and its temporal change. Hence, amendment of soil with maize straw may be used for experiments in which the partitioning of CH4 production derived from root versus straw decomposition is determined. Such determinations are even feasible under field conditions by measuring the δ13C in the total CH4 produced by comparing fields amended with either rice straw or maize straw. We are presently about doing such studies.
Conclusion
The present study showed that despite the involvement of different methanogenic populations in the different soils, pathways and temporal patterns of CH4 production were nearly identical between straw treatments. Hence, the temporal patterns and paths of CH4 production were mostly dependent on the soil and not on the straw. Our study also demonstrates a surprising constancy of system functioning, despite the qualitative and quantitative differences in soil methanogenic populations, including those from a previous upland soil, and differences in straw quality. The observed constancy indicates flexible adaptation of the different microbial communities in order to achieve similar methanogenic degradation of straw.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Peter Claus for help during analysis of δ13C, Roey Angel for advice concerning qPCR, and Yanli Yuan and Pancheewan Pomphangnag for sampling of the Fuyang and Suwan soil, respectively. This study is part of the ICON project financed by the German Research Foundation. We thank Tillmann Lüders, Kim Yrjälä and Sascha Krause for helpful comments.
References
- Amelung W., Brodowski S., Sandhage-Hofmann A., Bol R. (2008). Combining biomarker with stable isotope analyses for assessing the transformation and turnover of soil organic matter. Adv. Agron. 100, 155–250 10.1016/S0065-2113(08)00606-8 [DOI] [Google Scholar]
- Angel R., Claus P., Conrad R. (2012). Methanogenic archaea are globally ubiquitous in aerated soils and become active under wet anoxic conditions. ISME J. (in press). 10.1038/ismej.2011.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angel R., Matthies D., Conrad R. (2011). Activation of methanogenesis in arid biological soil crusts despite the presence of oxygen. PLoS ONE 6, e20453. 10.1371/journal.pone.0020453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balesdent J., Balabane M. (1992). Maize root-derived soil organic carbon estimated by natural C-13 abundance. Soil Biol. Biochem. 24, 97–101 10.1016/0038-0717(92)90264-X [DOI] [Google Scholar]
- Bilek R. S., Tyler S. C., Sass R. L., Fisher F. M. (1999). Differences in CH4 oxidation and pathways of production between rice cultivars deduced from measurements of CH4 flux and δ13C of CH4 and CO2. Global Biogeochem. Cycles 13, 1029–1044 10.1029/1999GB900040 [DOI] [Google Scholar]
- Cai Z. C., Tsuruta H., Rong X. M., Xu H., Yuan Z. P. (2001). CH4 emissions from rice paddies managed according to farmer’s practice in Hunan, China. Biogeochemistry 56, 75–91 10.1023/A:1011940730288 [DOI] [Google Scholar]
- Chidthaisong A., Chin K. J., Valentine D. L., Tyler S. C. (2002). A comparison of isotope fractionation of carbon and hydrogen from paddy field rice roots and soil bacterial enrichments during CO2/H2 methanogenesis. Geochim. Cosmochim. Acta 66, 983–995 10.1016/S0016-7037(01)00812-2 [DOI] [Google Scholar]
- Chidthaisong A., Watanabe I. (1997). Methane formation and emission from flooded rice soil incorporated with 13C-labeled rice straw. Soil Biol. Biochem. 29, 1173–1181 10.1016/S0038-0717(97)00034-5 [DOI] [Google Scholar]
- Chin K. J., Lukow T., Conrad R. (1999). Effect of temperature on structure and function of the methanogenic archaeal community in an anoxic rice field soil. Appl. Environ. Microbiol. 65, 2341–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad R. (1999). Contribution of hydrogen to methane production and control of hydrogen concentrations in methanogenic soils and sediments [review]. FEMS Microbiol. Ecol. 28, 193–202 10.1111/j.1574-6941.1999.tb00575.x [DOI] [Google Scholar]
- Conrad R. (2002). Control of microbial methane production in wetland rice fields [review]. Nutr. Cycl. Agroecosyst. 64, 59–69 10.1023/A:1021178713988 [DOI] [Google Scholar]
- Conrad R. (2005). Quantification of methanogenic pathways using stable carbon isotopic signatures: a review and a proposal. Org. Geochem. 36, 739–752 10.1016/j.orggeochem.2004.09.006 [DOI] [Google Scholar]
- Conrad R. (2007). Microbial ecology of methanogens and methanotrophs. Adv. Agron. 96, 1–63 10.1016/S0065-2113(07)96005-8 [DOI] [Google Scholar]
- Conrad R., Chan O. C., Claus P., Casper P. (2007). Characterization of methanogenic archaea and stable isotope fractionation during methane production in the profundal sediment of an oligotrophic lake (Lake Stechlin, Germany). Limnol. Oceanogr. 52, 1393–1406 10.4319/lo.2007.52.4.1393 [DOI] [Google Scholar]
- Conrad R., Klose M. (1999a). Anaerobic conversion of carbon dioxide to methane, acetate and propionate on washed rice roots. FEMS Microbiol. Ecol. 30, 147–155 10.1111/j.1574-6941.1999.tb00643.x [DOI] [PubMed] [Google Scholar]
- Conrad R., Klose M. (1999b). How specific is the inhibition by methyl fluoride of acetoclastic methanogenesis in anoxic rice field soil? FEMS Microbiol. Ecol. 30, 47–56 10.1111/j.1574-6941.1999.tb00643.x [DOI] [Google Scholar]
- Conrad R., Klose M. (2006). Dynamics of the methanogenic archaeal community in anoxic rice soil upon addition of straw. Eur. J. Soil Sci. 57, 476–484 10.1111/j.1365-2389.2006.00791.x [DOI] [Google Scholar]
- Conrad R., Klose M., Claus P. (2002). Pathway of CH4 formation in anoxic rice field soil and rice roots determined by 13C-stable isotope fractionation. Chemosphere 47, 797–806 10.1016/S0045-6535(02)00120-0 [DOI] [PubMed] [Google Scholar]
- Conrad R., Klose M., Claus P., Dan J. G. (2009). Activity and composition of the methanogenic archaeal community in soil vegetated with wild versus cultivated rice. Soil Biol. Biochem. 41, 1390–1395 10.1016/j.soilbio.2009.03.013 [DOI] [Google Scholar]
- Conrad R., Klose M., Noll M., Kemnitz D., Bodelier P. L. E. (2008). Soil type links microbial colonization of rice roots to methane emission. Glob. Chang. Biol. 14, 657–669 10.1111/j.1365-2486.2007.01516.x [DOI] [Google Scholar]
- Denier van der Gon H. A. C., Neue H. U. (1995). Influence of organic matter incorporation on the methane emission from a wetland rice field. Global Biogeochem. Cycles 9, 11–22 10.1029/94GB03197 [DOI] [Google Scholar]
- Erkel C., Kube M., Reinhardt R., Liesack W. (2006). Genome of Rice Cluster I archaea - the key methane producers in the rice rhizosphere. Science 313, 370–372 10.1126/science.1127062 [DOI] [PubMed] [Google Scholar]
- Farquhar G. D., Ehleringer J. R., Hubick K. T. (1989). Carbon isotope discrimination and photosynthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 40, 503–537 10.1146/annurev.pp.40.060189.002443 [DOI] [Google Scholar]
- Fey A., Claus P., Conrad R. (2004). Temporal change of 13C-isotope signatures and methanogenic pathways in rice field soil incubated anoxically at different temperatures. Geochim. Cosmochim. Acta 68, 293–306 10.1016/S0016-7037(03)00426-5 [DOI] [Google Scholar]
- Flessa H., Ludwig B., Heil B., Merbach W. (2000). The origin of soil organic C, dissolved organic C and respiration in a long-term maize experiment in Halle, Germany, determined by 13C natural abundance. J. Plant Nutr. Soil Sci. 163, 157–163 [DOI] [Google Scholar]
- Glissmann K., Conrad R. (2002). Saccharolytic activity and its role as a limiting step in methane formation during the anaerobic degradation of rice straw in rice paddy soil. Biol. Fertil. Soils 35, 62–67 10.1007/s00374-002-0442-z [DOI] [Google Scholar]
- Goevert D., Conrad R. (2009). Effect of substrate concentration on carbon isotope fractionation during acetoclastic methanogenesis by Methanosarcina barkeri and M. acetivorans and in rice field soil. Appl. Environ. Microbiol. 75, 2605–2612 10.1128/AEM.02680-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosskopf R., Janssen P. H., Liesack W. (1998). Diversity and structure of the methanogenic community in anoxic rice paddy soil microcosms as examined by cultivation and direct 16S rRNA gene sequence retrieval. Appl. Environ. Microbiol. 64, 960–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson P. J., Edwards N. T., Garten C. T., Andrews J. A. (2000). Separating root and soil microbial contributions to soil respiration: a review of methods and observations [review]. Biogeochemistry 48, 115–146 10.1023/A:1006244819642 [DOI] [Google Scholar]
- Hayes J. M. (1993). Factors controlling 13C contents of sedimentary organic compounds: principles and evidence. Mar. Geol. 113, 111–125 10.1016/0025-3227(93)90153-M [DOI] [Google Scholar]
- Holzapfel-Pschorn A., Conrad R., Seiler W. (1986). Effects of vegetation on the emission of methane from submerged paddy soil. Plant Soil 92, 223–233 10.1007/BF02372636 [DOI] [Google Scholar]
- Janssen P. H., Frenzel P. (1997). Inhibition of methanogenesis by methyl fluoride – studies of pure and defined mixed cultures of anaerobic bacteria and archaea. Appl. Environ. Microbiol. 63, 4552–4557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb S., Knief C., Dunfield P. F., Conrad R. (2005). Abundance and activity of uncultured methanotrophic bacteria involved in the consumption of atmospheric methane in two forest soils. Environ. Microbiol. 7, 1150–1161 10.1111/j.1462-2920.2005.00791.x [DOI] [PubMed] [Google Scholar]
- Krüger M., Eller G., Conrad R., Frenzel P. (2002). Seasonal variation in pathways of CH4 production and in CH4 oxidation in rice fields determined by stable carbon isotopes and specific inhibitors. Glob. Chang. Biol. 8, 265–280 10.1046/j.1365-2486.2002.00476.x [DOI] [Google Scholar]
- Krüger M., Frenzel P., Kemnitz D., Conrad R. (2005). Activity, structure and dynamics of the methanogenic archaeal community in a flooded Italian rice field. FEMS Microbiol. Ecol. 51, 323–331 10.1016/j.femsec.2004.09.004 [DOI] [PubMed] [Google Scholar]
- Krumböck M., Conrad R. (1991). Metabolism of position-labelled glucose in anoxic methanogenic paddy soil and lake sediment. FEMS Microbiol. Ecol. 85, 247–256 10.1111/j.1574-6968.1991.tb04731.x [DOI] [Google Scholar]
- Kuzyakov Y. (2006). Sources of CO2 efflux from soil and review of partitioning methods [review]. Soil Biol. Biochem. 38, 425–448 10.1016/j.soilbio.2006.04.002 [DOI] [Google Scholar]
- Kuzyakov Y., Cheng W. (2001). Photosynthesis controls of rhizosphere respiration and organic matter decomposition. Soil Biol. Biochem. 33, 1915–1925 10.1016/S0038-0717(00)00115-2 [DOI] [Google Scholar]
- Lueders T., Friedrich M. (2000). Archaeal population dynamics during sequential reduction processes in rice field soil. Appl. Environ. Microbiol. 66, 2732–2742 10.1128/AEM.66.7.2732-2742.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Xu H., Yagi K., Cai Z. C. (2008). Methane emission from paddy soils as affected by wheat straw returning mode. Plant Soil 313, 167–174 10.1007/s11104-008-9689-y [DOI] [Google Scholar]
- Ma K., Conrad R., Lu Y. (2012). Responses of methanogen mcrA genes and their transcripts to alternate dry wet cycle of paddy field soil. Appl. Environ. Microbiol. 78, 445–454 10.1128/AEM.06934-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma K., Qiu Q. F., Lu Y. H. (2010). Microbial mechanism for rice variety control on methane emission from rice field soil. Glob. Chang. Biol. 16, 3085–3095 10.1111/j.1365-2486.2009.01932.x [DOI] [Google Scholar]
- Mayer H. P., Conrad R. (1990). Factors influencing the population of methanogenic bacteria and the initiation of methane production upon flooding of paddy soil. FEMS Microbiol. Ecol. 73, 103–112 10.1111/j.1574-6968.1990.tb03930.x [DOI] [Google Scholar]
- Minoda T., Kimura M. (1994). Contribution of photosynthesized carbon to the methane emitted from paddy fields. Geophys. Res. Lett. 21, 2007–2010 10.1029/94GL01595 [DOI] [Google Scholar]
- Peng J. J., Lü Z., Rui J., Lu Y. H. (2008). Dynamics of the methanogenic archaeal community during plant residue decomposition in an anoxic rice field soil. Appl. Environ. Microbiol. 74, 2894–2901 10.1128/AEM.01442-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penning H., Claus P., Casper P., Conrad R. (2006a). Carbon isotope fractionation during acetoclastic methanogenesis by Methanosaeta concilii in culture and a lake sediment. Appl. Environ. Microbiol. 72, 5648–5652 10.1128/AEM.72.1.178-184.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penning H., Tyler S. C., Conrad R. (2006b). Determination of isotope fractionation factors and quantification of carbon flow by stable isotope signatures in a methanogenic rice root model system. Geobiology 4, 109–121 10.1111/j.1472-4669.2006.00075.x [DOI] [Google Scholar]
- Penning H., Conrad R. (2006). Effect of inhibition of acetoclastic methanogenesis on growth of archaeal populations in an anoxic model environment. Appl. Environ. Microbiol. 72, 178–184 10.1128/AEM.72.1.178-184.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penning H., Conrad R. (2007). Quantification of carbon flow from stable isotope fractionation in rice field soils with different organic matter content. Org. Geochem. 38, 2058–2069 10.1016/j.orggeochem.2007.08.004 [DOI] [Google Scholar]
- Peters V., Conrad R. (1996). Sequential reduction processes and initiation of CH4 production upon flooding of oxic upland soils. Soil Biol. Biochem. 28, 371–382 10.1016/0038-0717(95)00146-8 [DOI] [Google Scholar]
- Rochette P., Flanagan L. B., Gregorich E. G. (1999). Separating soil respiration into plant and soil components using analyses of the natural abundance of carbon-13. Soil Sci. Soc. Am. J. 63, 1207–1213 10.2136/sssaj1999.6351207x [DOI] [Google Scholar]
- Schütz H., Holzapfel-Pschorn A., Conrad R., Rennenberg H., Seiler W. (1989). A 3-year continuous record on the influence of daytime, season, and fertilizer treatment on methane emission rates from an Italian rice paddy. J. Geophys. Res. 94, 16405–16416 10.1029/JD094iD13p16405 [DOI] [Google Scholar]
- Singh S., Kushwaha B., Nag S., Mishra A., Bhattacharya S., Gupta P., Singh A. (2011). In vitro methane emission from Indian dry roughages in relation to chemical composition. Curr. Sci. 101, 57–65 [Google Scholar]
- Sugimoto A., Wada E. (1993). Carbon isotopic composition of bacterial methane in a soil incubation experiment: contributions of acetate and CO2/H2. Geochim. Cosmochim. Acta 57, 4015–4027 10.1016/0016-7037(93)90350-6 [DOI] [Google Scholar]
- Thongsaga K., Ranamukhaarachchi S. L., Jampatong S., Samarakoon L., Noomhorm A., Clemente R. S., Hannaway D. B. (2010). Comparison of crop simulation and field performance of maize under 20-day dry period imposed during selected critical growth periods in Nakhon Ratchasima province, Thailand. Recent Res. Sci. Technol. 2, 17–31 [Google Scholar]
- Tian G., Kang B. T., Brussaard L. (1992). Biological effects of plant residues with contrasting chemical-compositions under humid tropical conditions - decomposition and nutrient release. Soil Biol. Biochem. 24, 1051–1060 10.1016/0038-0717(92)90035-V [DOI] [Google Scholar]
- Timsina J., Jat M. L., Majumdar K. (2010). Rice-maize systems of South Asia: current status, future prospects and research priorities for nutrient management [review]. Plant Soil 335, 65–82 10.1007/s11104-010-0418-y [DOI] [Google Scholar]
- Valentine D. L., Chidthaisong A., Rice A., Reeburgh W. S., Tyler S. C. (2004). Carbon and hydrogen isotope fractionation by moderately thermophilic methanogens. Geochim. Cosmochim. Acta 68, 1571–1590 10.1016/j.gca.2003.10.012 [DOI] [Google Scholar]
- Watanabe A., Satoh Y., Kimura M. (1995). Estimation of the increase in CH4 emission from paddy soils by rice straw application. Plant Soil 173, 225–231 10.1007/BF00011459 [DOI] [Google Scholar]
- Watanabe A., Takeda T., Kimura M. (1999). Evaluation of origins of CH4 carbon emitted from rice paddies. J. Geophys. Res. 104, 23623–23629 10.1029/1999JC900097 [DOI] [Google Scholar]
- Watanabe A., Yoshida M., Kimura M. (1998). Contribution of rice straw carbon to CH4 emission from rice paddies using 13C-enriched rice straw. J. Geophys. Res. 103, 8237–8242 10.1029/98JD01679 [DOI] [Google Scholar]
- Weber S., Lueders T., Friedrich M. W., Conrad R. (2001). Methanogenic populations involved in the degradation of rice straw in anoxic paddy soil. FEMS Microbiol. Ecol. 38, 11–20 10.1111/j.1574-6941.2001.tb00877.x [DOI] [Google Scholar]
- Werth M., Kuzyakov Y. (2010). 13C fractionation at the root-microorganisms-soil interface: a review and outlook for partitioning studies. Soil Biol. Biochem. 42, 1372–1384 10.1016/j.soilbio.2010.04.009 [DOI] [Google Scholar]
- Wu L. Q., Ma K., Li Q., Ke X. B., Lu Y. H. (2009). Composition of archaeal community in a paddy field as affected by rice cultivar and N fertilizer. Microb. Ecol. 58, 819–826 10.1007/s00248-009-9554-4 [DOI] [PubMed] [Google Scholar]
- Yan X. Y., Yagi K., Akiyama H., Akimoto H. (2005). Statistical analysis of the major variables controlling methane emission from rice fields. Glob. Chang. Biol. 11, 1131–1141 10.1111/j.1365-2486.2005.00976.x [DOI] [Google Scholar]
- Yu Y., Lee C., Kim J., Hwang S. (2005). Group-specific primer and probe sets to detect methanogenic communities using quantitative real-time polymerase chain reaction. Biotechnol. Bioeng. 89, 670–679 10.1002/bit.20347 [DOI] [PubMed] [Google Scholar]