Abstract
Recent evidence suggests that post-transcriptional events that are executed in the cytoplasm can be predetermined during transcription. Here I speculate that this is a widespread mode of regulation and discuss the potential mechanisms, advantages and implications of such a regulatory strategy.
Key words: post-transcriptional regulation, coupling, transcription, mRNA degradation, nuclear-cytoplasmic shuttling
Introduction
Eukaryotic gene expression is typically portrayed as a series of sequential events starting with transcription, followed by mRNA processing, mRNA transport to the cytoplasm and eventually either translation or mRNA degradation. However, accumulating evidence demonstrates various cross-talks between these processes and, in particular, that each of these subsequent events is often linked to transcription. mRNA processing, including 5′-end capping, 3′-end polyadenylation, splicing and mRNA export, typically initiate (and often terminate) co-transcriptionally.1 More surprising, however, are observations that also post-transcriptional events that occur in the cytoplasm are linked to transcription in the nucleus.
Multiple studies have shown that the transport and intracellular localization of mRNAs are often determined during transcription, through RNA-binding proteins (RBPs) that bind to the nascent pre-mRNA and later guide the mature mRNA to its correct localization.2,3 mRNA splicing in the nucleus involves binding of the exon junction complex (EJC) to the pre-mRNA, which, at least in some cases, occurs co-transcriptionally, and upon export to the cytoplasm the EJC influences mRNA localization, translation and degradation.3 mRNA degradation and translation are further linked to transcription through the activity of Rpb4/7, two dissociable subunits of RNA polymerase II (RNAP II). These subunits bind the nascent pre-mRNA and dissociate from the core RNAP II during transcription, facilitate the export of the bound mRNA to the cytoplasm and then influence the mRNA's degradation and translation.4,5
These previous observations rely primarily on analysis of few specific genes and thus might reflect mechanisms of limited scope. As a result, cytoplasmic post-transcriptional regulation is still thought of as largely independent of transcription. However, a more prominent role of transcription in priming cytoplasmic events is possible3 and is supported by recent work. In particular, two recent studies focusing on mRNA degradation in yeast have shown a widespread association between cytoplasmic mRNA degradation and transcriptional activity. First, transcriptional induction of hundreds of yeast genes is associated with changes in degradation of these transcripts.6 Notably, a mutation that inhibits the association of Rbp4/7 with RNAP II abolished the changes in mRNA degradation, indicating that Rbp4/7 exerts a widespread influence on mRNA degradation through transcription. In another work, we showed that evolutionary changes in the transcription of yeast genes are coupled to changes in degradation of these transcripts.7 Both trans-mutations and cis-mutations were found to have a dual effect, on transcription and on degradation; trans-mutations were linked to Rbp4/7 and to several other regulators, while cis-mutations were linked to promoter sequences, suggesting again that transcription primes mRNAs for their degradation in the cytoplasm. Note that, since different yeast genes are regulated by distinct cis-regulatory sequences, coupling by cis-mutations at ∼140 genes reflect independent evolutionary events, thus supporting the widespread occurrence of transcriptional priming.
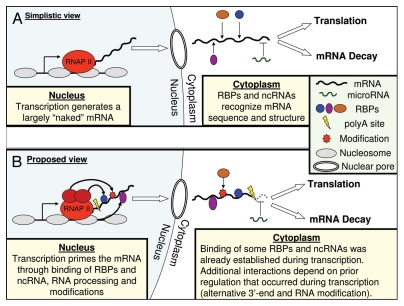
Taken together, transcription appears to prime the localization, mRNA degradation and translation of many genes. Here I hypothesize that this reflects a common strategy of gene regulation that occurs at high frequency in various organisms. Accordingly, a large proportion of all cytoplasmic post-transcriptional events are proposed to be (at least partially) predetermined during transcription, through both known and yet unknown mechanisms (Fig. 1). In the sections below I speculate about potential mechanisms, advantages and implications of this widespread transcriptional priming.
Figure 1.
Schemes of (A) simplistic view and (B) the proposed view for cytoplasmic regulation of gene expression, depicting a single mRNA whose proper regulation in the cytoplasm depends on interactions with three RBPs and on inhibition of the interaction with a microRNA. (A) These interactions occur in the cytoplasm independently of transcription, through recognition of the mRNAs sequence and/or structure. (B) These interactions are either established during transcription (blue and purple RBPs) or depend on prior mRNA regulation that occurred during transcription: RNA modification specifically recruits the brown RBP, and the use of an alternative polyadenylation site that is upstream to a microRNA binding site prevents the inclusion of that binding site and thereby the interaction with the microRNA. As indicated by black arrows, transcription-dependent binding and modifications of the mRNA may be directed through interactions with Pol II and its various associated factors (e.g., CTD-bound proteins), or through histone marks and associated chromatin factors. Other possibilities that are not shown include recruitment through promoter-bound transcription factors or through nuclear core complexes which are associated with actively transcribed genes (see main text).
Mechanisms of Transcriptional Priming
Co-transcriptional binding of RBPs to the nascent pre-mRNA followed by nuclear-cytoplasmic shuttling of the mRNA-bound proteins appears to be a common mechanism for transcriptional priming. This includes known examples (e.g., Rbp4/74 and She22) as well as additional suspected examples (e.g., Ccr4-Not affects both transcription and mRNA degradation and was recently shown to interact with the nuclear export machinery8). Co-transcriptional recruitment of RBPs, as well as of non-coding RNAs (ncRNAs9), could rely on interactions with the huge diversity of proteins in the vicinity of RNAP II, including transcription factors, the general transcription machinery and the RNAP II C-terminal domain (CTD),10 chromatin factors and modified histone tails, and the nuclear pore complexes.11 Binding to the transcriptional machinery may then be followed by loading onto the mRNA and export to the cytoplasm, promoted by the tight connection between transcription elongation and nuclear export.12 Notably, these latter steps may be the main target for regulation, as Rbp4/74 and She22 are recruited to the transcriptional machinery of many genes but shuttle with the mRNA only in a subset of these genes.
RNA structure may change during transcription, as the mRNA molecule is gradually extended and additional base-pairing are established. Some conformations of the nascent pre-mRNA are therefore transient and could define a window of opportunity for specific interactions during transcription that cannot be established once the mRNA reaches its final conformation in the cytoplasm. Furthermore, faster transcription elongation would limit this window of opportunity. Thus, regulation of elongation rates could provide a natural way to link transcriptional activity with the binding of specific factors to mRNA sites that are transiently exposed during transcription.13
In addition to loading of mRNA-binding factors during transcription, other means for altering the state of the transcribed mRNA could also be used to influence its future regulation in the cytoplasm. First, alternative selection of transcription start sites and 3′-polyadenylation (polyA) sites alters 5′ and 3′ UTR lengths and thus controls the inclusion of regulatory sequences in the mature mRNA. Recent work has shown that alternative 5′ and 3′ UTR lengths are a widespread phenomena that influences multiple post-transcriptional mechanisms.14,15 As selection of transcription start sites and polyA sites are coupled to transcription16 they reflect potential sources for widespread transcriptional priming.
Second, it is tempting to speculate that transcriptional priming may be executed by RNA editing and modifications. ADAR enzymes deaminate adenosine to inosine at thousands of loci.17 At least in some cases A-to-I editing is linked to transcription18 and can later affect the recognition of mRNAs by RBPs and ncRNAs17 (e.g., Tudor-SN specifically recognizes inosine-containing RNAs19). Interestingly, recent work suggested widespread RNA editing in human cells including both A-to-I editing and additional mechanisms.20 Finally, mRNAs are also altered by many types of covalent modifications (e.g., methylation) whose dynamics and functions are poorly understood,21 and these could potentially also be involved in transcriptional priming.
Potential Advantages of Transcriptional Priming
Regulation of gene expression is essentially an information processing activity, transforming the information about the state of a cell and its surroundings into decisions of the required activity of all proteins. A great deal of this information processing is performed during transcription, where each gene is bound by a variety of transcriptional regulators and chromatin factors that respond to regulatory signals and dictate the individual expression pattern of that gene. This gene-specific information is typically assumed to be lost once a mature mRNA is exported from the nucleus and is then subjected to another period of information processing, this time through binding of various cytoplasmic RBPs and ncRNA whose levels and activity are also tightly regulated. Instead of these multiple episodes of partial signal processing, each oblivious to the other, transcriptional priming may integrate the signals of transcriptional and post-transcriptional regulation, while their execution remains physically separated. This may allow more sophisticated processing of regulatory signals that could have important implications in various instances.
When a gene is transcriptionally induced, more of its mRNA molecules are generated and exported to the cytoplasm, yet the cytoplasmic regulation of these mRNAs is often unchanged. Transcriptional priming could mark the mRNAs of induced genes, for example if a transcriptional activator recruits an RBP to the mRNA, thereby signaling to the cytoplasmic regulators that these mRNAs are induced and possibly that their regulation should be altered accrodingly. The exact effect of transcriptional induction may be determined by the specific mark that is deposited on the mRNA, and several possibilities could be envisaged. First, an intuitive possibility is that a transcriptionally-induced gene would benefit from further induction by post-transcriptional regulation, thereby enhancing its upregulation. Second, as the time required for reaching a new steady-state of mRNA levels is proportional to the half-life of an mRNA, reduced half-lives could support a more dynamic expression pattern that rapidly adapts to environmental changes or that produces transient expression pulses.22 Consistent with this, increased transcription of yeast genes is often associated with reduced half-lives.6,7
Third, it may be beneficial to ensure that mRNAs of transcriptionally-induced genes are subject to error-free processing and translation. In fact, preferential investment in the fidelity of mRNA regulation of induced genes may have two complementary advantages: It increases the amount of correctly processed mRNAs, thereby facilitating the induced function (which often becomes highly important upon induction), and it may also reduce the load of damaged RNAs23 and translation-induced misfolded proteins.24 Even if only a small (and constant) fraction of mRNAs of a certain gene becomes damaged and toxic, either as mRNAs or later as proteins, the increased amount of these mRNAs upon induction of that gene would incur increased burden of their overall toxicity. This effect would favor any mechanism that reduces the accumulation of RNA damage in mRNAs of induced genes, for example by protecting these mRNAs from reactive oxygen species, enhancing the recruitment of RNA repair enzymes and shortening the half-lives of induced mRNAs. This latter possibility is again consistent with the recent observations that increased transcription is preferentially coupled to reduced half-lives.6,7 Note that this argument is an extension of the idea that the sequences of highly-expressed genes are more conserved due to selection to reduce their propensity to misfold.24 Accordingly, the burden of damaged RNA and misfolded protein toxicity may both impinge on the evolution of genes with constitutively high expression and on the transient regulation of genes with dynamic expression patterns.
Transcriptional priming may also promote specificity in mRNA-RBP interactions. Hundreds of RBPs compete for binding each mRNA in the cytoplasm, inevitably producing some spurious interactions. Yet during transcription only few RBPs may be directly recruited to the site of transcription through interactions with various transcription and chromatin factors. Furthermore, an RBP that is recruited to the transcriptional machinery may travel with the growing transcript and could be placed at specific orientations that promote mRNA interactions, including those that require transient mRNA structures that are only formed during transcription. Once an mRNA is exported to the nucleus its transcription-dependent marks could further induce specificity by interactions with relevant RBPs and ncRNAs.
Concluding Remarks
Transcription may reflect the ideal opportunity for integration of signals and could thus be used to regulate not only the production of mRNAs but also their subsequent processing and translation. Despite multiple studies demonstrating this possibility, transcription and post-transcriptional regulation are still typically regarded as independent processes (Fig. 1A). Here I hypothesize based on recent observations that transcriptional priming may be a widespread phenomenon and speculate about potential mechanisms and advantages (Fig. 1B). According to this hypothesis, the nucleus can be thought of as the cell's brain, where most of the signal processing and “decision making” is being done. Information about some of these decisions is being deposited on the mRNA and transmitted to the rest of the cell, where these decisions are finally executed. These imprinted mRNAs could also be transported to daughter or neighboring cells and thereby have long-distance epigenetic effects.25
Eukaryotic genomes encode hundreds of RBPs, yet RNA recognition motifs or structures are known only for a selected few.26 The hypothesis of widespread transcriptional priming suggests that RNA sequence/structure may not contain all the critical information for decoding mRNA-RBP interactions. These interactions could also be influenced by transcriptional complexes that recruit RBPs to the nascent mRNA, by transient RNA structures that are formed during transcription, by protein-protein interactions between the co-transcriptioally loaded RBPs and additional RBPs that later interact with the mRNA in the cytoplasm, and by additional modifications of the mRNA during transcription. As methods for identifying RBP-RNA interactions are improving,27 it would be interesting to test these possibilities by examining the impact of transcription on RBP-RNA interactions.
Traditionally, the study of gene expression has focused on transcriptional control based on the assumption that most regulation occurs at this step. In recent years, evidences of extensive post-transcriptional regulation have called for shifting much of the attention to regulation of mRNA processing, degradation and translation by RBPs and ncRNAs.28 The possibility that such regulation is partially predetermined at the time of transcription and orchestrated by RNAP II associated factors may suggest that the historical focus on transcription was in fact justified. Accordingly, post-transcriptional regulation should be examined in the context of transcription rather than as an independent layer of regulation.
Acknowledgments
I thank Ilya Soifer, Sagi Levy and Naama Barkai for critical reading. This work was supported by the Clore center at the Weizmann Institute of Science.
References
- 1.Hagiwara M, Nojima T. Cross-talks between transcription and post-transcriptional events within a ‘mRNA factory’. J Biochem. 2007;142:11–15. doi: 10.1093/jb/mvm123. [DOI] [PubMed] [Google Scholar]
- 2.Shen Z, St-Denis A, Chartrand P. Cotranscriptional recruitment of She2p by RNA pol II elongation factor Spt4-Spt5/DSIF promotes mRNA localization to the yeast bud. Genes Dev. 2010;24:1914–1926. doi: 10.1101/gad.1937510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giorgi C, Moore MJ. The nuclear nurture and cytoplasmic nature of localized mRNPs. Semin Cell Dev Biol. 2007;18:186–193. doi: 10.1016/j.semcdb.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 4.Goler-Baron V, Selitrennik M, Barkai O, Haimovich G, Lotan R, Choder M. Transcription in the nucleus and mRNA decay in the cytoplasm are coupled processes. Genes Dev. 2008;22:2022–2027. doi: 10.1101/gad.473608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harel-Sharvit L, Eldad N, Haimovich G, Barkai O, Duek L, Choder M. RNA polymerase II subunits link transcription and mRNA decay to translation. Cell. 2010;143:552–563. doi: 10.1016/j.cell.2010.10.033. [DOI] [PubMed] [Google Scholar]
- 6.Shalem O, Groisman B, Choder M, Dahan O, Pilpel Y. Transcriptome kinetics is governed by a genome-wide coupling of mRNA production and degradation: a role for RNA PolII. PLoS Genet. 2011;7:1002273. doi: 10.1371/journal.pgen.1002273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dori-Bachash M, Shema E, Tirosh I. Coupled evolution of transcription and mRNA degradation. PLoS Biol. 2011;9:1001106. doi: 10.1371/journal.pbio.1001106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kerr SC, Azzouz N, Fuchs SM, Collart MA, Strahl BD, Corbett AH, et al. The Ccr4-Not complex interacts with the mRNA export machinery. PLoS One. 2011;6:18302. doi: 10.1371/journal.pone.0018302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costa FF. Non-coding RNAs: Meet thy masters. Bioessays. 2010;32:599–608. doi: 10.1002/bies.200900112. [DOI] [PubMed] [Google Scholar]
- 10.Egloff S, Murphy S. Cracking the RNA polymerase II CTD code. Trends Genet. 2008;24:280–288. doi: 10.1016/j.tig.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 11.Casolari JM, Brown CR, Komili S, West J, Hieronymus H, Silver PA. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 2004;117:427–439. doi: 10.1016/S0092-8674(04)00448-9. [DOI] [PubMed] [Google Scholar]
- 12.Katahira J, Yoneda Y. Roles of the TREX complex in nuclear export of mRNA. RNA Biol. 2009;6:149–152. doi: 10.4161/rna.6.2.8046. [DOI] [PubMed] [Google Scholar]
- 13.Pinto PA, Henriques T, Freitas MO, Martins T, Domingues RG, Wyrzykowska PS, et al. RNA polymerase II kinetics in polo polyadenylation signal selection. EMBO J. 2011;30:2431–2444. doi: 10.1038/emboj.2011.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Giammartino DC, Nishida K, Manley JL. Mechanisms and Consequences of Alternative Polyadenylation. Mol Cell. 2011;43:853–866. doi: 10.1016/j.molcel.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pal S, Gupta R, Kim H, Wickramasinghe P, Baubet V, Showe LC, et al. Alternative transcription exceeds alternative splicing in generating the transcriptome diversity of cerebellar development. Genome Res. 2011;21:1260–1272. doi: 10.1101/gr.120535.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ji Z, Luo W, Li W, Hoque M, Pan Z, Zhao Y, et al. Transcriptional activity regulates alternative cleavage and polyadenylation. Mol Syst Biol. 2011;7:534. doi: 10.1038/msb.2011.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hundley HA, Bass BL. ADAR editing in double-stranded UTRs and other noncoding RNA sequences. Trends Biochem Sci. 2010;35:377–383. doi: 10.1016/j.tibs.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laurencikiene J, Kallman AM, Fong N, Bentley DL, Ohman M. RNA editing and alternative splicing: the importance of co-transcriptional coordination. EMBO Rep. 2006;7:303–307. doi: 10.1038/sj.embor.7400621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scadden AD. The RISC subunit Tudor-SN binds to hyper-edited double-stranded RNA and promotes its cleavage. Nat Struct Mol Biol. 2005;12:489–496. doi: 10.1038/nsmb936. [DOI] [PubMed] [Google Scholar]
- 20.Li M, Wang IX, Li Y, Bruzel A, Richards AL, Toung JM, et al. Widespread RNA and DNA sequence differences in the human transcriptome. Science. 2011;333:53–58. doi: 10.1126/science.1207018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He C. Grand challenge commentary: RNA epigenetics? Nat Chem Biol. 2010;6:863–865. doi: 10.1038/nchembio.482. [DOI] [PubMed] [Google Scholar]
- 22.Elkon R, Zlotorynski E, Zeller KI, Agami R. Major role for mRNA stability in shaping the kinetics of gene induction. BMC Genomics. 2010;11:259. doi: 10.1186/1471-2164-11-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wurtmann EJ, Wolin SL. RNA under attack: cellular handling of RNA damage. Crit Rev Biochem Mol Biol. 2009;44:34–49. doi: 10.1080/10409230802594043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drummond DA, Wilke CO. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell. 2008;134:341–352. doi: 10.1016/j.cell.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choder M. mRNA imprinting: Additional level in the regulation of gene expression. Cell Logist. 2011;1:37–40. doi: 10.4161/cl.1.1.14465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hogan DJ, Riordan DP, Gerber AP, Herschlag D, Brown PO. Diverse RNA-binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLoS Biol. 2008;6:255. doi: 10.1371/journal.pbio.0060255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanford JR, Wang X, Mort M, Vanduyn N, Cooper DN, Mooney SD, et al. Splicing factor SFRS1 recognizes a functionally diverse landscape of RNA transcripts. Genome Res. 2009;19:381–394. doi: 10.1101/gr.082503.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Plotkin JB. Transcriptional regulation is only half the story. Mol Syst Biol. 2010;6:406. doi: 10.1038/msb.2010.63. [DOI] [PMC free article] [PubMed] [Google Scholar]