Abstract
In past years, many efforts were invested to define epigenetic features associated with enhancers of transcription. We propose that both transcription initiation and the H3K4me3 histone modification are among the best hallmarks of active enhancers in several primary tissues and extend the concept of large transcription initiation platforms (TIPs).
Key words: RNA polymerase II, enhancers, transcription initiation, tissue-specificity, CpG islands
Introduction
Eukaryotic organisms are in constant need to respond to environmental and developmental queues. This is typically brought about via the alteration and tuning of the RNA polymerase II (RNAP II)-dependent transcriptional regulatory program. Control of gene expression requires a tight interplay between promoters, binding transcription factors (TFs) that are able to recruit RNAP II and general transcription factors (GTFs) to form the pre-initiation complex (PIC) and distant cis-regulatory elements such as enhancers.1 This interplay also provides important means to diversify the gene expression programs via dynamic promoter-enhancer interactions. Enhancers are not constitutively active and are known to be either active, inactive and also in a developmentally poised state.2 When active, they enhance the recruitment of RNAP II and GTFs at the target promoters in order to increase transcription levels. Recent genome-wide studies have shed light on the epigenetic signatures of enhancers and promoters.3–5 Most importantly, the recruitment of RNAP II and resulting local transcription has now been shown to be a global phenomenon in several primary cells, including neurons,6 macrophages7 and CD4+/CD8+ double positive thymocytes (DP).8 In the latter, we proposed that RNAP II recruitment to enhancers provides a mark not only of activity but also tissue-specificity of associated genes. We also proposed that large arrays of transcription initiation at promoters, which we termed transcription initiation platforms (TIPs), are hallmarks of highly tissue-specific gene expression. They overlap with a high density of transcription factor binding sites (TFBS) and CpG content. By analyzing published data, we extend or observations to other cell types and propose RNAP II enrichment as a means to isolate active, tissue-specific enhancers and promoters.
Enhancers
Enhancers were originally discovered in vitro as DNA elements that could activate and further strengthen promoter-dependent transcription on naked templates in an orientation independent manner and at great distances.2 These transient reporter assays, however, suffer from many conceptual shortcomings such as the cell type used for transfection, the lack of proper chromatin assembly on the plasmids or the heterogeneity in the overall design of experiments in various studies. Thus, validation of enhancer elements is a difficult task and generally involves genetic manipulation if to be tested in vivo. Their mode of action still remains somewhat elusive and is likely to differ depending upon location. Two generally accepted mechanisms are that of looping, tracking or a combination thereof.1 In the looping model a direct interaction between enhancers and promoters is believed to facilitate the increased recruitment of TFs or RNAP II. The tracking model instead proposes initial RNAP II recruitment to the enhancer and transcription toward the target gene promoter, thereby resulting in large areas of permissive chromatin such as via the recruitment of histone acetyltransferases (HATs). Genome-wide localization techniques like chromatin IP (ChIP) coupled to microarrays (ChIP-on-chip) or deep sequencing (ChIP-Seq) have made it possible to study specific chromatin states associated with enhancers or promoters. Following initial studies in human cervical cancer cells lines,3 it was proposed that enhancers display an H3K4me1high/H3K4me3absent/HAT combinatorial epigenetic mark. These findings were further validated in other tissues.4 Others have used only the recruitment of the HAT p300 as a criterion to isolate enhancers in the mouse embryo.9 More recently another layer of information has been added with the finding that the H3K27Ac mark is able to discriminate between active and poised but inactive enhancers.10,11 Many studies have since adopted such a combinatorial, the presence or absence of given histone marks or modifiers, as a criterion to isolate putative enhancers in the genome. However, the data are not always as unambiguous. Specifically in primary human CD4+ T cells, the “promoter mark” of H3K4me3 was enriched at many isolated enhancer regions.5,12 These results did not gain much attention, but were recently verified across several human tissues, showing that H3K4me3 is associated to very strong enhancer activity.13 Our laboratory recently validated these results in primary murine thymocytes, showing that this mark is indeed associated to active enhancers and that its gain or loss correlates with their activation and repression respectively.14 It therefore appears that from a strict epigenetic point of view, promoters and enhancers are not as easily distinguishable as previously thought.
Enhancer Transcription
In early studies, low levels of GTFs and RNAP II recruitment have been observed at a small subset of isolated enhancers,3 but until recently it was unknown whether this represents a more general phenomenon. In stimulated neurons, some 12,000 potential enhancers were isolated based on the H3K4me1high/H3K4me3absent/HAT combinatorial (as compared with 1,000 in unstimulated conditions). Many of these enhancers also recruited RNAP II, resulting in local transcription of non-polyadenylated enhancer RNAs (eRNAs). While not attributed to tracking due to their relatively small size, eRNA transcript levels correlated with those of nearby genes. Similar results were obtained in stimulated macrophages;7 however, RNAP II recruitment was mostly restricted to the relative proximity of transcription start sites (TSSs). The transcribed RNA was polyadenylated and essentially directional toward the genes, reminiscent of tracking. Interestingly, these RNAs were independent on transcriptional elongation and required only the presence of initiating RNAP II, phosphorylated at the serine 5 residue (Ser5P) of its C-terminal domain (CTD). The same epigenetic combinatorial was used to isolate enhancer regions in this study, except that a machine learning algorithm based on several hundred intergenic HAT binding sites was used, resulting in an H3K4me1high/H3K4melow/HAT putative enhancer profile. While possible direct or indirect functions of these RNAs cannot be excluded from either study, it was proposed that the mode of action lies in creating permissive chromatin structures around these cis-regulatory elements. Short abortive RNA transcripts were also detected at enhancer regions enriched for H3K27Ac in murine embryonic stem cells (ESC), indicating the recruitment and initiation of RNAP II at these regions.10 In stimulated human prostate cancer cells, global run-on assays (GRO-Seq) was used to show that the androgen receptor is able to recruit RNAP II to enhancer regions and that this in turn leads to local transcription.15 Besides the effect on local nucleosomal structure, it was proposed that the presence and levels of RNAP II and eRNA's at enhancers provides a more robust indicator of enhancer activity as compared with the previously used epigenetic combinatorial. We have shown similar genome-wide recruitment of Ser5P RNAP II to tissue-specific enhancers in primary murine T cells.8 Upon visual inspection of the data, we observed complete PIC recruitment and deposition of the H3K4me3 mark to known and well-described active enhancers, validated on the basis of genetic studies. Genome-wide isolation of putative enhancers based on the presence of H3K4me1/H3K4me3/TBP/Ser5P resulted in a highly tissue-specific selection of associated neighboring genes. When compared with the canonical combinatorial for isolation, we also observed a drastically improved tissue-restrictive expression profile, as exemplified by the expression level differences of selected genes in T cells as compared with other tissues. Whereas in neurons and macrophages either polyadenylated or non-polyadenylated RNA was observed, we showed the presence of both populations in our cells, including some well known enhancer regions. Regardless of the type of transcript however, another main discriminating feature between promoters and enhancers were the relative H3K4me1:H3K4me3 levels, being higher at enhancers as compared with promoters.
Enhancer-Transcription by RNAP II and Tissue-specificity
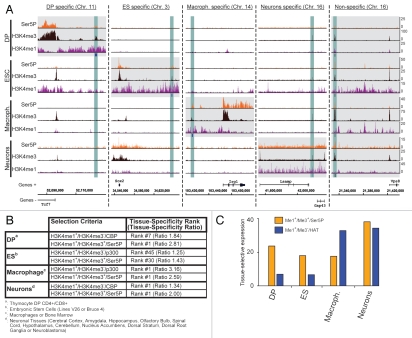
To strengthen and extend these observations in other cell types, we used previously published data for H3K4me1, H3K4me3, Ser5P and CBP/p300 from unstimulated neurons (GEO accession number GSE211616), unstimluated macrophages (GSE17631,16 GSE1955317 and GSE199917) and embryonic stem cells (ESC; GSE2053018 and GSE2416510) and performed similar comparisons of enhancer isolation strategies as described before in reference 8. Using the selection criteria of H3K4me1/H3K4me3/Ser5P (omitting TBP as it was not available in all data sets), we were able to isolate intergenic regions (IGRs) likely to represent enhancers controlling tissue-specific genes (Fig. 1A). These include the T-cell specific transcription factor 7 (Tcf7) in DP T cells, SRY-box 2 (Sox2) in ESC, immunoresponsive homolog 1 (Irg1) in macrophages and a region between the limbic system-associated membrane protein (Lsamp) and neuron growth-associated protein 43 (Gap43) genes in neurons. As expected, however, not all isolated regions were tissue-specific and we found a putative enhancer in all four cell types upstream of the vacuolar protein sorting 8 homolog (Vps8) gene. With the exception of macrophages, this selection criterion provided a more tissue-specific expression pattern as compared with H3K4me1high/H3K4me3absent/HAT (Fig. 1B). In this analysis, we compute the ratio of expression levels between the selected (neighboring the isolated enhancers) and all genes within one tissue. As a result, the higher the ratio, the more tissue-specific the gene selection is. It is also noteworthy that the most dramatic improvements were obtained in thymocytes and ES cells, as in macrophages and neurons the tissue in question was ranked first using all selection criteria, with changes only observed in ratios. Similar results were obtained when comparing the tissue selective expression patterns (Fig. 1C). In this case, we compute the median expression levels of selected genes in the tissues of interest and compare them to the median expression levels in all remaining tissues available in the BioGPS19 database. The higher the differential, the more tissue-selective the genes are expressed. In all cases except for macrophages again, H3K4me1high/H3K4me3absent/HAT provided the less tissue-restrictive expression pattern. These results were greatly improved for thymocytes, ES cells and neurons upon the inclusion of Ser5P in combination with H3K4me3. In three out of four cell-types, it is therefore apparent that the inclusion of Ser5P/H3K4me3 greatly improves the tissue-specific and tissue-restrictive expression pattern of genes associated to isolated enhancers.
Figure 1.
RNAP II and H3K4me3 are present at putative enhancers and increase the tissue-specific and -restrictive expression pattern of associated genes. (A) Examples of cell specific and non-specific RNAP II-bound enhancers in four different primary cell types. We used published data (see text) to isolate putative enhancers based on the presence of H3K4me1/H3K4me3/Ser5P at intergenic regions (±5 kb from any known annotation) in DP T cells, ESC, macrophages and neurons. The isolated regions are indicated as blue boxes underneath the H3K4me1 tracks. Using this strategy, we isolated tissue-specific enhancers (highlighted in blue) such as at Tcf7 in DP, Sox2 in ESC, Irg1 in macrophages, Lsamp/Gap43 in neurons as well as a conserved ones such as at Vps8 (from left to right, highlighted by gray boxes). (B) Tissue-specificity of putative enhancer selections. We assessed the tissue-specificity of genes associated to putative enhancers (neighboring upstream and downstream) by calculating the ratio of their expression level as compared with that of all remaining genes for all tissues available on BioGPS.19 Therefore, the higher ratios indicate higher tissue-specificity. The rank indicates the first occurrence of a tissue of interest (see legends A–D) when sorting the tissue-ratios from highest to lowest. With the exception of macrophages, either both the rank, but at least the ratio, increased when comparing a H3K4me1+/H3K4me3+/Ser5P to a H3K4me1+/H3K4me3−/HAT enhancer selection criteria. (C) Tissue-selective expression of genes associated with putative enhancer selections. To determine the tissue-selective expression of selected genes, we calculated the median expression level within our tissue(s) of interest for each cell type and plotted the differential to that of all remaining available tissues in the BioGPS database. Thus, the higher differentials represent the more tissue-selective expression pattern of the genes. Again, with the exception of macrophages, the H3K4me1+/H3K4me3+/Ser5P selection criteria (orange) improves the tissue-selective gene expression patterns as compared with H3K4me1+/H3K4me3−/HAT (blue).
TIPs and Tissue-specificity
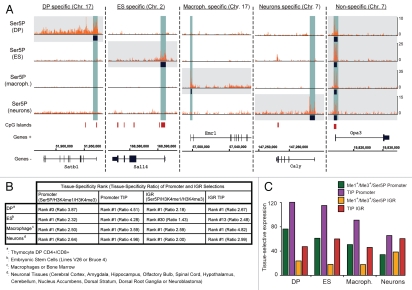
We previously proposed that TIPs drive tissue-specific gene expression in primary T cells and used the data described above to possibly extend this observation. We originally isolated TIPs based on TATA-binding protein (TBP) and initiating RNAP II recruitment areas. As for enhancer selection however, a TBP data set was not available for all cell types. We therefore instead defined TIPs based solely on the presence of a continuum of enriched initiating RNAP II within an area of greater than 400 bp. Using this approach we identified TIPs at the promoters of tissue-specific genes in all four data sets (Fig. 2A). These genes include special AT-rich sequence-binding protein 1 (Satb1) in DP, sal-like 4 (Sall4) in ESC, EGF-like module receptor 1 (Emr1) in macrophages and calcyon neuron-specific vesicular protein (Caly) in neurons. While we proposed that TIPs are enriched at tissue-specific genes, we do not exclude their presence at housekeeping genes as well. Not unexpectedly therefore, we also observed TIPs conserved across all four cell types such as at the more ubiquitously expressed optic atrophy 3 (Opa3) gene. As expected, three out of the five TIPs shown overlap with CpG islands at the promoters. Next, we wanted to test whether both the genes and putative enhancers in intergenic regions (IGRs) isolated via presence of TIPs increased the tissue-specific and -selective expression patterns. For the former, we compared the expression pattern of promoters isolated by the presence of H3K4me1/H3K4me3/Ser5P with those displaying TIPs. Across all four cell types, we observed an increased tissue-specificity as exemplified by the higher ratio between expression levels of our selected genes and all remaining ones within each tissue or group of tissues (Fig. 2B). In terms of tissue-expression signature, the highest ranked tissue was not significantly altered with the exception of the thymocyte selection, improving the rank from three to one. We obtained similar results in the case of our putative enhancer (IGR) selection. When compared with the selection via H3K4me1/H3K4me3/Ser5P, the presence of TIPs also appears to mark more tissue-specific enhancers. This is particularly evident in the case of ESC, where the tissue rank improved dramatically. Enhancers in these cells are likely to represent a special situation due to their pluripotent nature, in that many are poised for rapid activation upon differentiation and therefore mark genes of other tissues. Finally, genes isolated by the presence of TIPs either on their promoters or putative enhancers also greatly improved the tissue-restrictive pattern, as observed from increased differential expression levels between the tissues in question and all remaining ones (Fig. 2C). The presence of TIPs both at promoters and enhancers therefore increases both their tissue-specificity as well as tissue-restrictive expression pattern.
Figure 2.
TIPs at both promoters and enhancers increase the tissue-specific and -restrictive expression pattern of associated genes. (A) Examples of cell specific and non-specific promoter TIPs in four different primary cell types. We used the enrichment (at least two peaks with a minimal enrichment threshold across an area >400 bp) of RNAP II at promoters (−2/+1 kb) to isolate TIPs (highlighted in blue). Using this strategy, we isolated tissue-specific TIPs at Satb1 in DP, Sall4 in ESC, Emr1 in macrophages, Caly in neurons as well as conserved ones such as at Opa3 (from left to right, highlighted by gray boxes). The CpG islands track (red) was obtained from the UCSC browser25 and is shown as a reference. (B) Comparison of the tissue-specific expression of genes associated to promoters and putative enhancers associated to IGRs (H3K4me1+/H3K4me3+/Ser5P or TIPs). The tissue-specificity was calculated as described in Figure 1B. The presence of TIPs increased the tissue-specificity for both promoters and putative enhancers across all four tissues. The tissue-ratio rank either remained unchanged as first or increased in the case of DP and ESC. (C) Comparison of the tissue-selective expression of genes associated to promoters and putative enhancers (H3K4me1+/H3K4me3+/Ser5P or TIPs) as described in Figure 1C. The presence of TIPs either at promoters (purple) or putative enhancers (red) increased the tissue-restrictive expression pattern as compared with their isolation via H3K4me1+/H3K4me3+/Ser5P (green and orange respectively).
Discussion
The systematic and genome-wide isolation of enhancers based on epigenetic profiling has received much attention in recent years and was primarily based on early discoveries showing the differential deposition of the H3K4me3 mark between enhancers and promoters.3 As mentioned before, however, recent high-resolution genome-wide location studies have put these results in question12 and some studies propose the presence of H3K4me3 as a hallmark of (strong) enhancer activity.13,14 Furthermore, enhancers have now been shown to recruit RNAP II in several tissues and cell types,6,7,15 including the assembly of the complete PIC in murine thymocytes.8 It is therefore a very daunting question as to what really distinguishes enhancers from promoters. We and others,15 propose that the presence of RNAP II at these cis-regulatory regions provides a better mark of their activity as compared with the previously employed chromatin combinatorial. As shown here, isolation of enhancers based on the presence of both RNAP II and H3K4me3 generally provides both a more tissue-specific and tissue-selective expression signature of associated genes as compared with HAT and H3K4me1 only. The fact that many more enhancers were isolated in neurons and macrophages in stimulated conditions raises the possibility of discriminating induced from steady-state enhancers. We do not claim that the enhancers isolated in the presented studies are false-positives, but might instead represent a population that has been transiently activated and therefore lacks the deposition of further methyl groups on H3K4. Very strong, steady-state and active enhancers would instead carry this mark. Finally, we propose that isolating both promoters and enhancers based on the presence of TIPs results in a more tissue-specific and -selective gene expression pattern. They often overlap CpG islands, though to a lesser extent at intergenic regions.8 It would be interesting to investigate whether these genomic regions have a direct effect on PIC assembly. This could be mediated either via increased recruitment due to an overabundance of TFBS or by a thermodynamic environment that creates nucleosome depleted regions (NDRs) allowing for increased PIC recruitment.20 The observation that TIPs at putative enhancers also increases tissue-specificity, argues for the fact that the levels of RNAP II at these regions might be correlated with their activity. It will certainly be also of importance to further investigate the biological meaning and consequences of enhancer transcription. This phenomenon could be associated to the creation of a permissive open chromatin conformation. Alternatively, eRNAs could themselves exert enhancer functions directly21 or by binding to histone modifiers like many large intergenic non-coding RNAs (lincRNAs) 22 or Polycomb-associated RNAs.23 Finally, it was recently shown that paused RNAP II can act as an insulator24 in Drosophila, indicating its diverse roles in transcription regulation not only at the promoter or across gene bodies, but also indirectly via association to cis-regulatory elements. Overall, we propose the use of RNAP II recruitment as a means to isolate active and tissue-specific enhancers, and that this tissue-specificity both at promoters and enhancers is further increased via the presence of TIPs.
Acknowledgements
F.K. was supported by grants from Marie Curie Research Training Network “Chromatin plasticity” (MRTN-CT-2006-0357733), Association pour la Recherche sur le Cancer (ARC) and Agence Nationale de la Recherche (ANR “chromaTin”). We are thankful to the members of the P.F. laboratory for helpful discussions.
Abbreviations
- RNAP II
RNA polymerase II
- GTF
general transcription factor
- TF
transcription factor
- PIC
pre-initiation complex
- TFBS
transcription factor binding site
- TIP
transcription initiation platform
- HAT
histone acetyltransferase
- CTD
carboxy-terminal domain
- Ser5P
phosphorylation at serine5 of the CTD
- eRNA
enhancer RNA
- IGR
intergenic region
- TSS
transcription start site
References
- 1.Koch F, Jourquin F, Ferrier P, Andrau JC. Genome-wide RNA polymerase II: not genes only! Trends Biochem Sci. 2008;33:265–273. doi: 10.1016/j.tibs.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 2.Bulger M, Groudine M. Functional and mechanistic diversity of distal transcription enhancers. Cell. 2011;144:327–339. doi: 10.1016/j.cell.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 4.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 6.Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, et al. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8:1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koch F, Fenouil R, Gut M, Cauchy P, Albert TK, Zacarias-Cabeza J, et al. Transcription initiation platforms and GTF recruitment at tissue-specific enhancers and promoters. Nat Struct Mol Biol. 2011;18:956–963. doi: 10.1038/nsmb.2085. [DOI] [PubMed] [Google Scholar]
- 9.Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pekowska A, Benoukraf T, Zacarias-Cabeza J, Belhocine M, Koch F, Holota H, et al. H3K4 tri-methylation provides an epigenetic signature of active enhancers. EMBO J. 2011;30:4198–4210. doi: 10.1038/emboj.2011.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474:390–394. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Santa F, Narang V, Yap ZH, Tusi BK, Burgold T, Austenaa L, et al. Jmjd3 contributes to the control of gene expression in LPS-activated macrophages. EMBO J. 2009;28:3341–3352. doi: 10.1038/emboj.2009.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghisletti S, Barozzi I, Mietton F, Polletti S, De Santa F, Venturini E, et al. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity. 2010;32:317–328. doi: 10.1016/j.immuni.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 18.Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, et al. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramirez-Carrozzi VR, Braas D, Bhatt DM, Cheng CS, Hong C, Doty KR, et al. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell. 2009;138:114–128. doi: 10.1016/j.cell.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ørom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, et al. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chopra VS, Cande J, Hong JW, Levine M. Stalled Hox promoters as chromosomal boundaries. Genes Dev. 2009;23:1505–1509. doi: 10.1101/gad.1807309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, Cline MS, et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2011;39:876–882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]