Abstract
The hypomethylating agent 5-azacytidine (5AC) is widely used in patients at risk of invasive mycoses. We sought to determine whether 5AC affects the developmental competence and virulence of Aspergillus fumigatus. Incubation of A. fumigatus strain 293 with 5AC induced high-frequency conversion to a fluffy-variant (Af293FL). The conidiation defect was bypassed by exposing Af293FL to light during the initial 18 hours of growth on solid media. Transcriptional profiling revealed differential expression of multiple genes involved in G-protein signaling, including a putative G-protein coupled photoreceptor (opsin), suggesting that impaired signaling through a light-responsive pathway upstream of brlA is responsible for this phenotype. Af293FL was fully virulent in fruit fly and murine models of invasive aspergillosis. Moreover, Af293FL overexpressed aspergillopepsin F, had increased elastase activity and was more angioinvasive than the parental wild-type strain. The 5AC-induced A. fumigatus fluffy variant illustrates the potential effects of chemotherapeutic agents on the developmental and pathobiologic characteristics of opportunistic fungi.
Key words: azacytidine, aspergillus, G-protein, development, virulence
Introduction
Asexual reproduction in Aspergillus species requires a transition from multicellular, highly polarized structures (vegetative hyphae) to unicellular budding spores (conidia). This phenotypic switch is coordinated by a limited set of regulatory genes, which in turn activate hundreds of genes involved in morphogenesis. The principal regulatory genes were first characterized in the model fungus A. nidulans. Bristle (brlA) encodes a transcription factor that induces expression of the developmental regulatory genes Abacus (abaA) and wet-white conidia (wetA). Loss-of-function of brlA results in the formation of defective conidiophore-like structures (bristles) that fail to produce the specialized cell types required for conidiation.
Twenty-five years ago, Tamame et al.1,2 demonstrated that 5-azacytidine (5AC), a cytidine analog with DNA hypomethylating and mutagenic properties, can induce high-frequency conversion of A. nidulans and A. niger to a stable “fluffy” phenotypic variant that is characterized by severely impaired asexual sporulation and the formation of abundant vegetative hyphae. The authors further noted that fluffy variant colonies lacked contact inhibition and grew uncontrollably over other colonies. Complementation studies revealed that all 5AC-induced variants were defective in the same gene, leading the authors to postulate that a developmental gene is specifically targeted by 5AC at an early time point in development.1
Unlike A. nidulans, A. fumigatus is a common opportunistic pathogen that is associated with morbidity and mortality in immunocompromised patients, such as those with hematologic malignancies.3 Given the increasing use of hypomethylating agents in these patients,4 we sought to determine whether 5AC induces a developmental variant of A. fumigatus and whether this phenotype is associated with altered tissue invasiveness and pathogenicity. 5AC induced high-frequency conversion of A. fumigatus to a developmental mutant with impaired light-dependent conidiation. Whole-genome gene expression studies revealed early differential expression of an opsin-encoding gene and other genes with heterotrimeric G-protein regulatory functions [flbA (fluffy low brlA) and arrestin], followed by underexpression of brlA, suggesting that 5AC targets a light-sensing developmental pathway upstream of brlA. 5AC-induced A. fumigatus variants exhibited increased elastase activity and were fully pathogenic in fruit fly and murine model systems.
Results
5AC induces high-frequency conversion of A. fumigatus to the fluffy phenotype (Af293FL).
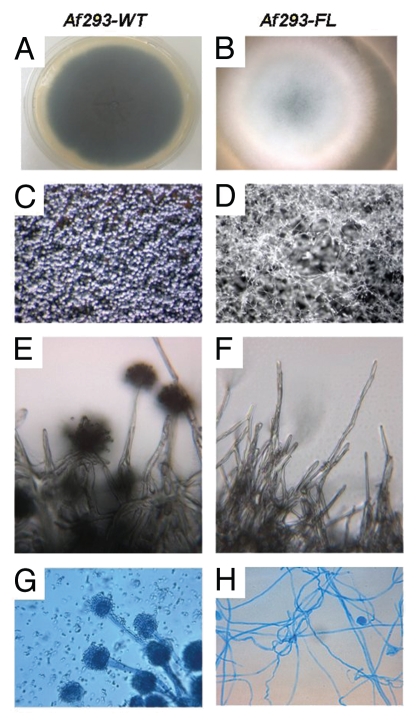
Exposure of submerged A. fumigatus cultures to 250 mM or 500 mM 5AC induced the fluffy phenotype in ∼10% of colonies. Af293FL was characterized by the complete absence of conidiation when incubated in the dark (Fig. 1). As previously described for 5AC-induced fluffy A. niger and A. nidulans variants,2 Af293FL formed extensive aerial hyphae that rose high above the surface of the solid medium. On microscopic examination, these aerial hyphae appeared as conidiophore-like structures that failed to differentiate into conidia-forming vesicles and grew indeterminately, a phenotype consistent with brlA silencing.16 Growth in proximity to a wild-type Af293 colony did not induce conversion of Af293FL to the wild-type phenotype, indicating that the fluffy variant does not result from deficiency of a diffusible factor such as FluG.17 Af293FL was similar to its parental wild-type strain with respect to its radial growth rate and susceptibility to antifungal drugs and the resistance of its conidia and hyphae to oxidative damage (data not shown).
Figure 1.
Morphologic characteristics of Af293FL. Af293FL (B, D, F and H) was compared with its parental wild-type strain (A, C, E and G). It demonstrated a lack of conidiophore formation and extensive proliferation of undifferentiated hyphae after 72 hours of incubation in the dark. Macroscopic colony appearance is shown in (A and B); appearance on dissecting microscopy (x40) is shown in (C and D); colony surface by light microscopy (x250) is shown in (E and F); and light microscopy (x250) of lactophenol cotton blue-stained tease slides is shown in (G and H).
Af293FL is a stable phenotype characterized by impaired light-dependent conidiation.
Repeated passages (>20) of Af293FL on solid medium did not abrogate the conidiation defect, indicating that this is a stable phenotype. Exposure of Af293FL cultures to light during the initial 18 hours of incubation induced conidiation equivalent to that of the wild-type strain. However, Af293FL cultures incubated in the dark for >18 hours failed to conidiate when transferred to light-exposed conditions, suggesting that light exposure is required during a critical timeframe before the acquisition of developmental competence. Light dependent conidiation was observed in all fluffy colonies tested (>10). Conidia collected from these strains invariably produced colonies exhibiting the fluffy phenotype.
Af293FL has preserved virulence in fly and murine models of aspergillosis.
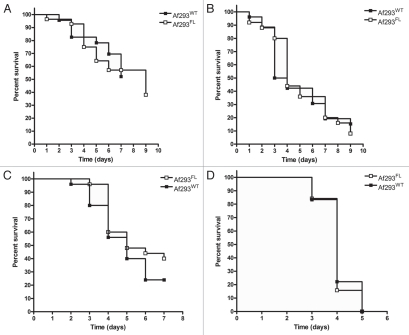
Inoculation with Af293FL conidia produced rapidly fatal infection in Tl-/- flies and neutropenic and non-neutropenic BALB/c mice. The survival rates of Af293FL-infected animals did not significantly differ from those of Af293WT-infected animals in any of the models evaluated (Fig. 2). In addition, pulmonary fungal burden was similar for Af293WT and Af293FL-infected neutropenic and non-neutropenic mice (median burden, 2.46 × 106 versus 1.64 × 106 conidial equivalents respectively; p = 0.72).
Figure 2.
Assessment of Af293FL virulence in fly and mouse models of invasive aspergillosis. The virulence of Af293FL relative to that of its parental wild-type strain was assessed in Toll-deficient flies using an inoculum suspension of 106 conidia/ml (A) or 107 conidia/ml (B), and in BALB/c mice immunosuppressed with a neutropenia-inducing drug regimen (cyclophosphamide plus cortisone acetate) (C) or a non-neutropenic regimen (cortisone acetate) (D). In each model system, survival rates were similar between animals infected with Af293FL and those infected with Af293WT (p = NS by the log-rank test).
Af293FL exhibits increased elastase activity.
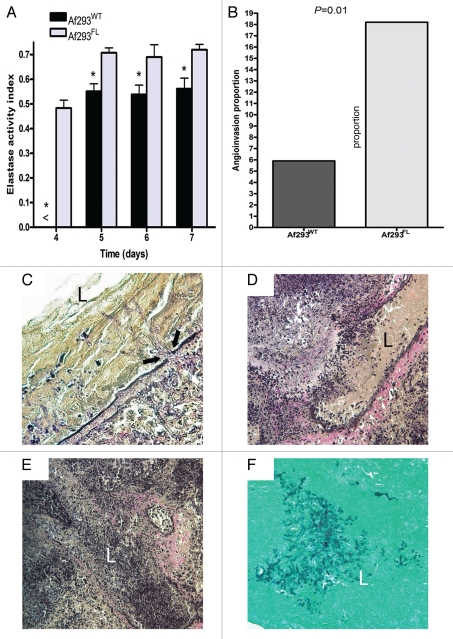
We measured the elastase activity of five independent Af293FL strains and compared it with that of Af293WT. Af293FL strains exhibited significantly higher elastase activity on elastin-Congo-red medium than did the parental wild-type strain. The mean (±standard error of the mean) elastase activity indices were 0.72 ± 0.04 for Af293FL and 0.53 ± 0.02 for Af293WT (p = 0.002; Fig. 3A). Moreover, an examination of EVG-stained lung sections from BALB/c mice infected with Af293FL revealed extensive angioinvasion and disruption of elastin fibers in the walls of invaded blood vessels (Fig. 3B). Angioinvasion was observed in 19 of 104 blood vessels (18.2%) in Af293FL-infected lungs versus 5 of 84 blood vessels (5.9%) in Af293WT-infected lungs (odds ratio, 3.5; p = 0.01 by Fisher's exact test).
Figure 3.
Elastase activity and angioinvasiveness. (A) The elastase activity indices (EAIs) of Af293FL and its parental wild-type strain were evaluated on elastin-Congo red agar according to the method described by Blanco et al.11 Values represent the means of experiments with five different Af293FL strains. No elastase activity was detected prior to day 4. *p < 0.01. (B) The proportion of blood vessels invaded by fungal hyphae was evaluated in tissue sections from mouse lungs stained with the EVG stain. Proportions were compared with Fisher's exact test. (C–E) Representative tissue sections from Af293FL-infected lungs: (C) penetration of the vascular lumen by hyphae is associated with thinning and disruption of elastin fibers (arrows mark site of penetration) (EVG x400 magnification), (D) complete disruption of vessel wall by a penetrating fungal mass (EVG x200), (E) vessel lumen packed with proliferating hyphae (EVG, x200), (F) same vessel as (E) observed in a GMS-stained specimen (x200). L, vascular lumen.
Af293FL differentially expresses opsin and G-protein regulatory genes.
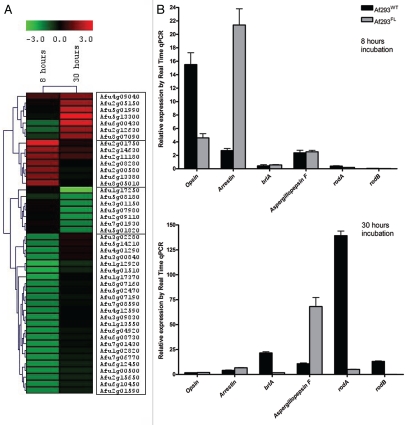
Forty-five genes were differentially expressed by a factor of 1.5 or more during submerged growth in Af293FL compared with the parental wild-type strain (Fig. 4A, Table 1). Hierarchal clustering revealed four distinct clusters among these 45 genes. The largest of these four groups included 24 genes that were underexpressed in Af293FL during precompetent growth. This cluster contained developmentally regulated genes such as heat-shock protein (HSP) Awh11,18 HSP 9/12,18 and NAD-dependent formate dehydrogenase AciA/Fdh.19 At the 30-hour time point, Af293FL underexpressed the developmentally regulated genes esdC, rodA and rodB but overexpressed genes encoding for cell wall-associated proteins AspF13, IgE-binding protein, and galactomannoprotein Mp2 as well as aspergillopepsin F, a secreted elastinolytic aspartic protease.20
Figure 4.
Whole-genome expression studies in Af293FL. Transcriptional analysis of Af293FL using cDNA microarrays revealed differential expression of four gene clusters (A); the expression of selected genes was confirmed by RT-PCR (B). After 8 hours of growth in submerged culture, Af293FL underexpressed the gene encoding opsin-related-protein as well as several developmentally regulated genes (HSP 9/12, HSP Awh11, and NAD-dependent formate dehydrogenase AciA/Fdh). The genes encoding arrestin and the RGS protein flbA were concurrently overexpressed. At the 30-hour time point, Af293FL underexpressed the key developmental regulator brlA and conidia-specific genes rodA and rodB, whereas aspergillopepsin F was overexpressed.
Table 1.
Af293FL differentially expresses opsin and G-protein regulatory genes
| Cluster | Gene name | ORF | Relative expression | |
| 8 hours | 30 hours | |||
| 1 | high affinity methionine permease | Afu4g09040 | 0.848774 | 1.513 |
| cell wall galactomannoprotein Mp2/allergen F17-like | Afu2g05150 | 0.143108 | 1.86916 | |
| BYS1 domain protein, putative | Afu5g01990 | −0.04796 | 2.342465 | |
| aspartic endopeptidase Pep1/aspergillopepsin F | Afu5g13300 | −0.13327 | 2.861234 | |
| IgE-binding protein | Afu6g00430 | −1.23993 | 2.657625 | |
| allergenic cerato-platanin Asp F13 | Afu2g12630 | −1.13578 | 1.781088 | |
| extracellular proline-serine rich protein | Afu8g07090 | −0.65047 | 1.7238 | |
| 2 | methionine aminopeptidase, type II, putative | Afu2g01750 | 2.730707 | 0.315456 |
| cell wall glycosyl hydrolase YteR, putative | Afu2g14630 | 1.760417 | 0.661531 | |
| developmental regulator FlbA | Afu2g11180 | 1.571861 | 0.837913 | |
| NADP-dependent malic enzyme MaeA | Afu2g08280 | 1.581181 | 0.007476 | |
| conserved hypothetical protein | Afu2g00580 | 1.584199 | −0.15622 | |
| arrestin (or S-antigen), N-terminal domain protein | Afu6g13380 | 1.538449 | −0.13847 | |
| C2H2 finger domain protein, putative | Afu8g05010 | 1.800728 | 0.011303 | |
| 3 | conidial hydrophobin RodB | Afu1g17250 | 0.112375 | −3.19989 |
| cell wall protein, putative | Afu5g08180 | −0.53011 | −1.66332 | |
| GPI anchored cell wall protein, putative | Afu3g01150 | 0.144695 | −1.66902 | |
| hypothetical protein | Afu5g07980 | 0.095115 | −1.77793 | |
| phenylalanine ammonia-lyase | Afu2g09110 | −0.05489 | −1.73881 | |
| GTP-binding protein EsdC | Afu7g01930 | 0.143474 | −1.5393 | |
| DUF221 domain protein, putative | Afu5g01820 | 0.042208 | −1.53861 | |
| 4 | alpha,alpha-trehalose glucohydrolase TreA/Ath1 | Afu3g02280 | −1.52994 | 0.386307 |
| glucose repressible protein Grg1, putative | Afu5g14210 | −1.51353 | 0.32186 | |
| endo-chitosanase, pseudogene | Afu4g01290 | −1.67239 | 0.274825 | |
| FAD-dependent oxygenase, putative | Afu3g00840 | −1.65143 | 0.259612 | |
| glycogen phosphorylase GlpV/Gph1, putative | Afu1g12920 | −2.10915 | −0.55088 | |
| C6 transcription factor, putative | Afu4g01510 | −2.02165 | −0.39924 | |
| HSP9/12 family heat shock protein | Afu1g17370 | −1.92717 | −0.02969 | |
| conserved hypothetical protein | Afu8g07160 | −1.6317 | −0.11533 | |
| thiamine biosynthesis protein (Nmt1), putative | Afu5g02470 | −1.60505 | −0.06664 | |
| conserved hypothetical protein | Afu8g07190 | −1.55362 | −0.15525 | |
| hypothetical protein | Afu7g08590 | −1.51399 | −0.09198 | |
| conserved hypothetical protein | Afu4g12590 | −1.61718 | 0.025716 | |
| conserved hypothetical protein | Afu3g09830 | −1.71358 | 0.020823 | |
| conserved hypothetical protein | Afu1g13550 | −1.70911 | −0.05334 | |
| NAD-dependent formate dehydrogenase AciA/Fdh | Afu6g04920 | −1.74368 | −0.15843 | |
| 6-phosphogluconate dehydrogenase, decarboxylating | Afu6g08730 | −1.70504 | −0.15849 | |
| opsin, putative | Afu7g01430 | −1.75658 | −0.11454 | |
| NADH-quinone oxidoreductase Pst2, putative | Afu1g02820 | −1.76404 | −0.22796 | |
| conserved hypothetical protein | Afu7g06770 | −1.72165 | −0.27497 | |
| chaperone/heat shock protein Awh11 | Afu6g12450 | −1.7315 | −0.3533 | |
| FMN dependent dehydrogenase, putative | Afu1g00500 | −1.70418 | −0.43737 | |
| N-methyltransferase, putative | Afu2g15650 | −1.57065 | −0.37734 | |
| conserved hypothetical protein | Afu6g10450 | −1.6392 | −0.36357 | |
| non-classical export protein Nce102, putative | Afu2g01590 | −1.59545 | −0.31464 | |
ORF, open reading frame.
Interestingly, two genes that are homologous to members of the rhodopsin light-responsive complex were differentially expressed in Af293FL in the precompetent phase. AFUA_7G01430, which encodes a predicted seven-helix transmembrane opsin-related protein,21 was underexpressed in Af293FL. The arrestin gene homolog AFUA_6G13380, which encodes an opsin regulator in vertebrates that blocks signal transduction by preventing the interaction between cytoplasmic domains and heterotrimeric G-proteins, was upregulated in Af293FL. In addition, the flbA gene (AFUA_2G11180), which encodes a regulator of G-protein signaling (RGS) protein that activates conidiation, was concurrently upregulated in Af293FL. brlA, whose expression is positively regulated by flbA,22 was underexpressed by Af293FL at the 30-hour time point. The significant differential expression of opsin, arrestin, brlA, aspergillopepsin F, rodA and rodB genes in Af293FL was confirmed with the use of RT-qPCR analysis (Fig. 4B).
Discussion
In this study, we determined the phenotypic characteristics and differential gene expression patterns of 5AC-induced A. fumigatus developmental mutants (Af293FL). A striking feature of Af293FL is its requirement for light exposure to achieve developmental competence, indicating a defect in a light-responsive signaling pathway. Af293FL was fully virulent in two disparate model host systems; moreover, this strain overexpressed the elastinolytic protease aspergillopepsin F, a putative virulence factor,20 and displayed enhanced elastase activity and angioinvasiveness.
In the 5AC-induced A. fumigatus strain Af293FL, we observed early differential expression of multiple genes encoding for proteins with known or predicted roles in G-protein signaling, including a putative G-protein-coupled photoreceptor (opsin), arrestin, flbA, and at a later time point, brlA and esdC, both developmentally important genes regulated by G-protein signaling. This pattern of gene expression suggests that altered G-protein signaling is responsible for the Af293FL phenotype.
G-protein signaling plays a crucial role in regulating development and growth in Aspergillus spp.22–24 and is required for vegetative growth.23,24 Inhibition of G-protein signaling, such as is mediated by the RGS protein flbA, results in upregulation of brlA, which leads to asexual sporulation, and esdC, which initiates sexual sporulation.23 Indeed, both inactivating mutations of flbA and activating mutations of fadA (encoding the Gα subunit) result in fluffy strains with uncontrolled vegetative growth and absent sexual and asexual sporulation.23,24
Opsins are photoreceptor proteins that use retinal as a chromophore; they are found in all major taxa and are involved in various light-driven processes.25 Arrestin proteins are a family of ubiquitous eukaryotic regulators of G-protein-coupled receptors, including photoreceptors.26 Thus, Af293FL differentially expresses genes that are homologous for both a seven-helix transmembrane receptor and arrestin, suggesting that the fluffy phenotype arises from impaired light-sensing through an opsin-like G-protein-coupled photoreceptor. Although intriguing, these results are preliminary and further work is required to characterize the role of opsin/G-protein signaling and its contribution to conidiation in A. fumigatus.
The failure of Af293FL to form conidiophores was coupled with exuberant proliferation of vegetative hyphae, similar to that reported by Tamame et al.1,2 for fluffy phenotypic variants of A. nidulans. Not surprisingly, the dysregulated proliferation of vegetative hyphae was accompanied by upregulation of genes that encode for cell-wall constituents, such as galactomannoprotein Mp2 and Asp F13, whereas conidia-specific genes such as the hydrophobins rodA and rodB were concurrently downregulated. Interestingly, aspergillopepsin F expression was increased in Af293FL mycelia, suggesting that the production and secretion of proteolytic enzymes is required to accommodate the expanding mycelial biomass. This notion was further supported by increased in vitro elastase activity of Af293FL compared with the parental wild-type strain. Elastase production by A. fumigatus was previously shown to be correlated with pulmonary invasion by hyphae and virulence, both in experimental models and the clinical setting.11,12 These observations are consistent with Af293FL's pathogenicity in fruit flies and immunosuppressed mice and increased angioinvasiveness in mouse lungs.
In spite of the increased proteolytic activity of Af293FL, this strain was not associated with reduced survival rates in model hosts. This paradox is consistent with the results of recent studies that failed to find an association between secreted protease production and A. fumigatus virulence.27,28 It is possible that the increased proteolytic activity seen in vitro does not reflect protease secretion and activity during infection. Protease inhibitors present within host lungs might attenuate the proteolytic activity of secreted A. fumigatus enzymes.29 Furthermore, given the multitude of proteolytic enzymes produced by A. fumigatus, the relative importance of aspergillopepsin F may be negligible. Alternatively, the animal models employed in the present study might have been insensitive for the detection of increments in virulence. Use of smaller A. fumigatus inoculums, resulting in a more subacute infection, might have uncovered subtle differences in virulence associated with the fluffy phenotype.
In summary, we demonstrated an interaction between a chemotherapeutic agent (5AC) and the pathogenic mold A. fumigatus that resulted in a distinct developmental variant. Whether such interactions have clinical relevance is unclear as the concentrations of 5AC required to induce the fluffy phenotype far exceed those achieved in patient's tissues during treatment.30 Of note, in addition to its use as an antineoplastic drug, 5AC has been used in recent years in epigenetic manipulations of crop plants.31 Moreover, an oral formulation of 5AC is currently being explored in preliminary clinical trials.32 Such applications could unintentionally expose saprophytic or mucosal colonizing molds to high concentrations of 5AC. Af293FL characteristics, such as increased protease secretion and upregulation of allergenic cell-wall components, indicate that 5AC-A. fumigatus interactions could have potentially important consequences for human health. Additionally, although Af293FL is a fully virulent A. fumigatus strain, its failure to conidiate at standard incubation conditions may delay its identification using standard microbiologic methods and may lead to it being considered a harmless saprophyte. The pleiotropic effects of chemotherapeutic agents on fungal biology add another level of complexity to the pathogenesis of invasive aspergillosis in immunocompromised hosts.
Materials and Methods
5-Azacytidine.
5-Azacytidine (Vidaza, Pharmion Corp., Boulder, CO) was reconstituted from 100-mg vials in 4 mL of sterile water and mixed by gentle rolling to yield a 25 mg/mL stock solution.
Induction of A. fumigatus developmental mutants.
A. fumigatus (strain Af293) was grown on yeast-extract agar glucose (YAG) plates for 72 hours. Conidia were collected in sterile 0.9% saline with 0.08% Tween-20 and resuspended at 105 cells/mL in liquid minimal medium (2% [v/v] 50x salts, 2% [w/v] glucose, 1.2% 1 M KPO4 [pH 6.8], and 0.1% trace elements) containing different 5AC concentrations (0 µM, 250 µM and 500 µM). Conidial suspensions were incubated at 37°C with shaking for 72 hours. Next, the mycelia were transferred onto glass microfiber filters (Whatman, Piscataway, NJ). Filters were placed on a monolayer of sterile glass beads immersed in liquid YAG and maintained in a humidified 37°C incubator until conidiation (48 to 72 hours). Conidia from several regions of each filter disc were collected with a swab, diluted in 100-µL saline, and spread on YAG plates. Plates were incubated at 37°C for an additional 48 hours. Colonies that exhibited fluffy morphologic characteristics (i.e., markedly reduced or absent conidiation and abundant formation of aerial hyphae) were counted and collected. In subsequent experiments, conidiation of Af293FL was induced when necessary by light exposure during the initial 18 hours of growth on YAG. To determine the stability of the fluffy phenotype, Af293FL was passaged repeatedly on solid medium. In each passage, conidia were collected by flooding the surface of the mycelium with 5 ml of sterile saline and lightly scraping with a Drigalsky spreader. The conidial suspension was aspirated, and conidia were washed three times in sterile normal saline by centrifugation. The resulting suspension was again plated on YAG.
Growth rate.
The radial growth rate was determined by spotting ∼1,000 conidia in 5 µL of saline in the center of a minimal medium agar plate. Plates were incubated at 37°C, and the colony diameter was measured daily. The radial growth rate, expressed as the slope of the colony diameter over time, was compared between the parental wild-type Af293 (Af293WT) and fluffy strains (Af293FL).
Analysis of fungal damage by exogenous H2O2.
Because resistance to oxidative stress is a virulence determinant of A. fumigatus,5 we compared the resistance of Af293FL and Af293WT to H2O2. Conidial susceptibility to oxidative damage was measured using the method described by Eisendle et al.6 Conidia (105/mL) were incubated with H2O2 at concentrations of 50 mM to 1.0 mM at 30°C for 30 minutes, with shaking. Conidial viability was determined by spreading 100 µL of the 50-fold diluted suspension on YAG and counting colonies after 18 hours of incubation at 37°C. Hyphal susceptibility to H2O2 was measured by spotting ∼1,000 conidia onto YAG with 4 mM H2O2 and peroxide-free YAG plates and measuring the growth rate at 37°C, as described above.
Antifungal drug susceptibility testing.
We determined the susceptibility of A. fumigatus strains to the antifungal drugs amphotericin B, voriconazole, caspofungin and nikkomycin Z using the broth dilution method, as outlined in document M38-A2 of the Clinical and Laboratory Standards Institute.7
Fruit fly model of invasive aspergillosis.
Toll-deficient (Tl-/-) Drosophila melanogaster has previously been shown to be a useful model host for assessing A. fumigatus virulence.8 To compare the virulence of Af293FL and Af293WT in this model, we infected Tl-/- flies with conidial suspensions of each strain and compared their survival rates. Two-four day-old female Tl-/- flies (25 flies per group) were inoculated with A. fumigatus by insertion of a needle (0.25 mm diameter) dipped in a suspension of freshly collected conidia. Experiments were performed in triplicate for each of two conidial inoculums, 106 and 107 cells/mL. After inoculation, the flies were housed at 29°C to maximize expression of the Tlr632 phenotype9 and transferred into fresh vials every three days. Fly survival was assessed daily over seven days. Flies that died within 3 hours after inoculation were excluded from the survival analysis.
Mouse model of invasive pulmonary aspergillosis.
To confirm our observations in fruit flies in a mammalian model host, we further assessed the virulence of Af293FL relative to Af293WT in immunosuppressed mice. Eight-week-old female BALB/c mice (weight, 18 to 20 gr, National Cancer Institute) were immunosuppressed with cortisone acetate alone (non-neutropenic model) or cortisone acetate in combination with cyclophosphamide (neutropenic model). In the non-neutropenic model, cortisone acetate (250 mg/kg, subcutaneously) was given 4 days and 1 day prior to inoculation and 2 days and 5 days after inoculation. In the neutropenic model, cyclophosphamide (100 mg/kg) was injected intraperitoneally 4 days and 1 day prior to inoculation, and a single cortisone acetate dose (250 mg/kg) was given 1 day prior to inoculation. Additional cyclophosphamide (100 mg/kg, intraperitoneally) was injected 2 days after inoculation to maintain neutropenia. Inoculation was performed under isoflurane-induced anesthesia by intranasal instillation of a 35-µL droplet containing 1.75 × 106 conidia. After inoculation, the animals were monitored for 7 days; any animal determined to be in a morbid state was euthanized by CO2-induced asphyxiation, and death was recorded as occurring 12 hours later. All procedures were performed according to the highest standards for humane handling, care, and treatment of research animals and were approved by the MD Anderson Cancer Center Institutional Animal Care and Use Committee.
Pulmonary fungal burden was determined in the lungs of mice using real-time-qPCR (RT-qPCR) as previously described.10 In brief, DNA samples isolated from homogenized lungs were assayed in duplicate using an ABI PRISM 7000 sequence detection system (Applied Biosystems) with primers and a dually labeled fluorescent hybridization probe specific for the Aspergillus 18S rRNA gene (Table 1).10 The cycle threshold of each sample was interpolated from a seven-point standard curve of cycle threshold values prepared by spiking uninfected mouse lungs with known amounts of conidia (101 to 107) from A. fumigatus 293. The results are reported as conidial equivalents of A. fumigatus DNA. Representative lung specimens from each infection group were fixed in 10% formalin and embedded in paraffin wax. Matched sections were stained with Grocott-Gomori methenamine-silver to visualize fungal elements and the elastic Van Gieson (EVG) stain, which enhances elastin and collagen. Angioinvasion, defined as the proportion of vessel lumens containing hyphae, was assessed in a blinded fashion in randomly chosen fields of each lung specimen. At least 80 vessels were evaluated per experimental group.
Elastase activity assay.
Elastase activity has been shown to correlate with the invasiveness of A. fumigatus isolates.11,12 We measured elastase activity in a solid medium containing 0.1% elastin-Congo-red (Sigma, St. Louis, MO), 0.05% yeast carbon base, 1.5% agar, and 0.05 M borate buffer, pH 7.6, as described by Kothary et al.12 Plates were inoculated centrally with ∼1,000 conidia and incubated at 37°C for 7 days. The colony diameters and elastase lysis zones were measured daily, and the elastase activity index was calculated as [elastase lysis diameter]/[growth diameter].11
Whole-genome gene expression analysis.
To analyze differential gene expression in the 5AC-induced A. fumigatus fluffy variant, we used A. fumigatus strain Af293 amplicon microarrays containing 9,516 genes.13 A. fumigatus conidia (5 × 107 cells/mL) were incubated in liquid minimal medium in the dark at 37°C, with shaking. Mycelia were collected by filtration through four layers of miracloth (EMD Chemicals, Gibbstown, NJ) at two time points: predevelopmental competence (8 hours incubation) and postdevelopmental competence (30 hours incubation). Gene expression for Af293FL was determined at each time point with a corresponding parental wild-type strain as the reference. RNA was extracted from Af293FL mycelia in three replicate experiments using TRIzol (Invitrogen) and pooled and hybridized with RNA extracted in parallel from Af293WT. cDNA microarrays were performed as described by the J. Craig Venter Institute Standard Operating Procedures (pfgrc.jcvi.org/index.php/microarray/protocols.html). In brief, cDNA was generated from 2 µg of total RNA by incubating for 18 hours at 42°C with Superscript II reverse transcriptase (Invitrogen), a dNTP mixture containing amino-allyl dTTP and primed with random hexamers. Unincorporated dNTPs and hexamers were removed by purification with a Qiagen polymerase chain reaction (PCR) purification column. Samples were then incubated for 18 hours at ambient temperature with Cy-3 or Cy-5 dye. After labeling, unincorporated dye was removed by purification with a Qiagen PCR purification column. Dye-coupled cDNA probes were evaporated and resuspended in hybridization buffer. Each sample was hybridized with a flip-dye replicate to account for labeling bias. Resuspended probes were mixed and spotted on the array for an 18-hour incubation. After postincubation washes, slides were scanned with a Genepix 6000B scanner and analyzed with Spotfinder.14 Exported fluorescence intensities were normalized in MIDAS14 using LOWESS, followed by flip-dye and in-slide replicate analyses. Heat maps and hierarchical expression clustering were performed using the MeV software program version 4.3.15
Real-time quantitative PCR.
cDNA was generated from 1 µg of total RNA using the Taqman reverse transcription kit (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions in a total volume of 100 µL. Twenty-microliter quantitative (qPCR) reactions were set up using 1 µL of cDNA, 10 µL of 2x SYBR green qPCR master mix (Applied Biosystems), 2 µL of 5 M Betaine (Sigma), and 4 pmol of each primer (Table 1). qPCR reactions were performed in 96-well plates in an ABI PRISM 7900HT Fast Real-Time thermocycler. The program used an initial 10-minute incubation at 95°C, followed by 40 cycles of 15 seconds at 95°C and 1 minute at 62°C. Primer specificity was verified using a denaturation step after the last amplification cycle. CT values were collected with a manual threshold of 0.2. Each target was tested in quadruplicate, and the mean of the four CT values was used for analysis. The mean CT was converted to an approximate transcript number by the equation n = 2(40-Ct). These values were then standardized to the determined value for actA and are reported as the number of target transcripts per actA transcript.
Statistical analysis.
Survival rates in animal models were calculated using the Kaplan-Meier method; the survival curves of Af293FL- and Af293WT-infected animals were compared using the log-rank test. Pulmonary fungal burdens were compared using the Mann-Whitney test. The elastase activity indexes of different A. fumigatus strains were compared using the unpaired Student's t-test. The angioinvasion scores (the proportion of blood vessels with fungal hyphae in their lumens) of different specimens were compared with Fisher's exact test. Gene expression by RT-qPCR, expressed as folds of actA transcript, was compared between strains using the unpaired Student's t-test. A two-tailed p value <0.05 was considered statistically significant. Calculations were performed using InStat and Prism 4.0 (GraphPad, San Diego, CA) software.
Acknowledgements
We thank Kim Nguyen for technical assistance and Ann Sutton for editorial assistance.
Footnotes
Previously published online: www.landesbioscience.com/journals/virulence/article/11750
Financial disclosure
This work was supported in part by the University of Texas M.D. Anderson Faculty E.N. Cobb Scholar award Research Endowment and the MD Anderson Cancer Center Core Grant (CA16672) from the University of Texas (to D.P.K.).
References
- 1.Tamame M, Antequera F, Santos E. Developmental characterization and chromosomal mapping of the 5-azacytidine-sensitive fluF locus of Aspergillus nidulans. Mol Cell Biol. 1988;8:3043–3050. doi: 10.1128/mcb.8.8.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tamame M, Antequera F, Villanueva JR, Santos T. High-frequency conversion to a “fluffy” developmental phenotype in Aspergillus spp. by 5-azacytidine treatment: evidence for involvement of a single nuclear gene. Mol Cell Biol. 1983;3:2287–2297. doi: 10.1128/mcb.3.12.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Segal BH. Aspergillosis. N Engl J Med. 2009;360:1870–1884. doi: 10.1056/NEJMra0808853. [DOI] [PubMed] [Google Scholar]
- 4.Garcia-Manero G. Demethylating agents in myeloid malignancies. Curr Opin Oncol. 2008;20:705–710. doi: 10.1097/CCO.0b013e328313699c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paris S, Wysong D, Debeaupuis JP, et al. Catalases of Aspergillus fumigatus. Infect Immun. 2003;71:3551–3562. doi: 10.1128/IAI.71.6.3551-3562.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eisendle M, Schrettl M, Kragl C, Muller D, Illmer P, Haas H. The intracellular siderophore ferricrocin is involved in iron storage, oxidative-stress resistance, germination and sexual development in Aspergillus nidulans. Eukaryot Cell. 2006;5:1596–1603. doi: 10.1128/EC.00057-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clinical and Laboratory Standards Institute, author. Reference method for broth dilution antifungal susceptibility testing of filamentous fungi Approved standard M38-A2. Wayne, PA: Clinical and Laboratory Standards Institute; 2008. [Google Scholar]
- 8.Lionakis MS, Lewis RE, May GS, et al. Toll-deficient Drosophila flies as a fast, high-throughput model for the study of antifungal drug efficacy against invasive aspergillosis and Aspergillus virulence. J Infect Dis. 2005;191:1188–1195. doi: 10.1086/428587. [DOI] [PubMed] [Google Scholar]
- 9.Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86:973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 10.Bowman JC, Abruzzo GK, Anderson JW, et al. Quantitative PCR assay to measure Aspergillus fumigatus burden in a murine model of disseminated aspergillosis: demonstration of efficacy of caspofungin acetate. Antimicrob Agents Chemother. 2001;45:3474–3481. doi: 10.1128/AAC.45.12.3474-3481.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blanco JL, Hontecillas R, Bouza E, et al. Correlation between the elastase activity index and invasiveness of clinical isolates of Aspergillus fumigatus. J Clin Microbiol. 2002;40:1811–1813. doi: 10.1128/JCM.40.5.1811-1813.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kothary MH, Chase T, Jr, Macmillan JD. Correlation of elastase production by some strains of Aspergillus fumigatus with ability to cause pulmonary invasive aspergillosis in mice. Infect Immun. 1984;43:320–325. doi: 10.1128/iai.43.1.320-325.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nierman WC, Pain A, Anderson MJ, et al. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature. 2005;438:1151–1156. doi: 10.1038/nature04332. [DOI] [PubMed] [Google Scholar]
- 14.Saeed AI, Sharov V, White J, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 15.Saeed AI, Bhagabati NK, Braisted JC, et al. TM4 microarray software suite. Methods Enzymol. 2006;411:134–193. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- 16.Yu JH, Mah JH, Seo JA. Growth and developmental control in the model and pathogenic aspergilli. Eukaryot Cell. 2006;5:1577–1584. doi: 10.1128/EC.00193-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee BN, Adams TH. The Aspergillus nidulans fluG gene is required for production of an extracellular developmental signal and is related to prokaryotic glutamine synthetase I. Genes Dev. 1994;8:641–651. doi: 10.1101/gad.8.6.641. [DOI] [PubMed] [Google Scholar]
- 18.Dutton JR, Johns S, Miller BL. StuAp is a sequence-specific transcription factor that regulates developmental complexity in Aspergillus nidulans. EMBO J. 1997;16:5710–5721. doi: 10.1093/emboj/16.18.5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chow CM, RajBhandary UL. Developmental regulation of the gene for formate dehydrogenase in Neurospora crassa. J Bacteriol. 1993;175:3703–3709. doi: 10.1128/jb.175.12.3703-3709.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee JD, Kolattukudy PE. Molecular cloning of the cDNA and gene for an elastinolytic aspartic proteinase from Aspergillus fumigatus and evidence of its secretion by the fungus during invasion of the host lung. Infect Immun. 1995;63:3796–3803. doi: 10.1128/iai.63.10.3796-3803.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li L, Wright SJ, Krystofova S, Park G, Borkovich KA. Heterotrimeric G protein signaling in filamentous fungi. Annu Rev Microbiol. 2007;61:423–452. doi: 10.1146/annurev.micro.61.080706.093432. [DOI] [PubMed] [Google Scholar]
- 22.Lee BN, Adams TH. Overexpression of flbA, an early regulator of Aspergillus asexual sporulation, leads to activation of brlA and premature initiation of development. Mol Microbiol. 1994;14:323–334. doi: 10.1111/j.1365-2958.1994.tb01293.x. [DOI] [PubMed] [Google Scholar]
- 23.Rosen S, Yu JH, Adams TH. The Aspergillus nidulans sfaD gene encodes a G protein beta subunit that is required for normal growth and repression of sporulation. EMBO J. 1999;18:5592–5600. doi: 10.1093/emboj/18.20.5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu JH, Wieser J, Adams TH. The Aspergillus FlbA RGS domain protein antagonizes G protein signaling to block proliferation and allow development. EMBO J. 1996;15:5184–5190. [PMC free article] [PubMed] [Google Scholar]
- 25.Spudich JL, Yang CS, Jung KH, Spudich EN. Retinylidene proteins: structures and functions from archaea to humans. Annu Rev Cell Dev Biol. 2000;16:365–392. doi: 10.1146/annurev.cellbio.16.1.365. [DOI] [PubMed] [Google Scholar]
- 26.Gurevich EV, Gurevich VV. Arrestins: ubiquitous regulators of cellular signaling pathways. Genome Biol. 2006;7:236. doi: 10.1186/gb-2006-7-9-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bergmann A, Hartmann T, Cairns T, Bignell EM, Krappmann S. A regulator of Aspergillus fumigatus extracellular proteolytic activity is dispensable for virulence. Infect Immun. 2009;77:4041–4050. doi: 10.1128/IAI.00425-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharon H, Hagag S, Osherov N. Transcription factor PrtT controls expression of multiple secreted proteases in the human pathogenic mold Aspergillus fumigatus. Infect Immun. 2009;77:4051–4060. doi: 10.1128/IAI.00426-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moutaouakil M, Monod M, Prevost MC, Bouchara JP, Paris S, Latge JP. Identification of the 33-kDa alkaline protease of Aspergillus fumigatus in vitro and in vivo. J Med Microbiol. 1993;39:393–399. doi: 10.1099/00222615-39-5-393. [DOI] [PubMed] [Google Scholar]
- 30.Pharmion Corporation, author. Azacitidine Investigator's Brochure. 5th ed. Boulder, CO: 2007. [Google Scholar]
- 31.Kumpatla SP, Teng W, Buchholz WG, Hall TC. Epigenetic transcriptional silencing and 5-azacytidine-mediated reactivation of a complex transgene in rice. Plant Physiol. 1997;115:361–373. doi: 10.1104/pp.115.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garcia-Manero G, Stoltz ML, Ward MR, Kantarjian H, Sharma S. A pilot pharmacokinetic study of oral azacitidine. Leukemia. 2008;22:1680–1684. doi: 10.1038/leu.2008.145. [DOI] [PubMed] [Google Scholar]