Abstract
The Ras, Raf, MEK and ERK proteins form an essential signal transduction pathway that is aberrantly activated in many human cancers. Kinase suppressor of Ras (KSR) is a conserved positive modulator of this pathway, and since its discovery, there has been a concerted effort to elucidate KSR function in both normal and aberrant Ras/ERK signaling. The KSR proteins possess a C-terminal region that is closely related to the Raf family kinase domain; however, mammalian KSR proteins lack a key catalytic residue, suggesting a role as a pseudokinase. Like many other pseudokinases, KSR has scaffolding activities and interacts with Raf, MEK and ERK to provide spatio-temporal regulation of ERK activation. Recently, significant advances have been made that further our understanding of how KSR proteins function in normal and oncogenic signaling. The newly solved KSR2/MEK1 structure has revealed important mechanistic details for how KSR regulates MEK activation and has raised questions regarding KSR kinase activity. In addition, KSR expression levels have been found to alter the effects of Raf inhibitors on oncogenic Ras/ERK signaling. Specifically, KSR1 competes with C-Raf for inhibitor-induced binding to B-Raf and in doing so attenuates the paradoxical activating effect of these drugs on ERK signaling.
Key words: KSR, Ras, B-Raf, C-Raf, Raf inhibitors, MEK, ERK cascade
The KSR Proteins: A Brief Introduction
The Ras GTPase is a critical regulator of many vital cellular processes including cell proliferation, differentiation and survival. The activation and GTP-loading of Ras is often initiated by ligand engagement of receptor tyrosine kinases on the cell surface, and an essential effector cascade functioning downstream of Ras is the ERK cascade, comprised of the Raf, MEK and ERK protein kinases.1 Once activated, ERK phosphorylates a variety of substrates throughout the cell that are needed to bring about a specific cellular response.2 The Ras/ERK signal transduction pathway also contributes to human disease, with various components of the pathway mutated in numerous human cancers and certain developmental disorders, including Noonan's and cardiofacio-cutaneous syndromes.3,4
Due to its fundamental role in cell signaling, considerable effort has been put forth in elucidating the molecular mechanisms that regulate components of the Ras/ERK pathway, in identifying additional proteins that might modulate the efficiency or level of signaling through the pathway, and in developing therapeutics to downregulate pathway signaling in the case of human disease. In particular, genetic studies conducted in the model organisms Drosophila and C. elgans have been extremely powerful at uncovering numerous modulators of the Ras/ERK pathway, one of which is Kinase suppressor of Ras (KSR).5
KSR is conserved from invertebrates to mammals and acts to positively regulate Ras/ERK signaling. The KSR proteins are most closely related to the Raf kinase family, and the C-terminal region of KSR contains many features of a protein kinase domain.6 Since its discovery, however, the question of whether KSR is a bona fide protein kinase or a pseudokinase has been a topic of debate. Nearly 10% of all proteins that contain a protein kinase domain have been classified as “pseudokinases” because they contain mutations in one or more residues required for catalytic activity.7 The mammalian KSR proteins lack the conserved arginine residue involved in phosphate transfer,6 and as is typical of many pseudokinases, demonstrate properties of a protein scaffold.8 Specifically, KSR associates with the kinase components of the ERK cascade, Raf, MEK and ERK and translocates to the cell surface in response to Ras activation. KSR interacts constitutively with MEK and is known to play an important role in co-localizing MEK with Raf at the plasma membrane.9 Recently, however, structure-function studies have revealed that some pseudokinases, such as STRADα and HER3, can serve as allosteric activators of their associated kinases in addition to their roles as scaffolds.10,11 Moreover, some pseudokinases still possess low kinase activity despite their lack of certain catalytic residues, and others, such as WNK1, have adopted compensatory mechanisms that enable them to be fully active.10,11 Thus, the mechanisms by which pseudokinases regulate signaling are more complex than originally envisioned. In the case of the Ras/ERK pathway, elucidating the full function of KSR has the added complications that Raf activation under normal conditions is intricate and poorly understood and that the extent to which KSR contributes to aberrant Ras/ERK signaling is unknown.
Although not every question regarding KSR function has been answered, significant research advances have been reported in 2011. These findings come from the reported structure of the KSR2/MEK1 complex12 and studies investigating the effect of Raf inhibitors on KSR1 function in Ras/ERK signaling.13 Here, the new insights learned from the inhibitor and structure studies will be discussed along with their potential significance to Ras/ERK signaling in human disease.
Insights from the KSR2/MEK Crystal Structure
In 2011, a major advancement in KSRology was made when the crystal structure of MEK1 in complex with the C-terminal kinase domain of the mammalian KSR2 protein (KSR2-KD) was published by Prof. David Barford's group at the Institute of Cancer Research UK.12 Not only does this elegant work provide important information regarding the structure of KSR, it also reveals how KSR and MEK interact. First and foremost, the structure shows that the C-terminal region of KSR2 assumes the conformation of a conventional protein kinase, capable of binding ATP and Mg2+. In the KSR2/MEK1 complex, the catalytic sites of MEK1 and KSR2 face one another, with binding mediated by their respective activation segments and C-lobe α-G-helices. KSR2-KD adopts an inactive kinase conformation due to the position of the α-C helix, and the activation segments of both proteins are constrained, thus precluding Raf from phosphorylating and activating MEK.
The structure studies also reveal that the KSR2/MEK1 complexes can form tetramers as a result of KSR's ability to homodimerize through conserved residues in what has been termed the “side-by-side dimer interface”.14 The dimer interface region is conserved in all KSR and Raf family members and is critical for dimerization of the Raf kinases as well as for binding between the KSR and Raf proteins, both of which occur in response to growth signaling or Raf inhibitor treatment.13,14 Although it has been known for some time that KSR plays a key role in delivering MEK to Raf at the cell surface,9 the intricacy of the interactions required for Raf to MEK signal transmission were not fully appreciated until the structures of B-Raf and the KSR2/MEK1 complex were solved.12,14,15 Structural analysis of the respective homodimer interface of KSR2 and B-Raf indicates that formation of a KSR2/B-Raf heterodimer would be accompanied by a shift in the KSR2 α-C helix to an active conformation, resulting in the release of the MEK activation segment.12,15 Interestingly, the B-Raf molecule that induces the shift in KSR2 conformation interacts with the backside of KSR2 and is not in a position to phosphorylate KSR2-bound MEK. Therefore, phosphorylation of the MEK activation segment must be accomplished by a second Raf molecule in trans.
Adding even more complexity to the process is the issue of KSR catalytic activity. Given that the KSR2 structure did not exclude the possibility that KSR proteins are functional kinases, Brennan and collaborators12 re-addressed the issue of KSR catalytic activity using in vitro kinase assays and a KSR2-KD mutant that utilizes an ATP analog, thus allowing the investigators to distinguish KSR2-catalyzed activity from that of a contaminating kinase. In these assays, weak phosphorylation of MEK1 was observed. When the phosphorylation sites were identified by mass spectrometry, the residues phosphorylated were not in the activation segment (S218 and S222), but were previously unidentified sites found in the MEK1 N-terminal region (S18, S24, S72 and T23). The stochiometry of KSR2-catalyzed phosphorylation was only 1.25%; however, the kinase activity attributed to KSR2 did increase when KSR2 was allowed to heterodimerize with B-Raf. Interestingly, a study by Hu et al.24 has also indicted that KSR1 demonstrates kinase activity toward MEK, but the sites of MEK reported to be phosphorylated differ from the sites identified by Brennan et al.12 Using immunoprecipitated KSR1 proteins in complex with inhibitor-treated C-Raf, Hu et al. found that KSR1 can weakly phosphorylate MEK on the activation segment S218 and S222 sites. However, because MEK phosphorylation was only evaluted using pS218/pS222 antibodies, it is possible that the sites identified by Brennan et al. might also be phosphorylated. Moreover, the assays conducted by Hu et al. contain C-Raf, which is a strong MEK pS218/pS222 kinase, and although Raf inhibitors were added to block C-Raf activity, weak phosphorylation of the MEK activation segment might reflect some residual C-Raf activity. Clearly, further experimentation is needed to assess the in vivo significance of KSR-mediated MEK phosphorylation and to determine whether KSR demonstrates catalytic activity toward any other substrate(s).
Insights from Raf Inhibitor Studies
Aberrant activation of the Ras/ERK pathway is a hallmark of many cancers, and there has been much interest in developing therapeutics that will inhibit pathway signaling. Moreover, the observation that the B-Raf kinase is highly mutated in a variety of human tumors16 has hastened the discovery of numerous ATP-competitive Raf kinase inhibitors.17,18 Although these inhibitors can block ERK activation by inhibiting the activity of the most prevalent and highly active V600E-B-Raf mutant, they paradoxically activate ERK by promoting dimerization of Raf family members in the presence of oncogenic or normally activated Ras.19–21 Activation of Ras, mediated by normal signaling events or by mutation, induces dimerization of the B-Raf and C-Raf kinases at the plasma membrane, which in turn stimulates Raf activity through a process that is not fully understood.22,23 In Raf-inhibitor-treated cells, binding of the drug appears to stabilize the B-Raf/C-Raf dimers at the membrane and in doing so enhances ERK activation.19,20
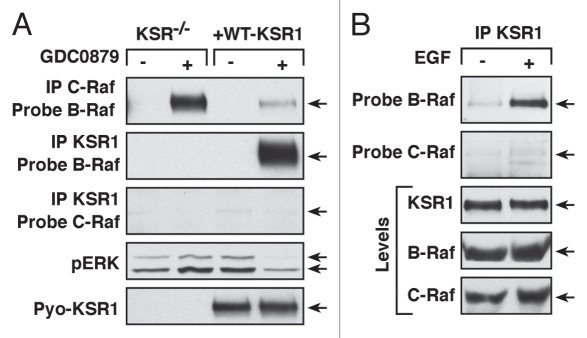
Raf dimerization is mediated by residues in the side-by-side dimer interface. Given that the dimer interface is conserved in the KSR proteins and that KSR interacts with Raf in response to Ras activation as well, it seemed plausible that Raf inhibitor treatment might also promote KSR/Raf binding. Utilizing Raf inhibitors that bind to either the active (Type I inhibitors, SB590885, L779450, PLX4720 and GDC0879) or inactive (Type II inhibitors, Sorafenib) kinase conformations of Raf, work from our laboratory found that this was indeed the case. Treatment with L779450, SB590885, GDC0879 or Sorafenib induced robust binding of KSR1 to endogenous B-Raf in cycling mouse embryonic fibroblasts (MEFs),13 (for GDC0879 effect see Fig. 1A). Another inhibitor tested, PLX4720, triggers a shift in the αC-helix of B-Raf that does not occur with the other inhibitors,19 and PLX4720 was the only Raf inhibitor that failed to promote the KSR1/B-Raf interaction.13 All the mammalian KSR proteins (KSR1, B-KSR1 and KSR2) were capable of forming inhibitor-induced complexes with B-Raf. Strikingly, however, drug treatment resulted in little to no interaction with C-Raf even when C-Raf was highly overexpressed, indicating that the inhibitors primarily promote KSR/B-Raf binding. Preferential binding between KSR1 and B-Raf does not appear to be related to the affinity of the inhibitors for B-Raf vs. C-Raf, in that Sorafenib, which has higher binding affinity for C-Raf, also promotes KSR/B-Raf complex formation and not KSR/C-Raf binding. Rather, this result seems to be consistent with an intrinsic preference for KSR to interact with B-Raf, which is also observed in growth factor-treated cells (Fig. 1B).
Figure 1.
Raf inhibitor and growth factor treatment induces KSR1/B-Raf binding. (A) KSR1 competes with C-Raf for inhibitor-induced binding to B-Raf. Cycling KSR-/- MEFs and those expressing WT-KSR1 were treated with 10 µM GDC0879 for 1 h. KSR1 or endogenous C-Raf complexes were isolated and examined for endogenous B-Raf. Lysates were analyzed for pERK and KSR levels. (B) Serum-starved HeLa cells were treated with EGF for 5 min. Endogenous KSR1 complexes were isolated and examined for endogenous B-Raf or C-Raf binding.
The Raf inhibitors are typically used in the context of oncogenic signaling; therefore, to evaluate the functional relevance of our findings, we next investigated whether inhibitor treatment induced KSR/Raf binding in cancer cell lines. In agreement with our studies in MEFs, we found that L779450 treatment induced robust binding between the endogenous KSR1 and B-Raf proteins, but little to no association of endogenous KSR1 and C-Raf in representative non-small cell lung carcinoma and melanoma lines.13 PLX4720 treatment again failed to induce complex formation between KSR1 and either the B-Raf or C-Raf proteins.13 It should be noted that in contrast to our findings (reviewed in ref. 13 and Fig. 1A), Hu et al.24 has reported that treatment with PLX4720 as well as GDC0879 induces KSR1/C-Raf binding; however, these studies were conducted under non-physiological conditions where both the C-Raf and KSR1 proteins were overexpressed in 293T cells. Co-overexpression of the KSR and Raf proteins has been shown to promote interactions that do not occur under endogenous conditions25 and may account for the different results.
Inhibitor-induced KSR1/B-Raf binding was observed in cancer lines that contained activated Ras (A549, HMCB), kinase-impaired G466A-B-Raf (Cal12T) or high activity V600E-B-Raf (A375),13 indicating that this interaction occurs in a variety of mutational backgrounds. Drug-induced binding of KSR1 to V600E B-Raf was further substantiated using a Flag-tagged V600E-B-Raf protein and provided the first indication that there might be important differences between inhibitor-induced binding of KSR1 and B-Raf vs. that of C-Raf and B-Raf. From previously published reports and our own studies characterizing Raf inhibitor-induced C-Raf/B-Raf dimerization, the following was known.13,19–21 First, Raf inhibitor treatment promotes binding of C-Raf to WT B-Raf or impaired activity B-Raf mutants, but not to the high activity V600E-B-Raf. Second, drug-induced binding is mediated by the side-by-side dimer interface, and this region must be intact in both Raf molecules. Third, the drug must bind to the ATP pocket of at least one Raf molecule within the dimer. Fourth, inhibitor-mediated C-Raf/B-Raf dimerization requires Ras binding and occurs at the plasma membrane.
Given our finding that the activity level of B-Raf had no effect on the drug-mediated interaction with KSR1, we went on to systematically compare the similarities and differences between Raf inhibitor-induced KSR/B-Raf and C-Raf/B-Raf complex formation.13 Among the similarities, we found that inhibitor-induced KSR1/B-Raf binding is mediated by the side-by-side dimer interface and could be disrupted when a conserved arginine residue was mutated to histidine in either the KSR1 (R615H) or B-Raf (R509H) interface region. Mutation of the B-Raf gatekeeper site (T529M) also prevented the drug-induced interaction with KSR1, indicating that the inhibitor must bind directly to B-Raf. Interestingly, drug treatment promoted little KSR1/KSR2 or KSR1/KSR1 interaction and mutation of the predicted gatekeeper site (T636M) in KSR1 reduced but did not eliminate B-Raf binding, suggesting that the Raf inhibitors may bind only weakly to the KSR proteins.
In sharp contrast to inhibitor-mediated C-Raf/B-Raf dimerization, drug-induced binding between KSR1 and B-Raf was found to be Ras-independent and could occur in the cytosol. More specifically, we observed that mutation of the Ras-binding region of B-Raf did not disrupt inhibitor-induced KSR1 binding, that KSR1 proteins unable to translocate to the cell surface were still competent to interact with B-Raf in drug-treated cells, and that inhibitor-induced KSR/B-Raf complexes were readily detectable in the cytoplasm of fractionated cells. Of note, these findings are also in contrast to growth factor mediated-KSR1/B-Raf binding, which like growth factor- and inhibitor-induced C-Raf/B-Raf dimerization occurs at the membrane in a Ras-dependent manner.13,25
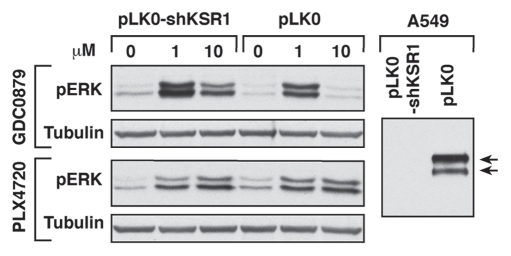
As a result of the above differences and because both KSR and C-Raf interact with B-Raf through the dimer interface, KSR1 was found to compete with C-Raf for inhibitor-induced binding to B-Raf. Utilizing MEFs that lack all KSR proteins (KSR-/- MEFs) and those reconstituted with WT-KSR1, we found that the presence of KSR1 significantly reduced complex formation between C-Raf and B-Raf when cells were treated with any drug capable of promoting the KSR1/B-Raf interaction13 (for GDC0879 effect see Fig. 1A). WT-KSR1 also blocked the inhibitor-induced membrane association of both B-Raf and C-Raf, and analysis of KSR1 mutant proteins revealed a correlation between the ability of a KSR1 mutant to bind B-Raf and the degree to which inhibitor-induced B-Raf/C-Raf binding was reduced. Most importantly, we found that the presence of KSR1 attenuated the activating effects of the inhibitors as measured by pERK levels.13 The decrease in ERK activation correlated with a decrease in inhibitor-induced Raf kinase activity and was only observed when cells were treated with drugs that promote KSR1/B-Raf binding. In agreement with previous reports that activation of C-Raf by Raf dimerization is critical for inhibitor-induced ERK activation,19–21 the presence of KSR1 severely reduced C-Raf activity in drug-treated cells. Further demonstrating the functional importance of these findings, the attenuating effect of KSR1 on inhibitor-induced ERK signaling was also observed in oncogenic cell lines expressing activated Ras (HMCB and A549). In this case, cells depleted of KSR1 exhibited higher inhibitor-induced pERK levels than were observed in control cells possessing KSR113 (GDC0789 treatment is shown in Fig. 2). This effect was not observed when cells were treated with PLX4720 (Fig. 2), which does not induce KSR/B-Raf binding, and in agreement with previous reports in references 19 and 20, was not observed in A375 cells where the inhibitors effectively block V600E-B-Raf activity.13 Together, the findings from our study indicate that KSR1 expression may impact the therapeutic effect of select Raf inhibitors. Thus, determining KSR1 protein levels will likely have important implications for the use of these inhibitors in cancer treatment.
Figure 2.
KSR1 attenuates ERK activation in GDC0879, but not PLX4720-treated cells. A549 cells expressing either the pLKO.1 vector or pLKO.1-KSR1 shRNA were treated as indicated for 1 hr. Lysates were analyzed for pERK and tubulin levels. Depletion of KSR1 is also shown.
Concluding Comments
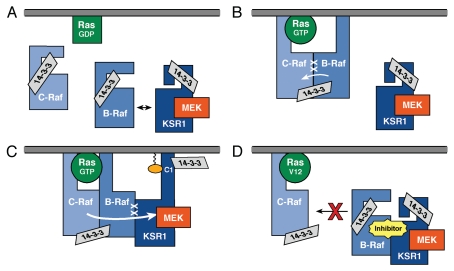
The recent KSR structure and Raf inhibitor studies have significantly expanded our knowledge of KSR function. They have further demonstrated the importance of KSR's scaffolding activity and have shown that signal transmission from Raf to MEK is a highly intricate process. In light of these new insights, a revised model for KSR function in MEK regulation is as follows (Fig. 3). Under quiescent conditions, C-Raf, B-Raf and the KSR/MEK complex are all present in the cytosol. KSR retains MEK in an inactive state by binding the MEK activation segment and preventing its access for phosphorylation. In addition, transient and unstable binding interactions appear to occur between the cytosolic KSR/MEK complex and B-Raf. Upon growth factor treatment, GTP-bound Ras recruits the Rafs to the cell surface where B-Raf may first dimerize with C-Raf and contribute to C-Raf activation. The KSR1/MEK complex is also recruited to the cell surface through a mechanism that involves the atypical C1 domain of KSR1.26 At the membrane, B-Raf then stably interacts with the KSR1/MEK complex through the dimer interface, inducing conformational changes that expose the MEK activation segment, which in turn is phosphorylated in trans by activated C-Raf. In the case of Raf inhibitor treatment, inhibitor binding stabilizes the cytosolic KSR/MEK/B-Raf complexes, thus sequestering B-Raf and antagonizing its ability to interact with C-Raf at the membrane and contribute to C-Raf activation. Notably, even though MEK is present in the cytosolic KSR/MEK/B-Raf inhibitor complex, it cannot be activated, given that B-Raf bound to KSR through the dimer interface is sterically unable to phosphorylate the MEK activation segment.
Figure 3.
Model for KSR function. (A) In a quiescent cell, C-Raf, B-Raf, and the inactive KSR/MEK complex are localized in the cytosol, with transient binding interactions occurring between B-Raf and the KSR/MEK complex. (B) Ras activation recruits the Rafs to the plasma membrane where B-Raf participates in C-Raf activation. (C) The KSR/MEK complex also localizes to the membrane and interacts with B-Raf, inducing conformational changes that expose the MEK activation segment for phosphorylation by C-Raf in trans. (D) Inhibitor-induced KSR1/B-Raf complexes can form in the cytosol, thus antagonizing the formation of C-Raf/B-Raf dimers at the membrane.
Numerous gaps in our understanding of KSR function have been filled; however, there are still outstanding questions. In particular, the door is left open as to whether kinase activity contributes to KSR's biological function. It is not uncommon for pseudokinases to retain some residual catalytic activity but determining the physiological relevance of that activity awaits the identification and validation of physiological substrates. Even in regard to KSR's scaffolding activity, it is unclear how dynamic the binding interactions at the membrane are and whether binding partners and binding affinities may change as a result of other interactions. Finally, KSR's full contribution to aberrant Ras/ERK signaling and whether targeting its scaffolding or kinase activity will be of therapeutic value awaits further investigation.
Acknowledgements
This work was funded by federal funds from the National Cancer Institute.
Abbreviations
- ATP
adenosine triphosphate
- GTP
guanosine triphosphate
- KSR
kinase suppressor of Ras
- pERK
phosphorylated ERK
- WT
wild-type
Extra View to: McKay MM, Ritt DA, Morrison DK. RAF inhibitor-induced KSR1/B-RAF binding and its effects on ERK cascade signaling. Curr Biol. 2011;21:563–568. doi: 10.1016/j.cub.2011.02.033.
References
- 1.McKay MM, Morrison DK. Integrating signals from RTKs to ERK/MAPK. Oncogene. 2007;26:3113–3121. doi: 10.1038/sj.onc.1210394. [DOI] [PubMed] [Google Scholar]
- 2.Shaul YD, Seger R. The MEK/ERK cascade: from signaling specificity to diverse functions. Biochim Biophys Acta. 2007;1773:1213–1226. doi: 10.1016/j.bbamcr.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 3.Roberts PJ, Der CJ. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene. 2007;26:3291–3310. doi: 10.1038/sj.onc.1210422. [DOI] [PubMed] [Google Scholar]
- 4.Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer. 2007;7:295–308. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- 5.Rubin GM, Chang HC, Karim F, Laverty T, Michaud NR, Morrison DK, et al. Signal transduction downstream from Ras in Drosophila. Cold Spring Harb Symp Quant Biol. 1997;62:347–352. [PubMed] [Google Scholar]
- 6.Therrien M, Chang HC, Solomon NM, Karim FD, Wassarman DA, Rubin GM. KSR, a novel protein kinase required for RAS signal transduction. Cell. 1995;83:879–888. doi: 10.1016/0092-8674(95)90204-X. [DOI] [PubMed] [Google Scholar]
- 7.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 8.Morrison DK, Davis RJ. Regulation of MAP kinase signaling modules by scaffold proteins in mammals. Annu Rev Cell Dev Biol. 2003;19:91–118. doi: 10.1146/annurev.cellbio.19.111401.091942. [DOI] [PubMed] [Google Scholar]
- 9.Müller J, Ory S, Copeland T, Piwnica-Worms H, Morrison DK. C-TAK1 regulates Ras signaling by phosphorylating the MAPK scaffold, KSR1. Mol Cell. 2001;8:983–993. doi: 10.1016/S1097-2765(01)00383-5. [DOI] [PubMed] [Google Scholar]
- 10.Boudeau J, Miranda-Saavedra D, Barton GJ, Alessi DR. Emerging roles of pseudokinases. Trends Cell Biol. 2006;16:443–452. doi: 10.1016/j.tcb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 11.Zeqiraj E, van Aalten DM. Pseudokinases-remnants of evolution or key allosteric regulators? Curr Opin Struct Biol. 2010;20:772–781. doi: 10.1016/j.sbi.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brennan DF, Dar AC, Hertz NT, Chao WC, Burlingame AL, Shokat KM, et al. A Raf-induced allosteric transition of KSR stimulates phosphorylation of MEK. Nature. 2011;472:366–369. doi: 10.1038/nature09860. [DOI] [PubMed] [Google Scholar]
- 13.McKay MM, Ritt DA, Morrison DK. RAF Inhibitor-Induced KSR1/B-RAF Binding and Its Effects on ERK Cascade Signaling. Curr Biol. 2011;21:563–568. doi: 10.1016/j.cub.2011.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rajakulendran T, Sahmi M, Lefrancois M, Sicheri F, Therrien M. A dimerization-dependent mechanism drives RAF catalytic activation. Nature. 2009;461:542–545. doi: 10.1038/nature08314. [DOI] [PubMed] [Google Scholar]
- 15.Wan PT, Garnett MJ, Roe SM, Lee S, Niculescu-Duvaz D, Good VM, et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855–867. doi: 10.1016/S0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 16.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 17.Dhomen N, Marais R. BRAF signaling and targeted therapies in melanoma. [ix.] Hematol Oncol Clin North Am. 2009;23:529–545. doi: 10.1016/j.hoc.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 18.McCubrey JA, Steelman LS, Abrams SL, Chappell WH, Russo S, Ove R, et al. Emerging Raf inhibitors. Expert Opin Emerg Drugs. 2009;14:633–648. doi: 10.1517/14728210903232633. [DOI] [PubMed] [Google Scholar]
- 19.Hatzivassiliou G, Song K, Yen I, Brandhuber BJ, Anderson DJ, Alvarado R, et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature. 2010;464:431–435. doi: 10.1038/nature08833. [DOI] [PubMed] [Google Scholar]
- 20.Heidorn SJ, Milagre C, Whittaker S, Nourry A, Niculescu-Duvas I, Dhomen N, et al. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell. 2010;140:209–221. doi: 10.1016/j.cell.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464:427–430. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rushworth LK, Hindley AD, O'Neill E, Kolch W. Regulation and role of Raf-1/B-Raf heterodimerization. Mol Cell Biol. 2006;26:2262–2272. doi: 10.1128/MCB.26.6.2262-72.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garnett MJ, Rana S, Paterson H, Barford D, Marais R. Wild-type and mutant B-RAF activate C-RAF through distinct mechanisms involving heterodimerization. Mol Cell. 2005;20:963–969. doi: 10.1016/j.molcel.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 24.Hu J, Yu H, Kornev AP, Zhao J, Filbert EL, Taylor SS, et al. Mutation that blocks ATP binding creates a pseudokinase stabilizing the scaffolding function of kinase suppressor of Ras, CRAF and BRAF. Proc Natl Acad Sci USA. 2011;108:6067–6072. doi: 10.1073/pnas.1102554108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McKay MM, Ritt DA, Morrison DK. Signaling dynamics of the KSR1 scaffold complex. Proc Natl Acad Sci USA. 2009;106:11022–11027. doi: 10.1073/pnas.0901590106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou M, Horita DA, Waugh DS, Byrd RA, Morrison DK. Solution structure and functional analysis of the cysteine-rich C1 domain of kinase suppressor of Ras (KSR) J Mol Biol. 2002;315:435–446. doi: 10.1006/jmbi.2001.5263. [DOI] [PubMed] [Google Scholar]