Abstract
Two multicentre genome-wide association (GWA) studies provided substantial evidence, implicating the complement receptor 1 gene (CR1) in Alzheimer disease (AD) genetic etiology. CR1 encodes a large transmembrane receptor with a crucial role in the immune complement cascade. We performed a genetic follow-up of the GWA CR1 association in a Flanders–Belgian cohort (n=1883), and investigated the effect of single-nucleotide polymorphisms (SNPs) located in the CR1 locus on AD risk and cerebrospinal fluid (CSF) biomarker levels. We obtained significant association (Padj<0.03; odds ratio (OR)=1.24 (95% confidence interval (CI): 1.02–1.51)) for one CR1 risk haplotype, and haplotype association was strongest in individuals carrying apolipoprotein E (APOE) ɛ4 alleles (Padj<0.006; OR=1.50 (95% CI: 1.08–2.09)). Also, four SNPs correlated with increased CSF amyloid Aβ1−42 levels, suggesting a role for the CR1 protein in Aβ metabolism. Moreover, we quantified a low-copy repeat (LCR)-associated copy number variation (CNV) in CR1, producing different CR1 isoforms, CR1-F and CR1-S, and obtained significant association in carriers of CR1-S. We replicated the CR1 CNV association finding in a French cohort (n=2003) and calculated in the combined cohorts, an OR of 1.32; 95% CI: 1.10–1.59 (P=0.0025). Our data showed that the common AD risk association may well be explained by the presence of CR1-S increasing the number of C3b/C4b and cofactor activity sites and AD risk with 30% in CR1-S carriers. How precisely the different functional role of CR1-S in the immune complement cascade contributes to AD pathogenesis will need additional functional studies.
Keywords: genetic risk, Alzheimer's disease, copy number variation, β-amyloid, biomarkers
Introduction
Two independent multicentre genome-wide association (GWA) studies, comprising over 25 000 individuals,1, 2 provided genome-wide significant evidence for Alzheimer disease (AD, MIM#104300) risk association of three novel genes, besides the apolipoprotein E gene (APOE). Apart from the clusterin gene (CLU, MIM*185430), which was identified in both the GWA cohorts, the Cardiff GWA study identified the phosphatidylinositol-binding clathrin assembly protein gene (PICALM, MIM*603025) and the French GWA study the complement receptor 1 (CR1, MIM*120620).1, 2 In CR1, the single-nucleotide polymorphism (SNP) rs6656401 showed the strongest association with an odds ratio (OR) of 1.19 (95% confidence interval (CI):1.12–1.28) and a P-value of 9.8 × 10−8.2 Further, AD risk association showed interaction with APOE genotype and was strongest in APOE ɛ4 carriers (OR=1.36 (95% CI: 1.20–1.53)).2 A second SNP, rs3818361, showed evidence for association only in the APOE ɛ4 stratum (OR=1.34 (95% CI: 1.19–1.51), P=1.5 × 10−6).
CR1, also known as CD35, immune adherence receptor or complement component (3b/4b) receptor 1, is a large type-I transmembrane glycoprotein. It is an important member of the family of regulators of complement activation and a crucial, multifunctional mediator of innate immunity. Its wide range of functions can predominantly be perceived as the result of its capacity to bind complement components C3b and C4b. CR1 functions as a major immune adherence molecule on erythrocytes, transporting complement-opsonized particles to liver and spleen for clearance from the blood. In other cells, CR1 binding to complement components induces phagocytosis of the opsonized complexes. Lastly, CR1 functions as an inhibitor of the complement cascade by exerting decay accelerating activity toward the C3/C5 convertases and/or cofactor activity for factor I-mediated cleavage of C3b/C4b.3, 4
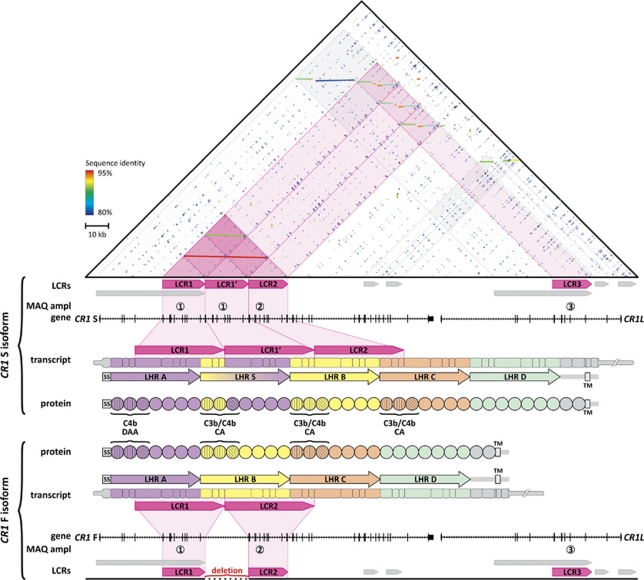
Different CR1 isoforms have been identified, varying in size by units of ∼30 kDa.3, 5, 6 The most common isoform, designated CR1-F (also CR1-A or CR1*1), has a frequency of 83%, and a molecular weight of 220 kDa, whereas the second most common isoform, CR1-S (CR1-B or CR1*2) has a frequency of 15% and a molecular weight of 250 kDa (Figure 1). In addition, two rare isoforms have also been described of 190 kDa (CR1-F′, CR1-C or CR1*3; frequency 1%) and 280 kDa (CR1-D or CR1*4; frequency <1%).3 The CR1 extracellular domain is entirely composed of tandem repeated units of a conserved domain of 60–70 amino acids, known as short consensus repeat (or complement control repeat or sushi domain).3 All short consensus repeats, except the last two, are organized into higher order repeat units composed of seven short consensus repeats, termed long homologous repeats (LHRs)3, 7 (Figure 1). Of the four LHRs (LHR-A to -D; Figure 1), all but LHR-D bind C3b and/or C4b, resulting in inhibition of complement activation through decay accelerating activity or cofactor activity.8, 9 CR1 is located at chromosome 1q32 in a regulators of complement activation gene cluster,3 and encompasses a variable number of 18-kb long low-copy repeats (LCRs)—LCR1 and LCR210, 11 (Figure 1). Both LCR1 and LCR2 are located in the CR1-coding region, and so is LCR1′, the extra LCR1-derived copy. The presence of LCR1′ explains the extra LHR, LHR-S in the long CR1-S isoform, creating additional C3b/C4b binding sites. The rare isoforms CR1-D and CR1-F′ result from either a triplication or deletion of one LCR1 copy. The third LCR, LCR3, is located in the neighboring CR1-like gene, CR1L.
Figure 1.
Genomic architecture of the CR1 locus in relation to the major CR1 isoforms. The upper triangle shows a dot plot of the self-alignment of the genomic region (5′ to 3′) encompassing the genes CR1 and CR1-like (CR1L). Dot plot colors represent sequence identity. Low-copy repeats (LCRs) are represented as horizontal arrows below the dot plot. The genomic segment is graphically annotated at different levels below the dot plot. Annotation is given for the two major CR1 isoforms CR1-S and CR1-F. LCRs: arrows indicate location and orientation of the LCRs. The LCR present at variable number of copies, resulting in the CR1 isoforms indicated with pink arrows. Multiplex amplicon quantification (MAQ) ampl: location of amplicons 1–4, used for the CR1 LCR dosage analysis. Gene: location and exon structure of CR1 and CR1L. Transcript: organization of the CR1 transcripts. Boxes represent exons with wide regions representing coding sequences and narrow regions untranslated sequences (UTRs). Arrows show the organization of the long homologous repeat regions (LHRs). The open box labeled TM denotes the region encoding the transmembrane domain. Protein: CR1 functional domains with each circle representing a separate short consensus repeat. The first three short consensus repeats in LHR A, S, B and C are required for complement factor binding and inhibitory activity (decay accelerating activity or cofactor activity), in which the first two determine factor-binding specificity and the type of inhibition, indicated by different hatching patterns.
Because of the myriad of functions ascribed to CR1, the protein and its potential role in disease has been intensively studied.4 Although biological evidence for a direct role of CR1 in AD etiology is lacking, CR1 might be involved at several levels. For instance, it has long been established that neuroinflammation and, more specifically, activation of the complement system, has an important role in pathological processes in AD brains.12, 13, 14 CR1 might be involved in amyloid β (Aβ) clearance from brain as fibrillar Aβ peptides were shown to activate the complement cascade and become covalently bound to C3b,15, 16, 17 and CR1 induces clearance and/or phagocytosis of C3b-opsonized particles. Interestingly, it has been demonstrated that in plasma, C3b mediates adherence of Aβ peptides to erythrocyte CR1, a process impaired early in the course of AD.18 Thus, CR1 may well contribute to AD by altering clearance of Aβ in the periphery (in line with the peripheral sink hypothesis stating that Aβ can be cleared from the brain by Aβ-binding molecules in the periphery). On the other hand, as CR1 is a potent regulator of the complement system, it could also participate in the disease process by inhibiting complement activation in brain, which in some studies of AD animal models had a detrimental rather than a beneficial effect.19, 20
In light of these findings, we initiated an in-depth follow-up of the GWA association finding in a Flanders–Belgian cohort of well-characterized AD patients and control individuals (n=1883). Part of this cohort had been included as replication sample in the French GWA study and showing a trend towards association with rs6656401 of OR=1.24 (95% CI: 0.99–1.24; P=5.6 × 10−2, n=1566) and rs3818361 of OR=1.05 (95% CI: 0.84–1.48; P=6.8 × 10−1, n=1408).2 Here, we examined in detail this risk association using multiple SNPs based on linkage disequilibrium (LD) and genetic variability within the CR1 locus. Also, we analyzed a potential correlation between CR1 SNPs and AD cerebrospinal fluid (CSF) biomarkers. Further, we quantified the CR1 LCRs by defining the different-sized CR1 isoforms S and F to test whether AD risk may be attributed to a specific CR1 isoform. To further strengthen our observations, we analyzed our genetic findings obtained in the Flanders–Belgian cohort in an independent French cohort.
Materials and methods
Study cohorts
All studies were approved by the medical ethics committees of the Hospital Network Antwerp (ZNA), University of Antwerp, the University Hospitals Leuven, Lille University Medical Center and Université de Lille. After written informed consent, blood samples were obtained for genomic DNA extraction, generation of EBV cell lines and plasma and serum collection.
The Flanders–Belgian AD group consisted of 1039 patients (mean onset age 74.1±9.1 years, 65.4% women) ascertained at the memory clinics of the ZNA Middelheim, Antwerpen, Belgium21, 22 and the University Hospitals Leuven, Leuven, Belgium,23 as previously described.23 Consensus diagnosis of possible or probable AD was given by at least two neurologists, based on the NINCDS/ADRDA criteria.24 The Flanders–Belgian control group consisted of 844 healthy, unrelated individuals from Belgium (mean age at inclusion 65.4±14.8 years, 57.0% women).23 The French cohort consisted of Caucasian individuals ascertained in the north of France,25, 26 and represents part of the study population used in the French GWAS.2 French AD patients (n=1393, mean onset age 69.5±8.0 years, 66.1% women) were diagnosed as probable AD according to the DSM-III-R and NINCDS/ADRDA criteria.24 The French control group consisted of n=610 healthy individuals (mean age at inclusion 72.7±8.1 years, 62.4% women). See Supplementary Methods for a detailed description of the study cohorts.
Biomarker profiling
Cerebrospinal fluid levels of β-amyloid protein1−42 (Aβ1−42), total tau (T-tau) and tau phosphorylated at threonine 181 (P-tau181P) were determined blinded and in duplicate, with commercially available single-parameter ELISA kits (Innogenetics, Gent, Belgium), following the instructions provided by the manufacturer (for details see Supplementary Methods). Biomarker profiles were available for 339 Flanders–Belgian AD patients (mean onset age 75.8±8.5 years, 63.2% women).
SNP selection and genotyping
TagSNPs (n=26) throughout the CR1 locus were selected for association analyses from the International HapMap Project (Rel24/phaseII27) using HaploView v4.2.28 Details regarding the SNP selection criteria are described in the Supplementary Methods section. SNP genotyping was performed in two multiplex assays by MassARRAY using iPLEX Gold chemistry (Sequenom, Hamburg, Germany), followed by MALDI-TOF mass spectrometry. PCR and extension primers were designed using MassARRAY Assay Design software v3.0.2.0 (Sequenom). Genotypes were called automatically using MassARRAY Typer software v4.0 (Sequenom) and were visually inspected by two researchers blinded for disease status. Samples that failed for >20% of the genotyping assays were disregarded for further analysis (n=35). The APOE genotype assay and data were previously described.29
Dosage analyses
Copy number status of the 18-kb long LCRs in CR1 and CR1L (Figure 1) was determined using multiplex amplicon quantification (MAQ) (Multiplicon SA, http://www.multiplicon.com), as described.30 Briefly, three fluorescently labeled test amplicons, targeting specifically LRCs 1–3 (Figure 1), and seven control amplicons, recognizing sequences outside the CR1 locus, were amplified in one multiplex PCR reaction and resolved on an Applied Biosystems 3730xl DNA analyzer (Applied Biosystems, Foster City, CA, USA). Peak areas of the test amplicons were normalized based on the peak areas of the control amplicons. From these normalized peak areas dosage quotients were calculated by dividing the ratios from the test individuals by that of reference individuals. Amplicon 1, designed to target LCR1, also recognizes LCR1′ because of their very high-sequence homology. Presence or absence of LCR1′ determines CR1-S (250 kDa) and CR1-F (220 kDa). Dosage analysis was performed using MAQ-S software (Multiplicon SA, Antwerpen, Belgium).
Western blotting
Cultured lymphoblast cells of patients and control individuals were lysed using NP-40 lysis buffer (50 m Tris-HCl, 150 m NaCl, 1% Nonidet P-40) supplemented with protease inhibitors (Complete protease inhibitor cocktail, Roche (Vilvoorde, Belgium)). Protein concentration was measured using the bicinchoninic acid method (Pierce BCA Protein Assay, Thermo Scientific, Erembodegem, Belgium) and 30 μg were used for electrophoresis. Protein samples were separated on a 3–8% Tris-acetate NuPage gel (Invitrogen, Merelbeke, Belgium) under reducing conditions. Resolved proteins were electroblotted onto a polyvinylidene difluoride membrane (Amersham Hybond-P PVDF Membrane, GE Healthcare, Diegem, Belgium). After blocking, membranes were immunoblotted with CD-35 (H-2) mouse monoclonal antibody (1:100, Santa Cruz Biotechnology, Heidelberg, Germany, sc-166329). Immunodetection was performed with a Horseradish peroxidase-coupled sheep anti-mouse secondary antibody (1:10 000, GE Healthcare) and Amersham ECL Plus Western Blotting Detection Reagents (GE Healthcare).
Statistical analyses
SNP associations
Deviations from Hardy–Weinberg equilibrium of genotype frequencies were determined using an exact Hardy–Weinberg equilibrium test (http://www.pharmgat.org/IIPGA2/Bioinformatics/exacthweform). Allele and genotype frequencies between AD patients and control individuals were compared using χ2-statistics in SPSS 16.0 (SPSS Inc., Chicago, IL, USA). Odds ratio (relative to the common genotype) and 95% CI were calculated, adjusted for onset/inclusion age, gender and APOE ɛ4 genotype (absence/presence of ɛ4 allele), using binary logistic regression analysis. LD between the 26 tagSNPs was calculated using Haploview v4.2, and LD blocks (n=5) were based on the solid spine of LD rule (Haploview, Supplementary Table S1). An LD plot with r2 as the LD measure was drawn using the LDheatmap v0.2.8 package.31 Haplotype analyses were performed with Haplo Stats v1.4.432 by means of haplotype tagging SNP, selected with Haploview v4.2. Using progressive insertion EM algorithms, haplotype frequencies were estimated and compared between patients and control individuals using score statistics (estimated frequency >0.01). Analyses were adjusted for onset/inclusion age, gender and APOE ɛ4 genotype. For significantly associated haplotypes, ORs and corresponding 95% CIs (corrected for age, gender and APOE ɛ4 genotype) were calculated in Haplo Stats v1.4.4. To avoid false-positive results due to multiple testing, we computed simulated P-values (1000 random permutations of patient and control labels).
Effect of SNP genotypes on biomarker profiles of patients was assessed using univariate analysis of variance analyses, adjusted for age and disease duration at lumbar puncture and gender, using SPSS. To assess the effect on CSF Aβ1−42 levels, APOE ɛ4 genotype was included as a covariate in the model, as this correlated significantly with Aβ1−42 levels. T-tau and P-tau181P levels were log10 transformed to approximate normality before analysis of variance.
Copy number variation associations
Association analyses of the CR1 LCR1 with disease status were performed using CNVassoc,33 and normalized signal intensities or dosage quotients obtained from MAQ analysis. Association analyses were performed through logistic regression (adjusted for age, gender and APOE ɛ4 genotype) taking into account copy number variation (CNV) calling uncertainty in a latent class model. To assess the effect of the different CNV classes we used a multiplicative model.
Conditional logistic regression was used to assess independence of the SNP and CNV associations. LD between the LCR1 CNV and the 26 tagSNPs in the CR1 locus was estimated by recoding CNV status into a biallelic polymorphism (two copies=A/A, three copies=A/B, four copies=B/B; introducing a slight bias as the allelic status of individuals with four copies cannot unambiguously be determined) and estimating LD (Haploview).
Meta-analyses
Pooled OR for rs4844610 and the LCR1 CNV in the Flanders–Belgian and French cohorts were calculated by random-effects meta-analysis (Der Simonian-Laird), based on the effect estimates obtained in the individual cohorts (adjusted for age, gender and APOE ɛ4 genotype). Analyses were performed and forest plots generated using the package rmeta in R (http://cran.r-project.org/web/packages/rmeta/index.html).
Results
CR1 SNP association in Flanders–Belgian cohort
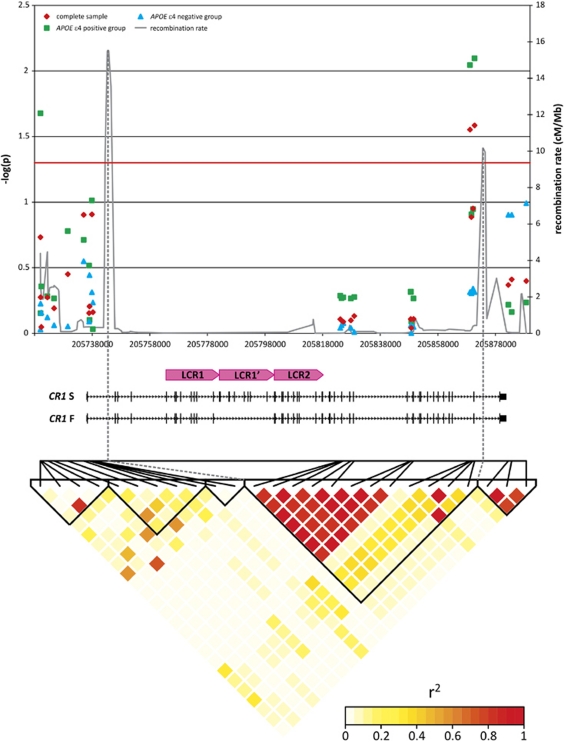
We genotyped 26 tagSNPs located across the CR1 locus. The selected SNPs had a minor allele frequency (MAF)>5%, were located outside the CR1 LCRs or other repeat sequences, were in Hardy–Weinberg equilibrium in the control group and captured 87% of the genetic variability within the locus (Supplementary Table S1). SNPs rs4844610 and rs1408077 showed significant allelic association with AD risk (Padj<0.03) (Figure 2, Supplementary Table S2). LD was very high between the two associated SNPs (r2=0.97), as well as between the two associated SNPs in our study and the two SNPs rs6656401 and rs3818361 (0.87<r2<0.90), that were associated with AD in the French GWA study.2 Our SNP selection criteria did not include the two French GWA SNPs as they are located in CR1 LCRs or repeat sequences. Stratification for APOE ɛ4 genotype indicated that the AD risk association was predominantly observed in APOE ɛ4 carriers (allelic association Padj<0.01) (Figure 2, Supplementary Table S2).
Figure 2.
LD structure of the CR1 locus and single single-nucleotide polymorphism (SNP) allelic association in AD. Allelic associations corrected for age (onset/inclusion), gender and APOE ɛ4 genotype (for analyses not stratified for APOE ɛ4 genotype) are plotted as −log10(P). SNPs are placed in the graph according to their position in CR1 shown below the graph. The lower panel of the figure shows the pairwise linkage disequilibrium pattern between SNPs as measured by their r2 values.
On the basis of LD structure in the CR1 locus, the 26 tagSNPs could be assigned to five LD blocks (Supplementary Table S1, Figure 2). Haplotype association analyses using haplotype tagging SNPs per LD block showed significant association only in LD block 4. This block 4 is located between the two recombination hotspots and spans nearly the entire gene CR1, except for the first and last exons (130 kb, Supplementary Table S3, Figure 2). The association resulted from the overrepresentation in patients of one haplotype (HapB; ORHapB=1.24, 95% CI: 1.02–1.51; Padj=0.027). This risk haplotype harbored the risk allele of both the associated SNPs rs4844610 (A-allele) and rs1408077 (A-allele). In accordance with the single SNP analyses, haplotype association was also more evident in APOE ɛ4 carriers (ORHapB=1.50, 95% CI: 1.08–2.09; Padj=0.006).
CR1 SNP correlation with CSF biomarkers
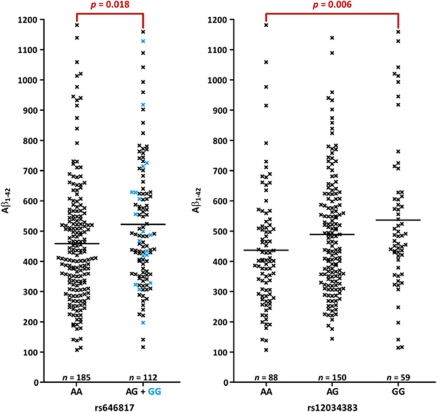
We assessed the effect of CR1 SNPs on AD biomarkers, considering CSF levels of Aβ1−42, T-tau or P-tau181P in AD patients as a quantitative trait. There was no evidence for an effect of rs4844610 or rs1408077 on CSF Aβ1−42, T-tau or P-tau181P levels. In contrast, we observed overall significant increased Aβ1−42 levels for carriers of the minor allele at four SNPs in LD block 4 (Supplementary Table S4, Figure 3), that is, rs646817, rs1746659, rs11803956 and rs12034383, of which the former two and the latter two represent the same signals because of high pairwise LD (r2⩾0.99). As the number of homozygous carriers of the minor allele was small for rs646817 (and, given the high r2, for rs1746659 (n=18)), we also calculated the effect of this SNP on Aβ1−42 levels under a dominant genotype model, showing further evidence for association (Poverall=0.018). Haplotype analysis in LD block 4 showed HapC (Padj=0.016) and HapD (Padj<0.001) to be significantly associated with increased levels of CSF Aβ1−42. Both haplotypes contain the associated alleles of the four SNPs (Supplementary Table S3).
Figure 3.
Association of SNP genotypes with Aβ1−42 cerebrospinal fluid (CSF) levels. CSF Aβ1−42 levels (pg ml−1) measured in patients carrying different single-nucleotide polymorphism (SNP) genotypes. Because of high r2 values between rs646817 and rs1746659, and between rs12034383 and rs11803956, only results for rs646817 and rs12034383 are shown. Significant pairwise comparisons are indicated, that is, AA vs AG+GG for rs646817 and AA vs GG for rs12034383, with P-values corrected for age at lumbar puncture, disease duration at lumbar puncture, gender and APOE ɛ4 genotype. For rs646817, results of the association test under a dominant model are depicted.
CR1 CNV association in Flanders–Belgian cohort
To determine whether a specific protein CR1 isoform could be associated with risk for AD, we determined copy number status of the three LCRs (LCR1, LCR2 and LCR3) located in the CR1 locus using a MAQ assay. We designed three test amplicons, 1 to 3, specifically targeting these LCRs (Figure 1). Test amplicon 1 also recognizes LCR1′, whereas LCR2 is targeted by amplicon 2 and LCR3 by amplicon 3.
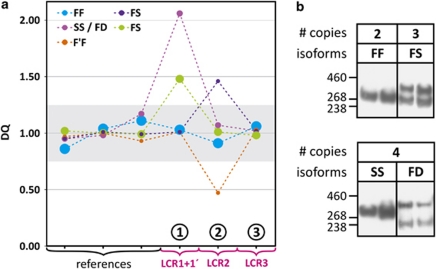
Quantitative dosage analysis of the target amplicons versus control amplicons showed that nearly all variations resulted from test amplicon 1. Copy numbers varied between 2 and 4 for individuals homozygous LCR1/LCR1, heterozygous LCR1/LCR1, LCR1′ and homozygous LCR1, LCR1′/LCR1and LCR1′ (Table 1). To confirm that the copy numbers of the CR1, LCR1 CNV correlated with the predicted protein isoforms F (one copy of LCR1) and S (two copies, LCR1 and LCR1′), we performed CR1 immunoblotting of lymphoblast cell lysates (Figure 4). In majority of the individuals, the LCR1 CNV copy number corresponded with the predicted lengths of CR1-F (220 kDa, one LCR1 copy) and CR1-S (250 kDa, two LCR1 copies). Also, the genotype frequencies of the LCR1 CNV were in agreement with previously reported frequencies of CR1-F (83%) and CR1-S (15%) (Table 1). However, in three out of eight tested individuals carrying four copies of the LCR1 CNV (37.5%), western blotting detected a 280 kDa band corresponding to CR1-D and resulting from a triplication of LCR1 on one allele, together with the 220 kDa CR1-F. As there were 84 individuals carrying four LCR1 copies, the number of individuals with F/D genotype may well be 32, that is, 1.8% of the total cohort of 1717 individuals. This is in line with the predicted frequency (∼1.7%) based on the reported frequencies of CR1-F (83%) and CR1-D (<1%). Further, in 3.5% of the individuals (n=60), amplicon 2 dosage variation instead of amplicon 1 was observed (Figure 4). Here, copy number varied between 3 (heterozygous for CR1-S, 1.7%) and 1 (heterozygous for CR1-F′, 1.8%), of which the latter is in agreement with the predicted F/F′ genotype frequency (1.66%). No dosage variation was observed for LCR3 in CR1L.
Table 1. Association of AD with LCR1 CNV.
| LCR1 CNV | CR1 isoform |
Patients |
Controls |
P | P-adj | OR | 95% CI | ||
|---|---|---|---|---|---|---|---|---|---|
| n | n | % | n | % | |||||
| Belgian sample | |||||||||
| 2 | F/F | 595 | 62.3 | 525 | 68.9 | 0.013 | 0.083 | ref. | |
| 3 | F/S | 312 | 32.7 | 201 | 26.4 | 0.003 | 0.028 | 1.32 | 1.03–1.69 |
| 4 | S/S or F/D | 48 | 5.0 | 36 | 4.7 | 0.574 | 0.583 | 1.16 | 0.69–1.94 |
| French sample | |||||||||
| 2 | F/F | 793 | 64.9 | 406 | 70.5 | 0.084 | 0.049 | ref. | |
| 3 | F/S | 354 | 30.0 | 142 | 24.7 | 0.032 | 0.039 | 1.33 | 1.02–1.74 |
| 4 | S/S or F/D | 74 | 6.1 | 28 | 4.9 | 0.500 | 0.456 | 1.44 | 0.87–2.40 |
Abbreviations: AD, Alzheimer disease; CI, confidence interval; CNV, copy number variation; CR1, complement receptor 1 gene; LCR, low-copy repeats; OR, odds ratio.
Note: significant associations (P<0.05) are indicated in bold; P-adj represent P-values adjusted for age, gender and APOE ɛ4 genotype.
Figure 4.
CR1 LCR dosage and CR1 isoform analysis. (a) Dosage analysis of the low-copy repeats (LCRs) underlying CR1 isoforms in selected individuals. Colored dashed lines connect dosage quotient values obtained with multiplex amplicon quantification (MAQ) amplicons specific for the CR1 LCRs (amplicons 1–3) (Figure 1) referred to by encircled numbers and three representative reference amplicons. The graph legend shows the isoforms predicted to be encoded by the CR1 copy number variation (CNV) genotype. The size of the symbols indicates the relative abundance of the respective CNV genotype. (b) Anti-CR1 immunoblot analysis of selected individuals illustrating the correlation between CNV genotype and protein isoforms. Lanes are grouped per number of copies of LCR1 present (Table 1).
To assess whether the common LCR1 CNV contributes to AD risk, we performed an association analysis with disease status. Individuals carrying three LCR1 copies had a 1.3 times increased risk for AD compared with carriers of two copies (Padj=0.028; Table 1). The CNV was in LD with the two associated SNPs, with an estimated r2=0.78 and r2=0.76 between CNV and rs4844610 and rs1408077, respectively. Conditional logistic regression showed that the associations of both the LCR1 CNV and the SNPs with AD risk represented the same association signal.
CR1 CNV replication in French cohort
To replicate our association findings, we genotyped SNP rs4844610 and LCR1 CNV in an independent French cohort and calculated the association with AD risk. Association with rs4844610 was nonsignificant in the French cohort (ORCA vs CC=1.20, 95% CI: 0.95–1.52; Padj=0.134), but the meta-analysis in the combined Flanders–Belgian and French cohorts provided significant results with a summary ORCA vs CC=1.31 (95% CI: 1.11–1.55; P=0.0015; heterogeneity P-value=0.317) (Supplementary Figure S1). The LCR1 CNV association results in the French cohort were comparable to those obtained in the Flanders–Belgian cohort with individuals carrying three copies of the LCR1 CNV having a 30% increased risk for AD (Table 1). Meta-analysis in the combined cohorts further confirmed these observations with and OR of 1.32 (95% CI: 1.1–1.59; P=0.0025; heterogeneity P-value=0.975) (Supplementary Figure S1).
Discussion
In this study, we focused on examining the role of CR1 in AD genetic risk in an extended Flanders–Belgian cohort by assessing single-nucleotide variations (SNV or SNPs) as well as CNV derived from LCR rearrangements in the CR1 locus. Also, as CR1 is presumably involved in Aβ clearance from brain, we investigated whether genetic variation in CR1 affected CSF Aβ1−42 levels, as well as levels of T-tau and P-tau181P. Finally, to replicate our SNP and CNV findings, we analyzed genetic association in an independent French cohort.
In an association analysis using 26 SNPs across the CR1 locus, two SNPs (rs4844610 and rs1408077), we showed significant association with AD in a Flanders–Belgian cohort. Both SNPs showed high pairwise LD (r2=0.97), and thus recognized the same association signal. They were also in strong LD (r2>0.87), with the two associated SNPs in the French GWA study, suggesting that they mimic the CR1 GWA cohort signal. Analysis of rs4844610 in the French cohort showed a similar, though nonsignificant trend. A combined cohort analysis provided an OR of 1.31 (95% CI: 1.11–1.55; P=0.0015). Haplotype analysis identified significant association (OR=1.24 (95% CI: 1.02–1.51; Padj=0.027)) with one haplotype (HapB) within a 130 kb LD-block containing CR1 except for its first and last exon. In agreement with the French GWA study,2 single SNP and haplotype associations were strongest in APOE ɛ4 carriers. Because CR1 is potentially involved in clearance of fibrillar Aβ peptides from the brain after complement activation and opsonization of Aβ peptides with C3b fragments,34 we investigated whether genetic variability at CR1 could affect CSF Aβ1−42 levels in patients. We calculated a significant association not with the two AD-associated SNPs (rs4844610, rs1408077), but with four other SNPs from within the associated CR1 region (rs646817/rs1746659 and rs11803956/rs12034383, showing strong pairwise LD). CSF Aβ1−42 levels increased with ∼20% in carriers homozygous for the rare allele compared with carriers of the common allele (Figure 3). Seemingly, the increased CSF Aβ1−42 contradicts with the decreased CSF Aβ1−42 levels that are considered indicative of AD in dementia patients and predictive in elderly people with or without mild cognitive impairment.35 However, in our study, the increased Aβ1−42 levels were not associated with the risk haplotype HapB but with haplotypes HapC and HapD. One possible explanation might be that functional variants are located on HapC and HapD that correspond to the so-called ‘intronic or exonic density polymorphisms', which have been correlated with number of CR1 molecules present on erythrocytes.4, 36 In plasma, C3b mediates adherence of Aβ peptides to erythrocyte CR1, a process, which is impaired early in the disease.18 However, it is doubtful that these variants can explain the observed association as genotype analysis of rs3811831, a SNP in high LD with the erythrocyte density polymorphisms, was not associated with AD or with CSF Aβ levels (data not shown). Additional studies of CR1 association and CSF Aβ1−42 levels in AD will be needed to analyze a potential influence of CR1 genetic variability.
Using a MAQ dosage assay, we were able to show that the LCR variability in the CR1 locus was driven nearly exclusively by variability of the coding LCR1 in CR1. Association analysis of LCR1 copy number with AD indicated that individuals carrying an extra LCR1 copy had ∼30% increased risk (Padj=0.028, OR=1.32 (95% CI: 1.03–1.69)). Presence of an extra copy of LCR1 (i.e., LCR1′) defines CR1-S (allelic frequency 15%) that carries an extra set of C3b/C4b binding sites compared with the more frequent shorter CR1-F (allelic frequency 83%) (Figure 1). Further, we showed that the LCR1 CNV was in high LD with SNPs rs4844610 and rs1408077 (r2⩾0.76), indicating that the association with CR1 can be explained by the intragenic CNV underlying CR1-S and CR1-F and, thus, number of C3b/C4b binding sites in CR1. Moreover, the observation that analysis of rs4844610 in the replication sample only showed a trend toward association, whereas the LCR1 CNV showed significant association in both study populations, independently, further strengthens the hypothesis that the CR1 CNV likely explains the association initially observed in the GWAS. It should be noted, however, that our MAQ dosage assay could not distinguish among carriers of four LCR1 copies those homozygous for CR1-S or heterozygous CR1-D/CR1-F. Also, our analysis did not include LCR2 variability (e.g., CR1-F′). However, in accordance with the reported frequencies, the respective isoforms are rare (1.7–1.8%) in our cohort, therefore unlikely to have wrongly affected the observed CR1 CNV association.
How the CR1 CNV genetic variations affect the biological processes that eventually culminate into AD remains unclear. One explanation could be that the longer CR1-S, with its additional C3b binding site (Figure 1), increases Aβ clearance from brain either through attacking Aβ plaques directly or by functioning as an Aβ-sink in the periphery, mediated by erythrocyte CR1. The observed association with AD with CR1-S, however, contradicts this hypothesis. Further, we did not observe an association between CR1-S and CSF Aβ1−42, although we cannot exclude that the lack of association results from the use of levels of soluble Aβ1−42,35 whereas CR1-S perhaps only affects aggregated Aβ1−42.16, 17 Nevertheless, we did obtain evidence of association between other genetic CR1 variants and CSF Aβ1−42, indicating CR1 may have a role in AD pathogenesis through an effect on Aβ. Another explanation for the association with CR1-S might lie in the enhancement of the proteins' inhibitory potential (cofactor activity) towards complement activation, through its gained C3b site (Figure 1). One of the functions of CR1 is controlling amplification of the complement cascade. By binding to C3b-opsonized particles, CR1 will function as cofactor of Factor I, which converts C3b into its inactivated form C3b. The latter is unable to sustain or amplify the complement cascade, ultimately dampening the immune response and minimizing bystander tissue damage.37 Therefore, CR1-S might represent a more potent inhibitor of complement cascade amplification than CR1-F, resulting in a shorter-lasting complement reaction and reducing the levels of C3b fragments that can opsonize Aβ aggregates. Thus, individuals carrying one or two CR1-S alleles might exhibit an elevated inhibition of a potentially neuroprotective process (e.g., clearance of Aβ aggregates from the brain), which is in agreement with several observations in AD mouse models, in which inhibition of complement activation was shown to correlate with enhanced Aβ plaque deposition and loss of neuronal integrity.34, 38 On the other hand, complement activation can elicit a potent immune reaction in the brain, which might kill healthy bystander neurons in the process. In this light, inhibition of complement activation could have a more beneficial outcome, as was likewise observed in AD mouse models in which the classical complement cascade was inhibited.19, 20 However, the fact that the longer CR1-S with more binding sites that can inhibit complement activation through enhanced cofactor activity, increases risk for developing AD argues against this idea.
In summary, our data strongly support a role for CR1 in the pathogenesis of AD. We were able to replicate the genetic association with AD risk and extended the genetic findings by fine mapping the SNP association to a 130 kb genomic region comprising mostly of CR1. Further, we showed that the association might likely be explained by an intragenic CNV that translates into the two major CR1 isoforms differing in number of C3b/C4b and cofactor activity sites, and thus different functional roles in the complement cascade. Lastly, we were able to replicate our genetic findings in an additional French cohort, further implying that an extra LCR1 because of duplication at the intragenic CR1 CNV increases risk for AD. However, how exactly these genetic and biological findings contribute to AD pathogenesis remains to be determined.
Acknowledgments
The work was made possible by the generous participation of the Flanders–Belgian and French control individuals, patients and their families. We further acknowledge the contribution of the personnel of the VIB Genetic Service Facility (http://www.vibgeneticservicefacility.be), the Biobank of the Institute Born-Bunge (http://www.bornbunge.be/Home/index_en.shtml) and the department of Neurology/Memory Clinic of ZNA Middelheim and ZNA Hoge Beuken hospitals and the University Hospitals of Leuven. We thank Marie-Eve Maturski for her technical help in preparing the French DNA samples for analyses and Jill Luyckx for technical help with the CSF biomarker analyses. The Antwerp site was in part supported by the Special Research Fund of the University of Antwerp (http://www.ua.ac.be/main.aspx?c=.ENGLISH), the Fund for Scientific Research-Flanders (FWO-V, http://www.fwo.be/Default.aspx); the Foundation for Alzheimer Research (SAO/FRMA, http://www.alzh.org/), the Interuniversity Attraction Poles (IAP) program P6/43 of the Belgian Federal Science Policy Office (http://www.belspo.be/belspo/iap/index_en.stm), the Antwerp Medical Research Foundation and Neurosearch, and a Methusalem Excellence Grant of the Flemish Government (http://www.ewi-vlaanderen.be), Belgium. KS and NB are postdoctoral fellows and KB and NLB PhD fellows of the FWO-V. The Lille site was funded in part by the National Foundation for Alzheimer's disease and related disorders, the Institut Pasteur de Lille and INSERM.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
Disclaimer
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Supplementary Material
References
- Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat Genet. 2009;41:1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- Krych-Goldberg M, Atkinson JP. Structure-function relationships of complement receptor type 1. Immunol Rev. 2001;180:112–122. doi: 10.1034/j.1600-065x.2001.1800110.x. [DOI] [PubMed] [Google Scholar]
- Khera R, Das N. Complement receptor 1: disease associations and therapeutic implications. Mol Immunol. 2009;46:761–772. doi: 10.1016/j.molimm.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykman TR, Cole JL, Iida K, Atkinson JP. Polymorphism of human erythrocyte C3b/C4b receptor. Proc Natl Acad Sci USA. 1983;80:1698–1702. doi: 10.1073/pnas.80.6.1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong WW, Wilson JG, Fearon DT. Genetic regulation of a structural polymorphism of human C3b receptor. J Clin Invest. 1983;72:685–693. doi: 10.1172/JCI111018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klickstein LB, Wong WW, Smith JA, Weis JH, Wilson JG, Fearon DT. Human C3b/C4b receptor (CR1). Demonstration of long homologous repeating domains that are composed of the short consensus repeats characteristics of C3/C4 binding proteins. J Exp Med. 1987;165:1095–1112. doi: 10.1084/jem.165.4.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klickstein LB, Bartow TJ, Miletic V, Rabson LD, Smith JA, Fearon DT. Identification of distinct C3b and C4b recognition sites in the human C3b/C4b receptor (CR1, CD35) by deletion mutagenesis. J Exp Med. 1988;168:1699–1717. doi: 10.1084/jem.168.5.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong WW, Farrell SA. Proposed structure of the F′ allotype of human CR1. Loss of a C3b binding site may be associated with altered function. J Immunol. 1991;146:656–662. [PubMed] [Google Scholar]
- Wong WW, Cahill JM, Rosen MD, Kennedy CA, Bonaccio ET, Morris MJ, et al. Structure of the human CR1 gene. Molecular basis of the structural and quantitative polymorphisms and identification of a new CR1-like allele. J Exp Med. 1989;169:847–863. doi: 10.1084/jem.169.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLure CA, Dawkins RL, Williamson JF, Davies RA, Berry J, Natalie LJ, et al. Amino acid patterns within short consensus repeats define conserved duplicons shared by genes of the RCA complex. J Mol Evol. 2004;59:143–157. doi: 10.1007/s00239-004-2609-8. [DOI] [PubMed] [Google Scholar]
- Eikelenboom P, Veerhuis R, Scheper W, Rozemuller AJ, van Gool WA, Hoozemans JJ. The significance of neuroinflammation in understanding Alzheimer's disease. J Neural Transm. 2006;113:1685–1695. doi: 10.1007/s00702-006-0575-6. [DOI] [PubMed] [Google Scholar]
- McGeer EG, Klegeris A, McGeer PL. Inflammation, the complement system and the diseases of aging. Neurobiol Aging. 2005;26 (Suppl 1:94–97. doi: 10.1016/j.neurobiolaging.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Sleegers K, Lambert JC, Bertram L, Cruts M, Amouyel P, Van BC. The pursuit of susceptibility genes for Alzheimer's disease: progress and prospects. Trends Genet. 2010;26:84–93. doi: 10.1016/j.tig.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Rogers J, Cooper NR, Webster S, Schultz J, McGeer PL, Styren SD, et al. Complement activation by beta-amyloid in Alzheimer disease. Proc Natl Acad Sci USA. 1992;89:10016–10020. doi: 10.1073/pnas.89.21.10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradt BM, Kolb WP, Cooper NR. Complement-dependent proinflammatory properties of the Alzheimer's disease beta-peptide. J Exp Med. 1998;188:431–438. doi: 10.1084/jem.188.3.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster S, Bradt B, Rogers J, Cooper N. Aggregation state-dependent activation of the classical complement pathway by the amyloid beta peptide. J Neurochem. 1997;69:388–398. doi: 10.1046/j.1471-4159.1997.69010388.x. [DOI] [PubMed] [Google Scholar]
- Rogers J, Li R, Mastroeni D, Grover A, Leonard B, Ahern G, et al. Peripheral clearance of amyloid beta peptide by complement C3-dependent adherence to erythrocytes. Neurobiol Aging. 2006;27:1733–1739. doi: 10.1016/j.neurobiolaging.2005.09.043. [DOI] [PubMed] [Google Scholar]
- Fonseca MI, Zhou J, Botto M, Tenner AJ. Absence of C1q leads to less neuropathology in transgenic mouse models of Alzheimer's disease. J Neurosci. 2004;24:6457–6465. doi: 10.1523/JNEUROSCI.0901-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Fonseca MI, Pisalyaput K, Tenner AJ. Complement C3 and C4 expression in C1q sufficient and deficient mouse models of Alzheimer's disease. J Neurochem. 2008;106:2080–2092. doi: 10.1111/j.1471-4159.2008.05558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelborghs S, Dermaut B, Goeman J, Saerens J, Marien P, Pickut BA, et al. Prospective Belgian study of neurodegenerative and vascular dementia: APOE genotype effects. J Neurol Neurosurg Psychiatry. 2003;74:1148–1151. doi: 10.1136/jnnp.74.8.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelborghs S, Dermaut B, Marien P, Symons A, Vloeberghs E, Maertens K, et al. Dose dependent effect of APOE epsilon4 on behavioral symptoms in frontal lobe dementia. Neurobiol Aging. 2006;27:285–292. doi: 10.1016/j.neurobiolaging.2005.02.005. [DOI] [PubMed] [Google Scholar]
- Bettens K, Brouwers N, van MH, Gil A, Engelborghs S, De Deyn PP, et al. Follow-up study of susceptibility loci for Alzheimer's disease and onset age identified by genome-wide association. J Alzheimers Dis. 2010;19:1169–1175. doi: 10.3233/JAD-2010-1310. [DOI] [PubMed] [Google Scholar]
- McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- Dreses-Werringloer U, Lambert JC, Vingtdeux V, Zhao H, Vais H, Siebert A, et al. A polymorphism in CALHM1 influences Ca2+ homeostasis, Abeta levels, and Alzheimer's disease risk. Cell. 2008;133:1149–1161. doi: 10.1016/j.cell.2008.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapuis J, Hot D, Hansmannel F, Kerdraon O, Ferreira S, Hubans C, et al. Transcriptomic and genetic studies identify IL-33 as a candidate gene for Alzheimer's disease. Mol Psychiatry. 2009;14:1004–1016. doi: 10.1038/mp.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Brouwers N, Sleegers K, Engelborghs S, Bogaerts V, Serneels S, Kamali K, et al. Genetic risk and transcriptional variability of amyloid precursor protein in Alzheimer's disease. Brain. 2006;129:2984–2991. doi: 10.1093/brain/awl212. [DOI] [PubMed] [Google Scholar]
- Kumps C, Van RN, Heyrman L, Goossens D, Del-Favero J, Noguera R, et al. Multiplex amplicon quantification (MAQ), a fast and efficient method for the simultaneous detection of copy number alterations in neuroblastoma. BMC Genomics. 2010;11:298. doi: 10.1186/1471-2164-11-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J-H, Blay S, McNeney B, Graham J.LDheatmap: an R function for graphical display of pairwise linkage disequilibria between single nucleaotide polymorphisms J Stat Soft 200616code snippet 3. [Google Scholar]
- Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70:425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez JR, Subirana I, Escaramis G, Peraza S, Caceres A, Estivill X, et al. Accounting for uncertainty when assessing association between copy number and disease: a latent class model. BMC Bioinformatics. 2009;10:172. doi: 10.1186/1471-2105-10-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyss-Coray T, Yan F, Lin AH, Lambris JD, Alexander JJ, Quigg RJ, et al. Prominent neurodegeneration and increased plaque formation in complement-inhibited Alzheimer's mice. Proc Natl Acad Sci USA. 2002;99:10837–10842. doi: 10.1073/pnas.162350199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelborghs S, De Vreese K, Van de CT, Vanderstichele H, Van Everbroeck B, Cras P, et al. Diagnostic performance of a CSF-biomarker panel in autopsy-confirmed dementia. Neurobiol Aging. 2008;29:1143–1159. doi: 10.1016/j.neurobiolaging.2007.02.016. [DOI] [PubMed] [Google Scholar]
- Herrera AH, Xiang L, Martin SG, Lewis J, Wilson JG. Analysis of complement receptor type 1 (CR1) expression on erythrocytes and of CR1 allelic markers in Caucasian and African American populations. Clin Immunol Immunopathol. 1998;87:176–183. doi: 10.1006/clin.1998.4529. [DOI] [PubMed] [Google Scholar]
- Dunkelberger JR, Song WC. Complement and its role in innate and adaptive immune responses. Cell Res. 2010;20:34–50. doi: 10.1038/cr.2009.139. [DOI] [PubMed] [Google Scholar]
- Maier M, Peng Y, Jiang L, Seabrook TJ, Carroll MC, Lemere CA. Complement C3 deficiency leads to accelerated amyloid beta plaque deposition and neurodegeneration and modulation of the microglia/macrophage phenotype in amyloid precursor protein transgenic mice. J Neurosci. 2008;28:6333–6341. doi: 10.1523/JNEUROSCI.0829-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.