Background: Clostridium perfringens enterotoxin (CPE) binds to a subset of claudin tight junction proteins.
Results: The molecular interface of the CPE-claudin interaction was mapped.
Conclusion: Claudin-3 and -4 interact with CPE in the same orientation but in different modes.
Significance: The mechanistic insights might advance design of CPE-based claudin modulators to improve paracellular drug delivery or to target claudin-overexpressing tumors.
Keywords: Molecular Modeling, Mutagenesis, Protein Domains, Structural Biology, Tight Junctions, Clostridium perfringens Enterotoxin, Claudin, Structure-Function Study
Abstract
Claudins (Cld) are essential constituents of tight junctions. Domain I of Clostridium perfringens enterotoxin (cCPE) binds to the second extracellular loop (ECL2) of a subset of claudins, e.g. Cld3/4 and influences tight junction formation. We aimed to identify interacting interfaces and to alter claudin specificity of cCPE. Mutagenesis, binding assays, and molecular modeling were performed. Mutation-guided ECL2 docking of Cld3/4 onto the crystal structure of cCPE revealed a common orientation of the proposed ECL2 helix-turn-helix motif in the binding cavity of cCPE: residues Leu150/Leu151 of Cld3/4 bind similarly to a hydrophobic pit formed by Tyr306, Tyr310, and Tyr312 of cCPE, and Pro152/Ala153 of Cld3/4 is proposed to bind to a second pit close to Leu223, Leu254, and Leu315. However, sequence variation in ECL2 of these claudins is likely responsible for slightly different conformation in the turn region, which is in line with different cCPE interaction modes of Cld3 and Cld4. Substitutions of other so far not characterized cCPE residues lining the pocket revealed two spatially separated groups of residues (Leu223, Asp225, and Arg227 and Leu254, lle258, and Asp284), which are involved in binding to Cld3 and Cld4, albeit differently. Involvement of Asn148 of Cld3 in cCPE binding was confirmed, whereas no evidence for involvement of Lys156 or Arg157 was found. We show structure-based alteration of cCPE generating claudin binders, which interact subtype-specific preferentially either with Cld3 or with Cld4. The obtained mutants and mechanistic insights will advance the design of cCPE-based modulators to target specific claudin subtypes related either to paracellular barriers that impede drug delivery or to tumors.
Introduction
Clostridium perfringens enterotoxin (CPE)3 causes the gastrointestinal symptoms of one of the most common foodborne illnesses in the United States and Europe (1). CPE binds to claudin-3 and -4, initially defined as CPE receptors (2), and some other members of the claudin (Cld) family (3). Claudins form the backbone of tight junctions and regulate paracellular permeability in epithelia and endothelia (4, 5). After CPE binding to claudins via the C-terminal domain (cCPE), pore formation in the plasma membrane of the host mucosa cells is meditated by the N-terminal domains, leading to cell death (6, 7). However, cCPE is not cytotoxic and was suggested to be a promising claudin modulator (8, 9). It increases paracellular permeability (10) and could be used to improve drug delivery across tissue barriers. Furthermore, CPE constructs can be used to target claudin-overexpressing tumors (11–14) because deregulation of claudin expression and function is associated with tumor proliferation/growth (15). Functional domain mapping of cCPE demonstrated that Tyr306, Tyr310, Tyr312, and Leu315 are involved in binding to Cld4 (16, 17). The crystal structure of cCPE194–319 (18) and recently of full-length CPE (19, 20) revealed a globular, nine-stranded β-sandwich of the claudin binding domain and that Tyr306, Tyr310, Tyr312, and Leu315 form part of a surface loop and an adjacent β-strand.
CPE and cCPE bind to the different claudin subtypes with distinct affinities: to Cld3, 4, 6, 7, and 8 with high affinity (KD between 1 × 106 m−1 and 1.1 × 108 m−1) (10, 21), to Cld1, 2, and 14 with low affinity (22), and they do not interact with Cld5, 10, 11, 12, 13, 15, 16, 18, 19, 20, and 22 (3, 23). Despite the fact that cCPE (GST-CPE194–319) does not bind to Cld5, weak binding was obtained for Cld5 with GST-CPE116–319 (23). Claudins are tetraspan transmembrane proteins with two extracellular loops (ECL). cCPE binds to the ECL2 but not to the ECL1 of Cld3 or Cld4 (21, 22). Previously, we identified the motif 148NPLVP152 in the turn region of the ECL2 of Cld3 to be involved in binding to cCPE (23). Here, we revealed the molecular interface between cCPE and ECL2 of its high affinity receptors, Cld3 and Cld4. Furthermore, we achieved a clear shift of claudin subtype specificity by structure-based mutations of cCPE. These modifications might advance claudin targeting for improvement of paracellular drug delivery (9) or tumor treatment (15, 24, 25).
EXPERIMENTAL PROCEDURES
Plasmids
For construction of plasmid encoding GST-CPE194–319 (GST-cCPE) fusion protein, cDNA of CPE (kindly provided by Dr. Y. Horiguchi, Osaka, Japan) was amplified by PCR and cloned into pGEX-4T1 (GE Healthcare) using EcoRI and SalI (23). Plasmids encoding GST-CPE194–319 with single or multiple mutations (L223A, D225A, R227A, L254A, S256A, K257A, I258A, D284A, D284N, Y306A, Y310F) were generated by site-directed mutagenesis of pGEX-4T1-CPE194–319 (23). Plasmids based on pEYFP-N1/pECFP-N1 encoding murine Cld3WT, Cld3N148D, Cld3L150A, Cld3E153V, Cld3Q155E, Cld3WT-YFP (23), murine Cld5-YFP (26), and human Cld1-YFP (27) have been described previously. Cld3R157Y, Cld3L150F-YFP, and Cld3K156A-YFP were generated by site-directed mutagenesis of Cld3WT or Cld3WT-YFP, respectively. pEGFP-Cld4 encoding human Cld4 with N-terminal GFP was kindly provided by Dr. W. Hunziker (Singapore). GFP-Cld4L151F and GFP-Cld4A153P/S154E/G155A were generated by site-directed mutagenesis of pEGFP-Cld4.
Antibodies
Rabbit anti-Cld3, rabbit anti-Cld4, goat HRP-anti-rabbit, goat HRP-anti-mouse were from Invitrogen. Mouse anti-GST was from Sigma-Aldrich. Phycolink® anti-GST R-phycoerythrin-conjugated antibodies were from Europa Bioproducts Ltd., Cambridge, UK. Mouse anti-GFP/YFP was from Clontech.
Expression and Purification of cCPE Constructs
CPE194–319 with N-terminal GST fusions as well as GST (control) were expressed in Escherichia coli BL21 strain. For use of GST fusion proteins, see supplemental Comment. Bacteria were grown to A600 = 0.6–0.8, and expression was induced by addition of 1 mm isopropyl-β-d-thiogalactopyranoside. Three h later bacteria were harvested (10 min, 20,000 × g, 4 °C) and lysed in PBS with 1% (v/v) Triton X-100, 0.1 mm PMSF, 1 mm EDTA, protease inhibitor mixture (Sigma-Aldrich) and sonicated with Vibra CellTM model 72434 (BioBlock Scientific, Strasbourg, France) by 10 × 1-s pulses. To remove insoluble cell debris, lysates were centrifuged at 20,000 × g for 1 h at 4 °C. The proteins were purified from supernatants using glutathione-agarose (Sigma-Aldrich), and eluted proteins were dialyzed against PBS. Protein concentration was determined with the BCA Protein kit (Thermo Scientific).
Cell Culture and Transfection
MDCK I cells were maintained in Minimum Essential Medium with Earle's salts (MEM) supplemented with 10% fetal calf serum, 100 units/ml penicillin, 100 μg/ml streptomycin, and 1% l-alanyl-l-glutamine. HEK293 cells (HEK cells) were maintained and transfected as described (28).
Pulldown Assay
The assay was performed as described previously (23). Briefly, transient or stable transfected HEK cells or MDCK I cells were lysed with 1% Triton X-100, EDTA-free protease inhibitor mixture (Roche Applied Science) in PBS. The 10,000 × g supernatant was incubated with GST-cCPE constructs bound to glutathione-Sepharose beads (GE Healthcare) for 2 h on a shaker at 4 °C. Beads were washed three times with PBS containing 0.5% Triton X-100 and bound proteins eluted with Laemmli buffer and analyzed by SDS-PAGE and Western blotting with anti-Cld3, anti-Cld4, or anti-GFP antibodies. After stripping, membranes were incubated with anti-GST antibodies to verify that similar amounts of GST-cCPE were bound to the beads.
Cellular cCPE Binding Assay
Two to 3 days after transient transfection or 1 day after plating of stable lines, HEK cells expressing claudin constructs were incubated with 0.5 μg/ml GST-CPE constructs (30 min, 37 °C) in 12- or 24-well plates (Techno Plastic Products AG, Trasadingen, Switzerland). Cells were washed with ice-cold PBS (with Ca2+ and Mg2+), scraped, harvested (300 × g, 5 min, 4 °C), and lysed with radioimmuneprecipitation assay buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 1.0 mm EDTA, 1.0% (v/v) Nonidet P-40, 0.5% (w/v) deoxycholate, 0.1% SDS, EDTA-free protease inhibitor mixture (Roche Applied Science)) for 10 min on ice. After centrifugation (10,000 × g, 5 min, 4 °C), supernatants were analyzed for the amount of bound GST-cCPE by SDS-PAGE and Western blotting. Alternatively, for detection of bound GST-cCPE with fluorescence plate reader (Tecan), cells were washed twice with ice-cold PBS (with Ca2+ and Mg2+), fixed (10 min with 2.4% (w/v) paraformaldehyde in PBS) followed by quenching (20 min with 100 mm glycine in PBS) and blocking (10 min with 1% (w/v) BSA, 0.05% (v/v) Tween 20 in PBS). Cells were incubated with PhycoLink® anti-GST-R-phycoerythrin conjugate in 1% (w/v) BSA, 0.05% (v/v) Tween 20, 2 μm Hoechst 33258 in PBS for 1 h and washed three times with PBS. Bound GST-cCPE was detected via fluorescence intensity of PhycoLink® α-GST at λex 545 ± 12 nm/λem 578 ± 12 nm; claudins were detected at λex 506 ± 5 nm/λem 525 ± 5 nm for YFP-tagged, or λex 488 ± 5 nm/λem 510 ± 5 nm for GFP-tagged claudins. Cell number was monitored with Hoechst dye at λex 365 ± 12 nm/λem 480 ± 12 nm. Untransfected HEK cells were used as a negative control (fluorescence intensity of GST-cCPE bound to Cld3WT was 15.4 ± 0.8 times higher than the negative control). Fluorescence intensity of bound cCPE was normalized to amount of claudins (YFP or GFP intensity) or cell number (Hoechst intensity).
Measurements of Transepithelial Resistance (TER)
Subconfluent cultures of MDCK I cells, expressing both Cld3 and Cld4 endogenously, were seeded in Millicell® culture inserts (Millipore). TER of cell monolayers was measured every 3 h by using a CellZscope® (NanoAnalytics). Normalization of TER values by area of the cell monolayers was done automatically by CellZscope. When TER values reached the plateau, indicating a stable tight junction barrier, MDCK I monolayers were treated with GST (negative control), GST-cCPEWT or its mutants (R227A, L254A/S256A/I258A/D284A, Y306A) on both basal and apical sides of the chamber. The background (value of blank Millicell® culture insert) was subtracted from the measured TER. TER values 18 h after the addition of cCPE were normalized to the TER values immediately before the cCPE addition.
Structural Bioinformatics and Molecular Modeling
Homology models for murine Cld3-ECL2134–164 and human Cld4-ECL2135–165 were created based on the model for murine Cld5-ECL2135–165 as described previously (28). Due to sequence and binding differences of Cld3 and Cld4 in the N-terminal region (153–158, 154–159, respectively) of the C-terminal helix an extensive search for further templates was conducted by performing sequence similarity searches (FASTA program) of the Protein Data Bank (PDB). For murine Cld3-ECL2 parts of the PDB structure 1W5C (Photosystem II from Thermo-synechococcus elongatus) were used as template for the N-terminal helix cap region of ECL2 helix 2. For human Cld4-ECL2, no further sufficient template was found (cCPE crystal structure used as deposited by Van Itallie et al. PDB 2QUO (18)). All manual reciprocal dockings, manipulations, optimizations of ECL2 models, and calculations of hydrophobic and electrostatic potentials on the molecular surfaces were performed with Sybyl X1.2 (Tripos, Inc.). Models were energetically minimized using the AMBER7 FF99 force field.
Statistics
Unless stated otherwise, results are shown as means ± S.E. Statistical analyses were performed using Prism version 5.0 (GraphPad, San Diego, CA). First, normality tests were performed (D'Agostino and Pearson omnibus, Shapiro-Wilk and Kolmogorov-Smirnov test). Data sets showing normal distribution were analyzed using an unpaired, one-tailed Student's t test. Data sets not showing normal distribution were analyzed using Wilcoxon Signed Rank test. p < 0.05 was taken as significant.
RESULTS
cCPE
Mapping Claudin Binding Pocket on Surface of cCPE Structure
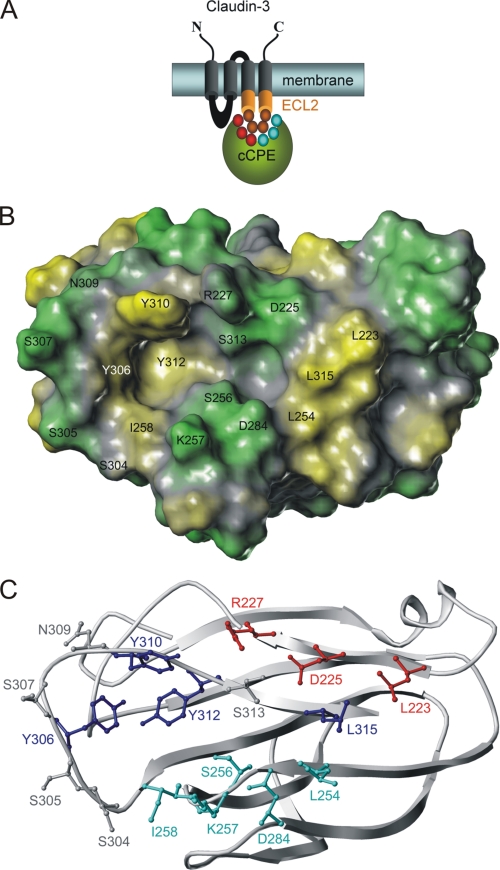
Inspection of the crystal structure of cCPE (PDB 2QUO (18)) showed a two-pit cavity nonuniformly surrounded by residues previously described for Cld4 binding. Ala substitution of the cCPE residues Tyr306, Tyr310, Tyr312, and Leu315 crucially inhibits Cld4 binding (16). Mapping these positions (blue sticks in Fig. 1C) onto the surface of the cCPE structure reveals that they are located close to, or in, the potential binding cavity for claudins. The cavity is formed by two pits and surrounded on the upper rim by the residues Leu223, Asp225, Arg227, and Ser313, on the lower rim by Leu254, Ser256, Lys257, Ile258, and Asp284 (Fig. 1C). Displaying the calculated hydrophobic potential on the cCPE surface indicates a deep, large, and strong hydrophobic pit (yellow in Fig. 1B) lined by the triple Tyr motif (Tyr306, Tyr310, and Tyr312) and a smaller, less deep and weaker hydrophobic or nonpolar pit (Fig. 1B, gray) surrounded by the triple Leu motif to the left (Leu223, Leu254, and Leu315). This second pit is less hydrophobic because it is flanked by polar amino acids (Fig. 1B, green) on both the upper (Asp225, Arg227, and Ser313) and the lower rim (Ser256, Lys257, and Asp284).
FIGURE 1.
Putative binding pocket for claudins on surface of cCPE structure (PDB 2QUO (18)). A, scheme of cCPE (green) binding to ECL2 (orange) of Cld3 or Cld4. Residues investigated are shown as circles (red and cyan, in cCPE; orange, in ECL2). B, hydrophobic potential calculated for surface of cCPE structure showing polar (green) and hydrophobic (yellow) areas for residues (labeled) that surround putative binding pocket for claudins. C, cCPE structure showing positions of residues (ball and stick) in more detail (blue, previously described positions for which Ala substitution inhibits binding; gray, previously described positions for which Ala substitution increases binding (16); red/cyan, reported in this study).
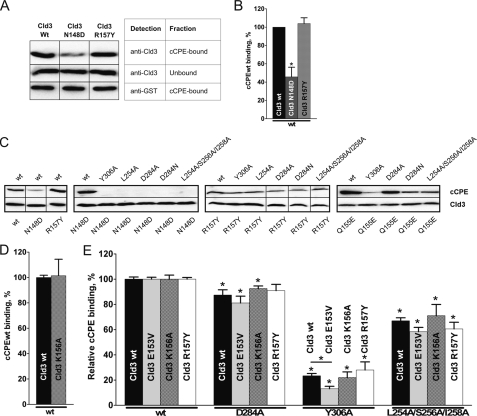
Amino Acid Substitutions around Putative Claudin Binding Pocket Affect cCPE-Cld3 Interaction
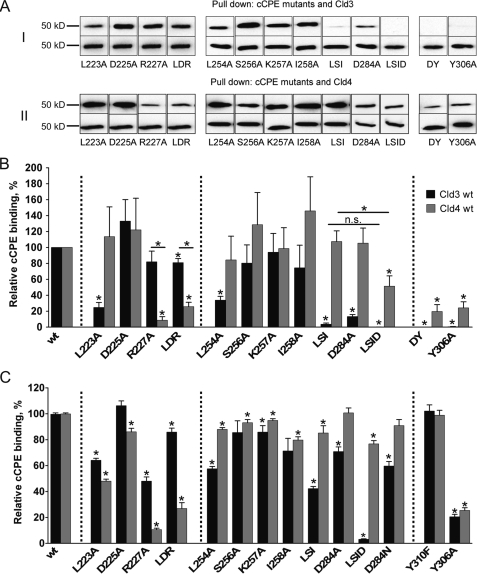
The above mentioned cCPE residues were substituted with Ala to test the contribution of the side chains to claudin binding. Mutants of GST-CPE194–319 (GST-cCPE) were analyzed by previously established pulldown assay (23) using lysates of claudin-transfected HEK cells. Since Y306A in cCPE was shown to inhibit binding to Cld4, it was used as a positive control. As expected, Y306A inhibited binding of full-length Cld4 to GST-cCPE in pulldown assays (Fig. 2, A and B). In addition, Y306A inhibited binding of full-length Cld3 to GST-cCPE (Fig. 2, A and B). First, residues on the upper rim of the putative binding pocket were analyzed. L223A in GST-cCPE strongly reduced binding of Cld3. D225A had no effect, and R227A slightly weakened binding to Cld3. However, the latter did not reach statistical significance. Surprisingly, the corresponding triple substitution L223A/D225A/R227A affected Cld3 binding less than L223A alone. On the lower rim, L254A showed strong reduction of Cld3 binding. S256A and I258A resulted in a slightly weakened, but not statistically significant, reduction of Cld3-cCPE interaction. However, the triple substitution L254A/S256A/I258A reduced binding more strongly than each of the three single substitutions. D284A severely affected Cld3 binding whereas K257A showed no clear reduction. L254A/S256A/I258A/D284A blocked Cld3 binding completely. Taken together, the pulldown data indicate involvement of Leu223, Leu254, Asp284, and possibly Ser256 and/or Ile258 of cCPE in the interaction with Cld3 (Fig. 2, A and B).
FIGURE 2.
cCPE-binding to full-length Cld3 and Cld4 is affected differently by amino acid substitutions in cCPE. A, lysates of HEK cells transfected with Cld3WT or Cld4WT used for pulldown assays with GST-cCPE constructs. Bound fractions were analyzed by SDS-PAGE and Western blotting. Top rows of I and II show bands of Cld3 or Cld4 bound to particular GST-cCPE mutants, and bands in bottom rows show Cld3 or Cld4 bound to corresponding GST-cCPEWT (used as internal standard). Substitutions in upper (left panel) or lower (middle panel) rim of cCPE binding pocket and D284A/Y306A or Y306A (right panel) are shown. LDR, L223A/D225A/R227A; LSID, L254A/S256A/I258A/D284A; DY, D284A/Y306A. B and C, quantification of pulldown (B) and cellular binding assay (C). Results reflect mean ± S.E. (error bars); n ≥ 4. *, p < 0.05 to GST-cCPEWT. Dotted lines separate different groups of substitutions. For L254A/S256A/I258A/D284A and Cld3 in C, n = 2; additional data were quantified by Western blotting relative cCPE binding = 1.0% ± 0.9%, n = 8.
Substitutions in cCPE Affect Binding to Cld3 and Cld4 Differently
To compare binding of cCPE to Cld3 with that of Cld4, GST-cCPE pulldown assays were also performed with lysates of Cld4-transfected HEK cells (Fig. 2, A and B). On the upper rim of the putative binding pocket D225A had no effect on Cld4 binding, as found for Cld3. In contrast to Cld3, L223A did not affect Cld4 binding, but R227A strongly reduced Cld4 binding. In addition, L223A/D225A/R227A weakened Cld4 binding much more strongly than observed for Cld3. In contrast to Cld3, none of the single substitutions on the lower rim strongly reduced Cld4 binding. Furthermore, the triple substitution L254A/S256A/I258A did not influence the Cld4-cCPE interaction. However, L254A/S256A/I258A/D284A clearly reduced the Cld4 binding, but not as strongly as Cld3 binding. The data indicate that binding of cCPE to Cld3 and Cld4 is affected differently by amino acid substitutions in cCPE.
Cellular Binding Assays Verify Different Involvement of Leu223, Arg227, Leu254, Ile258, and Asp284 of cCPE in Their Interaction with Cld3 and/or Cld4
Pulldown assays depend on detergent solubilization of the transmembranal claudins. To investigate binding of GST-cCPE to native Cld3 and Cld4 on the surface of living cells, the previously established cellular binding assay was optimized. Here, Cld3- or Cld4-transfected HEK cells were incubated with GST-cCPE constructs, and their binding was detected after fixation using R-phycoerythrin-conjugated anti-GST antibodies in a fluorescence plate reader. Similar to the results from the pulldown assay, L223A and R227A, but not D225A (upper rim of the cCPE pocket), affected binding to Cld3, and L223A/D225A/R227A showed weaker reduction than L223A or R227A alone (Fig. 2C). Likewise, the results for R227A also correspond to the pulldown assay, with Cld4 binding being more strongly reduced than Cld3 binding. In contrast to the pulldown assay, Cld4 binding was strongly inhibited by L223A and weakly inhibited by D225A. On the lower rim of the pocket, binding to Cld3 was reduced in the order L254A/S256A/I258A, L254A, I258A, S256A, similar to the pulldown data. In addition, all of these substitutions together with D284A and D284N affected binding to Cld3 more strongly than that to Cld4. Furthermore, like in pulldown assay, L254A/S256A/I258A/D284A blocked Cld3 binding nearly completely, whereas Cld4 binding was only weakly inhibited. K257A only slightly reduced binding of GST-cCPE to Cld3 and Cld4. Taken together, the data verify that amino acid substitutions around the putative claudin binding pocket of cCPE affect binding to Cld3 and Cld4 differently. In particular, substitutions in the upper rim (L223A, R227A) affect Cld4 binding more strongly than Cld3 binding, whereas those in the lower rim (L254A, D284A) mainly reduce Cld3 binding.
Effect on TER Is Weakened by Particular Substitutions
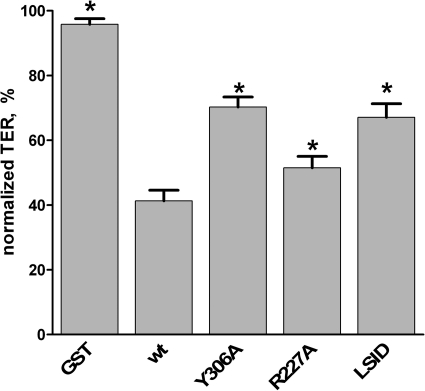
Incubation of epithelial monolayers with cCPE decreases the TER due to cCPE-triggered destabilization of tight junctions (10). To test whether substitutions in GST-cCPE that inhibit binding to Cld3 and/or Cld4 also impede TER reducing activity, MDCK I cells were used. These cells are a standard model to investigate paracellular barriers (10). In addition, GST-cCPE binds to endogenous Cld3 and Cld4 of MDCK I cells (supplemental Fig. S6). As shown in Fig. 3, treatment of cells with GST-cCPEWT reduced TER to 41.3 ± 3.3% of the TER values before application. This demonstrates that the recombinantly expressed and purified GST-cCPEWT is capable of increasing paracellular permeability. Incubation with GST-cCPEY306A reduced TER to 70.3 ± 3.1% of the TER values before application, verifying the inhibitory effect of Y306A on cCPE activity (17). Incubation with GST-cCPER227A or GST-cCPEL254A/S256A/I258A/D284A resulted in TER values significantly higher than that for GST-cCPEWT (51.5 ± 3.5% and 67.1 ± 4.2% relative to values of TER before application, respectively, Fig. 3). These data demonstrate that the cCPE substitutions R227A and L254A/S256A/I258A/D284A inhibit the cCPE-mediated decrease of TER.
FIGURE 3.
Substitutions L254A/S256A/I258A/D284A (LSID) and R227A in cCPE inhibit cCPE-mediated decrease in TER. MDCK I cells were cultured on Millicell® cell culture inserts, and TER was monitored by CellZscope. Cells were incubated for 18 h with 3.5 μg/ml GST or GST-cCPE constructs. TER values relative to the values before cCPE application were calculated. Results are means ± S.E. (error bars); n = 5. *, p < 0.05 to GST-cCPEWT.
Claudins
Cld3-mimicking Residues Introduced into Cld4 Led to Partial Cld3-like Binding
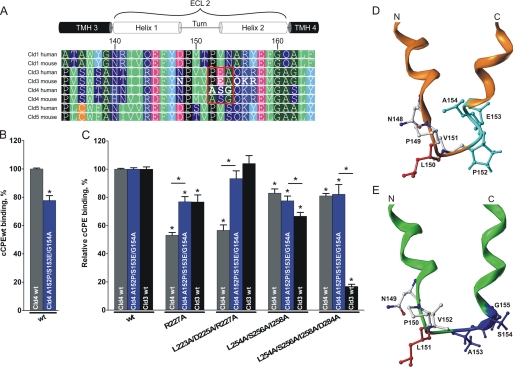
To investigate the differences between Cld3 and Cld4 binding of cCPE in more detail, we compared the amino acid sequences of ECL2 for these claudins (Fig. 4A). The most striking difference is 152PEA154 in Cld3 instead of the corresponding 153ASG155 in Cld4 (Fig. 4A, box). Hence, Cld4A153P/S154E/G155A was generated, and its interaction with GST-cCPE was tested using cellular binding assays. Compared with Cld4WT, the Cld4 triple mutant showed 22.3 ± 3.4% less binding of GST-cCPEWT (Fig. 4B). Next, binding of GST-cCPE mutants to Cld3WT-, Cld4WT-, and Cld4A153P/S154E/G155A-expressing cells was compared. The binding of a given GST-cCPE mutant to a particular claudin construct was quantified relative to the respective binding of GST-cCPEWT (relative binding). Interestingly, the relative binding of GST-cCPER227A and GST-cCPEL223A/D225A/R227A to Cld4A153P/S154E/G155A was significantly higher than to Cld4WT. In fact, the binding was as high as the relative binding of these GST-cCPE mutants to Cld3WT (Fig. 4C). In contrast, the relative binding of GST-cCPEL254A/S256A/I258A and GST-cCPEL254A/S256A/I258A/D284A to Cld4A153P/S154E/G155A was not significantly different from that to Cld4WT (Fig. 4C). These data indicate that Cld3-mimicking substitutions in Cld4 (A153P/S154E/G155A) induce Cld3-like sensitivity of Cld4 toward substitutions in upper but not lower rim of binding pocket of cCPE.
FIGURE 4.
Cld3-mimicking substitutions in Cld4 (A153P/S154E/G155A) decrease Cld4-related strong inhibitory effect of R227A and L223A/D225A/R227A in cCPE. A, sequence alignment of ECL2 of Cld1, 3, 4, and 5 (ClustalW2) is displayed using Geneious Pro version 5.3.4. Position numbering is according to murine Cld3. Red frame, striking difference between Cld3 and Cld4; green, bulky hydrophobic; dark green, small hydrophobic; dark blue, polar uncharged; blue, basic; magenta, acidic; cyan, Tyr; black, Pro; orange, Cys; bold letters, substituted residues; TMH, transmembrane helix. B and C, HEK cells expressing Cld3WT (black columns), Cld4WT (gray columns), or Cld4A153P/S154E/G155A (blue columns) were incubated with 0.5 μg/ml GST-cCPEWT or GST-cCPE mutant. Bound cCPE was detected using anti-GST antibodies in a plate reader. B, GST-cCPEWT binds to the Cld3-mimicking Cld4 mutant (Cld4A153P/S154E/G155A) more weakly than to Cld4WT. Results are mean ± S.E. (error bars); n ≥ 4. *, p < 0.05. C, quantification, normalized to GST-cCPEWT (relative binding) for each claudin construct. A153P/S154E/G155A induces a Cld3-like sensitivity of Cld4 to R227A and L223A/D225A/R227A mutants of cCPE. Results are mean ± S.E.; n ≥ 5. *, p < 0.05 to GST-cCPEWT. D and E, differences in the homologous helix-turn-helix model for ECL2 of Cld3 (orange) and Cld4 (green) are illustrated. Front view shows turn-flanking residues Leu150/Leu151 highlighted in red and the differing N-terminal cap conformation of the C-terminal helix in cyan (Cld3; based on PDB code 1W5C) or blue (Cld4; based on PDB code 2BDV).
Homology Models of ECL2 in Cld3 and Cld4 Indicate Similar but Not Identical Conformations
To explain the differences in binding of cCPE to Cld3 and Cld4, homology models of ECL2 were generated for both claudins (Fig. 4, D and E). The ECL2 conformation is suggested as a helix-turn-helix motif, which is similar to the ECL2 model of Cld5 previously predicted by us based on homologous helix-turn-helix motif structure (29). The only noticeable difference between Cld3 and Cld4 in ECL2 is a triple sequence (152PEA154 in Cld3 and 153ASG155 in Cld4). Consequently, comparative modeling of ECL2 in Cld3 and Cld4 resulted in a helix-turn-helix segment for both proteins. However, they differ in the N-terminal cap conformation of the respective C-terminal helix (helix 2) precisely at the variant triple sequence (Fig. 4, D and E, cyan and dark blue, respectively), which is also involved in differential cCPE binding. Consequently, the tilt of the C-terminal helix of ECL2 is likely different in both claudins.
In Cld3 N148D but Not Q155E, K156A, or R157Y Affects Binding to cCPE
We showed previously that N148D in ECL2 of Cld3 inhibits the binding of GST-cCPE (23). Others suggested that the pI of the ECL2 of claudins contributes to the affinity to CPE (22). To investigate this in more detail, additional Cld3-ECL2 mutants (E153V, Q155E, K156A, and R157Y) with altered pI (supplemental Table S1) were analyzed. In contrast to Cld3N148D, Cld3R157Y bound to GST-cCPE as strongly as to Cld3WT (Fig. 5A, B, and C, left panel). Furthermore, we tried to enhance the effects of substitutions in Cld3 by combining them with substitutions in GST-cCPE. Indeed, substitutions in GST-cCPE (Y306A, L254A, D284A, D284N, and L254A/S256A/I258A) that were shown to inhibit binding to Cld3WT (Fig. 2) completely blocked binding of GST-cCPE to Cld3N148D-transfected cells (Fig. 5C, middle left panel). However, such an additive effect was not obtained for Cld3R157Y-transfected cells because the GST-cCPE mutants bound to these cells (Fig. 5C, middle right panel). The results verify involvement of Asn148 in Cld3 in the interaction with cCPE but do not support participation of Arg157.
FIGURE 5.
Effect of substitutions in Cld3 on binding of cCPE and mutants thereof. HEK cells transfected with Cld3WT or Cld3 mutant were used for pulldown (A and B) or cellular binding assays (C–E). For pulldown assays, representative blots (A) and quantification (B) are shown. Cld3N148D interacts with GST-cCPEWT more weakly than Cld3WT, whereas R157Y in Cld3 does not affect binding. Results are mean ± S.E. (error bars); n = 4. *, p < 0.05 to Cld3WT. C, cellular binding assays analyzed by Western blotting verified inhibition of cCPE binding by N148D but not R157Y (left panel). N148D in Cld3 blocks binding of cCPE mutants (middle left panel) whereas R157Y does not enhance the effects of substitutions in cCPE (middle right panel). D and E, cellular binding assays were analyzed using a fluorescence plate reader. GST-cCPEWT binds to Cld3K156A as well as to Cld3WT. D, additive effect of changed charge in ECL2 of Cld3 (E153V, K156A, R157Y) and substitutions in cCPE was tested. E, binding of GST-cCPE mutants to Cld3 construct was quantified relative to GST-cCPEWT binding (relative binding). Substitutions K156A and R157Y do not change the effect of substitutions in cCPE considerably. E153V slightly decrease relative binding of GST-cCPEY306A. Results show mean ± S.E. (error bars); n ≥4). *, p < 0.05 to GST-cCPEWT.
Next, Cld3Q155E was analyzed. Although Cld3Q155E showed GST-cCPE binding similar to Cld3WT in pulldown assays (23) we looked for additive effects after combining Q155E in Cld3 with substitutions in cCPE. GST-cCPE mutants (Y306A, L254A, D284A, and D284N) bound more weakly to Cld3Q155E-transfected cells than GST-cCPEWT did (Fig. 5C, right panel). However, binding was not blocked, showing that an additive effect, as found for Cld3N148D, was not obtained. This finding indicates that Gln155 in Cld3 does not contribute to CPE binding.
In addition, K156A in Cld3 did not inhibit interaction with GST-cCPEWT (Fig. 5D). Again, we looked for additive effects by combining K156A in Cld3 with substitutions in cCPE (Fig. 5E). The binding of each GST-cCPE mutant to a Cld3 construct was quantified relative to the binding of GST-cCPEWT to the same Cld3 construct (relative binding). As observed for Cld3R157Y, no significant difference in the relative binding of GST-cCPE mutants (D284A, Y306A, and L254A/S256A/I258A) with Cld3K156A was found compared with Cld3WT. The relative binding of GST-cCPE mutants to Cld3E153V compared with Cld3WT was slightly reduced, although this reduction reached statistical significance only for GST-cCPEY306A. In total, the results do not indicate involvement of Gln155, Lys156, or Arg157 of Cld3 in cCPE binding. However, the data suggest that Glu153 has a minor contribution to the interaction.
Claudin-cCPE Interaction: Orientation of ECL2 within the Binding Pocket of cCPE
According to our ECL2 models (Fig. 4, D and E), Leu150 of Cld3 and corresponding Leu151 of Cld4 could either fit in the deep triple Tyr pit or in the less deep triple Leu pit of cCPE (Fig. 6A, Ia and IIa). To clarify the orientation of the ECL2 of claudins within the binding pocket of cCPE, we substituted Leu150/Leu151 with Phe, a bulkier and larger residue (Cld3L150F, Cld4L151F), because this would fit better in the deeper triple Tyr pit (Fig. 6A, Ib) than in the less deep triple Leu pit (Fig. 6A, IIb). Both Cld3L150F and Cld4L151F interacted with GST-cCPEWT and showed no significant change in binding compared with the corresponding wild-type claudin (Fig. 6B). This indicates that Leu150/Leu151 might bind into the triple Tyr pit. Additionally, this is also supported by the interaction models of cCPE with ECL2 of Cld3WT (Fig. 6D) and Cld4WT (supplemental Fig. S1), where in both cases the shape of side chain Leu150/Leu151 is complementary to the shape of the triple Tyr pit. Furthermore, the interaction model for the L150F/L151F mutants indicate that the substituted side chains can only fit properly in same orientation with the triple Tyr pit and not the triple Leu pit (supplemental Fig. S2).
FIGURE 6.
L150F in Cld3 and L151F in Cld4 do not block binding to cCPE but rescue the inhibitory effect of Y306A in cCPE. A, scheme of two possible orientations of ECL2 in Cld3 and Cld4 within the putative binding pocket of cCPE, which has two pits differing in shape. As shown on the scheme, the turn flanking residues Leu150/Leu151 (L) and Pro152/Ala153 (P/A) of Cld3WT or Cld4WT could fit in both pits of the cCPE binding pocket (Ia and IIa). However, for the bulkier L150F/L151F mutants of Cld3/4 only one orientation (Ib) could fit. C, N, C-/N-terminal tails. B, binding assay results of cells transfected with Cld3WT (black columns) and Cld3L150F (green columns) or with Cld4WT (gray columns) and Cld4L151F (red columns). Bound GST-cCPE was detected by use of a fluorescence plate reader. Substitution L150F in ECL2 of Cld3 and L151F of Cld4 did not influence interaction with GST-cCPEWT. C, binding of a given cCPE mutant to cells expressing a particular claudin construct quantified relative to the respective cCPEWT binding (relative binding). L150F in Cld3 and L151F in Cld4 rescued the inhibitory effect of Y306A but not other substitutions in cCPE. Results are mean ± S.E. (error bars); n ≥ 4. *, p < 0.05 to GST-cCPEWT. The binding data support suggested orientation I (A) and the interaction models. D, L150 in ECL2 of Cld3 fitting perfectly into deeper pit of cCPE binding pocket encircled by Tyr306, Tyr310, and Tyr312. In cross-section of the interaction model, cCPE is illustrated (white) with the surface displayed (green, polar/charged; gray, unpolar; yellow, hydrophobic) and the pocket-defining residues in ball and stick. Cld3 is shown in orange, with the residues of the turn region as ball and stick (white, C and H; blue, N; red, O) and the surface of Leu150 (orange mesh). E, interaction model for Cld3L150F with cCPEY306A. Y306A (magenta) widens the triple Tyr pit, and Phe150 resembles the aromatic ring structure of Tyr306 in cCPE thereby re-establishing the hydrophobic core of the triple Tyr pit.
To substantiate orientation I further, binding of a given GST-cCPE mutant to the Cld3 and Cld4 constructs was quantified relative to the binding of GST-cCPEWT to the respective claudin construct (relative binding, Fig. 6C). As shown above (Fig. 2, B and C), binding of GST-cCPEY306A to Cld3WT-transfected HEK cells was significantly decreased compared with GST-cCPEWT (Fig. 6C). Strikingly, binding of GST-cCPEY306A to Cld3L150F or Cld4L151F was only slightly weaker than binding of GST-cCPEWT (Fig. 6C). In contrast, binding of other GST-cCPE mutants (cCPEL254A, cCPED284A, cCPEL254A/S256A/I258A, and cCPEL223A/D225A/R227A) was not increased but rather partly decreased by the L150F/L151F substitution (Fig. 6C). The data show that L150F in Cld3 and L151F in Cld4 specifically rescue the inhibitory effect of Y306A in cCPE, but not that of the other substitutions in cCPE. These results strongly support binding in orientation I (Fig. 6A).
DISCUSSION
Here, we aimed to answer the question of how the interface between CPE and the ECL2 of Cld3 and Cld4 is arranged at the molecular level. Novel mechanistic insights into the interaction were identified by combining structural information, molecular modeling, different binding assays, and mutagenesis. None of the analyzed mutations prevented the ability of the respective claudin construct to reach the plasma membrane (supplemental Fig. S5). This indicates that the substitutions did not induce misfolding of the protein which would interfere with the binding assays.
As a well established method to analyze claudin-cCPE interaction, a pulldown assay was applied (17, 23). However, since detergent solubilization might change the structure of transmembrane proteins, additional cellular binding assays were used to analyze claudins in their native membrane environment. In general, similar results were obtained with the different binding assays (summarized in supplemental Table S1). The only noticeable difference was observed for the substitution L223A in cCPE. We propose that this could be due to the above mentioned solubilization having a different effect on the structure of Cld3 compared with Cld4. This interpretation is consistent with the idea that the ECL2 structure of Cld3 and Cld4 partly differs (Fig. 4, D and E).
Comprehensive Mapping of the Binding Pocket for Claudins in cCPE
Previously, it has been shown that mutation of the hydrophobic residues Tyr306, Tyr310, Tyr312, and Leu315 disturb Cld4 binding (16). Inspecting the surface of the cCPE structure revealed that these residues are distributed in a fragmentary pattern around a rather hydrophobic cavity. Because several amino acids that surround this cavity had not yet been investigated, we performed site-directed mutagenesis of these remaining residues (Fig. 7). Our results clearly show that the residues Leu223, Arg227, Leu254, Ile258 (and/or Ser256) and Asp284 of cCPE, which are flanking the cavity on both sides, are involved in binding to Cld3 and/or Cld4. The structure of cCPE used here (PDB code 2QUO) (18) shows no relevant differences in the region of the binding pocket compared with the recently published structure of full-length CPE (PDB code 3AM2) (19).
FIGURE 7.
Superimposed interaction models of ECL2 for Cld3 and Cld4 bound to the surface of the cCPE structure. cCPE residues identified by mutation as binding sensitive preferentially for Cld3 (cyan) or for Cld4 (red) are discrete located at the lower and upper rim of the binding pocket. The ECL2 helix-turn-helix models demonstrate common (N-terminal helix) but also partly differing binding modes (C-terminal helix) between Cld3 (orange) and Cld4 (green). This opposing helix tilt is caused by the variant helix capping sequence motifs 152PEA154 and 153ASG155, of Cld3 and Cld4, respectively (side chains not visualized for clarity, see Fig. 4D) Thus, the C-terminal ECL2 helix of Cld3 is tilted toward residues showing stronger mutational effects on Cld3 binding (cyan), whereas that of Cld4 is tilted to residues exhibiting stronger effects on Cld4 binding (red). Residues previously shown to participate in Cld4 binding are labeled in blue.
ECL2 Orientation in the Pocket of cCPE
Leu150 of Cld3 and L151 of Cld4 bind to the triple Tyr pit of cCPE
Our results indicate that Cld3L150F or Cld4L151F can only interact with cCPE in orientation I (Fig. 6A, Ib) because the bulkier Phe side chain can be placed into the triple Tyr pit but not into the less deep triple Leu pit (Fig. 6A, IIb), supporting that ECL2 of Cld3WT and Cld4WT is oriented within the binding pocket of cCPE in orientation I (Fig. 6A, Ia). This orientation was further substantiated by three other findings. First, Cld3L150F and Cld4L151F rescue the impaired binding of GST-cCPEY306A but not other mutants (Fig. 6C). Not a bulk reduction by the L150A mutation in Cld3 but the larger phenyl ring introduced by L150F in Cld3 or L151F in Cld4 is likely to compensate for the lack of an aromatic ring in the triple Tyr pit caused by the Y306A substitution in cCPE (Fig. 6E). Second, contrariwise at the triple Leu pit of cCPE the decrease in Cld binding by Ala substitutions such as by L254A cannot be rescued by Cld3L150F. Third, the alternative orientation II, where the N-terminal ECL2 helix is located close to the triple Leu pit of cCPE (Fig. 6A, IIa), is ruled out by testing a potential interaction between Asn148 in Cld3 and Asp284 in cCPE (Asn148 ↔ Asp284). Initially, we suspected an interaction such as that predicted in orientation II because N148D in Cld3 (23) and corresponding N149D in Cld4 (30) and D284A in cCPE disturbed the binding. This disturbance might be caused either by polar repulsion (Asp148 ↔ Asp284) or missing H-bond acceptors (Asn148 ↔ Ala284). However, after enabling the possibility of H-bond interactions to form between the two complementary mutated positions (Asp148 ↔ Asn284), we found that D284N in cCPE did not rescue the inhibition of cCPE-Cld3 interaction caused by N148D in Cld3 (Fig. 5C, middle left panel). Accordingly, the substitution D284N in cCPE also did not enable or enhance cCPE binding to Cld5 or Cld1 (supplemental Fig. S3), which also both have an Asp in the position corresponding to Asn148 in Cld3. This fits in with our suggested orientation I of cCPE-bound ECL2 in which Asn148/Asn149 is placed close to the triple Tyr pit, and thus the accessibility to allow interaction with D284 is spatially prevented.
Hence, all of these results strongly support binding in orientation I, where the triple Tyr pit of cCPE interacts with large and bulky hydrophobic residues (Leu150/Leu151 in mouse Cld3 and human Cld4; Met151 in mouse Cld4; Fig. 4A), and the triple Leu pit interacts with less bulky hydrophobic residues (Pro152/Ala153). Thereby the turn region (PLVP/A) of ECL2, previously shown to be important for interaction with cCPE (23), binds along the cavity.
The strong dependence of the claudin-cCPE binding on hydrophobic interactions is underlined by improved Cld4 binding of cCPE mutants containing Ala at positions of polar residues (Ser304, Ser305, Ser307, Asn309, Ser313) close to the binding pocket (16). The fact that multiple mutations at these six positions partially affect Cld4 binding (31) is consistent with our model too.
Others have suggested electrostatic attraction between positively charged residues in the ECL2 of CPE-sensitive claudins and negatively charged counterparts at CPE (22) (see supplemental Fig. S4 for electrostatic potential map). We found that single substitutions of basic residues in the ECL2 of Cld3 (K156A, R157Y) did not significantly reduce the Cld3-cCPE interaction. Therefore, we propose that the basic residues in the ECL2, at least for Cld3, are not significantly involved in binding. However, the findings of Kimura et al. are consistent with electrostatic interaction mediated by Asn148 in Cld3 (22), verified in this study, and Asn149 in Cld4 (30).
Claudin Subtype-specific Differences in Structure of ECL2
For claudins, no crystal structures are currently available. Combining mutagenesis studies and molecular modeling enables insights into the structural characteristics of transmembrane proteins. Based on an experimentally supported homology model of Cld5 (28), we propose here similar structures (helix-turn-helix motif) for ECL2 of Cld3 and Cld4. Molecular docking of ECL2 models using the crystal structure of cCPE proved to be a suitable tool to validate the ECL2 models of claudins. Despite a predicted general similarity in conformation (Refs. 23, 28 and this study), we also describe structural differences between the ECL2 of Cld3 and Cld4. (Figs. 4 and 7).
Cld3 and Cld4 Show in Part a Different Mode of Binding cCPE
Cld3-mimicking substitutions of the helix-capping residues in Cld4 (A153P/S154E/G155A) lead to Cld3-like cCPE binding functionality, suggesting a different binding behavior (Fig. 4C). These results are not only consistent with the predicted slightly different ECL2 conformations for Cld3 and Cld4 (Fig. 4, D and E) but are also concordant with the proposed different binding mode for Cld3 and Cld4 with cCPE (Fig. 7).
Whereas the N-terminal ECL2 helices (helix 1) of both Cld3 and Cld4 interact in a similar way with the triple Tyr pit of cCPE, the C-terminal ECL2 helix (helix 2) of Cld3 is tilted closer to the lower rim of the triple Leu pit of cCPE than the C-terminal ECL2-helix of Cld4, which is closer to the upper rim (Fig. 7). This corresponds with the finding that substitution of the residues located on the upper rim of the cCPE binding pocket has a stronger influence on the binding to Cld4 (Fig. 7, red highlighted residues), whereas substitution of the residues on the lower rim of the cCPE binding pocket has a stronger influence on the binding to Cld3 (Fig. 7, cyan highlighted residues).
We previously demonstrated binding of GST-cCPE116–319 to synthetic mouse Cld3 but not to mouse Cld4 peptides (23). A152P (together with M150L) enabled Cld3-like binding of Cld4 peptides to GST-cCPE116–319 (23). This is in line with the Cld3-like binding characteristics described here of full-length Cld4 containing the Cld3-mimicking 152PEA154. The multiple substitution in the upper rim (L223A/D225A/R227A) had a weaker effect than R227A alone. This indicates that removal of the negative charge of Asp225 partially rescues the effect of removal of the positive charge at Arg227. As a consequence, GST-cCPEL223A/D225A/R227A showed only a minor reduction in Cld3 binding but strong inhibition of Cld4 binding. In contrast, GST-cCPEL254A/S256A/I258A/D284A shows strong binding to Cld4 but weak binding to Cld3. Hence, along with our mechanistic analysis we obtained GST-cCPE mutants with shifted claudin subtype specificity.
In summary, we propose claudin subtype-specific structural differences in the ECL2, which could contribute to the subtype-specific barrier properties. In addition to the ECL1 (4, 32), ECL2 also contributes to paracellular tightening (33, 34). Hence, our data are relevant for mechanistic understanding of paracellular barrier formation (27). Furthermore, we have mapped the cCPE-Cld3/4 interaction sites comprehensively and present a mutation-guided docking of the ECL2 of Cld3 and Cld4 onto the cCPE structure by discriminating between two possible binding orientations. Our data strengthen a previously suggested helix-turn-helix conformation for claudin ECL2 and improve the molecular understanding of these tight junction proteins with yet unknown structure. Along with distinct binding data and slightly different ECL2 sequences partly differing binding modes between Cld3 and Cld4 were identified.
However, by our major finding, we prove that claudin subtype specificity of cCPE can be altered by structure-based molecular modification of cCPE. This facilitates the improved targeting of specific claudins. Here, as an initial step, we narrowed the specificity by generating claudin binders preferentially either for Cld3 or for Cld4. Several carcinomas overexpress particular claudins (including Cld3 or Cld4) in a subtype-specific manner (15, 24, 25). Targeting these tumors could be enhanced by use of cCPE-variants with optimized claudin subtype specificity (31, 35). Moreover, the prospective design of cCPE-variants binding to claudin subtypes that are not recognized by cCPEWT might be achieved. cCPE variants that bind to Cld5 or Cld1 could either improve modulation of tight junctions and in turn paracellular drug delivery across the blood brain barrier (9, 37) or mask Cld1 as a cofactor of hepatitis C virus entry and therefore inhibit virus infection (36).
Supplementary Material
Acknowledgments
We thank Dr. Y. Horiguchi (Osaka, Japan) for the cDNA of CPE and Dr. W. Hunziker (Singapore) for a plasmid encoding GFP-tagged human Cld4.
This work was supported by Deutsche Forschungsgemeinschaft Grants KR 1273/3-2 and PI 837/2-1.

This article contains supplemental Figs. S1–S6, Table S1, a Comment, and additional references.
- CPE
- C. perfringens enterotoxin
- cCPE
- domain I of CPE
- Cld
- claudin
- ECL
- extracellular loop
- MDCK
- Madin-Darby canine kidney
- PDB
- Protein Data Bank
- TER
- transepithelial resistance.
REFERENCES
- 1. McClane B. A. (2001) The complex interactions between Clostridium perfringens enterotoxin and epithelial tight junctions. Toxicon 39, 1781–1791 [DOI] [PubMed] [Google Scholar]
- 2. Katahira J., Sugiyama H., Inoue N., Horiguchi Y., Matsuda M., Sugimoto N. (1997) Clostridium perfringens enterotoxin utilizes two structurally related membrane proteins as functional receptors in vivo. J. Biol. Chem. 272, 26652–26658 [DOI] [PubMed] [Google Scholar]
- 3. Veshnyakova A., Protze J., Rossa J., Blasig I. E., Krause G., Piontek J. (2010) On the interaction of Clostridium perfringens enterotoxin with claudins. Toxins 2, 1336–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Angelow S., Ahlstrom R., Yu A. S. (2008) Biology of claudins. Am. J. Physiol. Renal Physiol. 295, F867–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Krause G., Winkler L., Mueller S. L., Haseloff R. F., Piontek J., Blasig I. E. (2008) Structure and function of claudins. Biochim. Biophys. Acta 1778, 631–645 [DOI] [PubMed] [Google Scholar]
- 6. Kokai-Kun J. F., McClane B. A. (1997) Deletion analysis of the Clostridium perfringens enterotoxin. Infect. Immun. 65, 1014–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Smedley J. G., 3rd, Uzal F. A., McClane B. A. (2007) Identification of a prepore large-complex stage in the mechanism of action of Clostridium perfringens enterotoxin. Infect. Immun. 75, 2381–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Takahashi A., Kondoh M., Suzuki H., Yagi K. (2011) Claudin as a target for drug development. Curr. Med. Chem. 18, 1861–1865 [DOI] [PubMed] [Google Scholar]
- 9. Kondoh M., Takahashi A., Fujii M., Yagi K., Watanabe Y. (2006) A novel strategy for a drug delivery system using a claudin modulator. Biol. Pharm. Bull. 29, 1783–1789 [DOI] [PubMed] [Google Scholar]
- 10. Sonoda N., Furuse M., Sasaki H., Yonemura S., Katahira J., Horiguchi Y., Tsukita S. (1999) Clostridium perfringens enterotoxin fragment removes specific claudins from tight junction strands: evidence for direct involvement of claudins in tight junction barrier. J. Cell Biol. 147, 195–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Saeki R., Kondoh M., Kakutani H., Tsunoda S., Mochizuki Y., Hamakubo T., Tsutsumi Y., Horiguchi Y., Yagi K. (2009) A novel tumor-targeted therapy using a claudin-4-targeting molecule. Mol. Pharmacol. 76, 918–926 [DOI] [PubMed] [Google Scholar]
- 12. Kominsky S. L., Tyler B., Sosnowski J., Brady K., Doucet M., Nell D., Smedley J. G., 3rd, McClane B., Brem H., Sukumar S. (2007) Clostridium perfringens enterotoxin as a novel-targeted therapeutic for brain metastasis. Cancer Res. 67, 7977–7982 [DOI] [PubMed] [Google Scholar]
- 13. Casagrande F., Cocco E., Bellone S., Richter C. E., Bellone M., Todeschini P., Siegel E., Varughese J., Arin-Silasi D., Azodi M., Rutherford T. J., Pecorelli S., Schwartz P. E., Santin A. D. (2011) Eradication of chemotherapy-resistant CD44+ human ovarian cancer stem cells in mice by intraperitoneal administration of Clostridium perfringens enterotoxin. Cancer 117, 5519–5528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Walther W., Petkov S., Kuvardina O. N., Aumann J., Kobelt D., Fichtner I., Lemm M., Piontek J., Blasig I. E., Stein U., Schlag P. M. (October 6, 2011) Novel Clostridium perfringens enterotoxin suicide gene therapy for selective treatment of claudin-3- and -4-overexpressing tumors. Gene Ther. 10.1038/gt.2011.136 [DOI] [PubMed] [Google Scholar]
- 15. Turksen K., Troy T. C. (2011) Junctions gone bad: claudins and loss of the barrier in cancer. Biochim. Biophys. Acta 1816, 73–79 [DOI] [PubMed] [Google Scholar]
- 16. Takahashi A., Komiya E., Kakutani H., Yoshida T., Fujii M., Horiguchi Y., Mizuguchi H., Tsutsumi Y., Tsunoda S., Koizumi N., Isoda K., Yagi K., Watanabe Y., Kondoh M. (2008) Domain mapping of a claudin-4 modulator, the C-terminal region of C-terminal fragment of Clostridium perfringens enterotoxin, by site-directed mutagenesis. Biochem. Pharmacol. 75, 1639–1648 [DOI] [PubMed] [Google Scholar]
- 17. Harada M., Kondoh M., Ebihara C., Takahashi A., Komiya E., Fujii M., Mizuguchi H., Tsunoda S., Horiguchi Y., Yagi K., Watanabe Y. (2007) Role of tyrosine residues in modulation of claudin-4 by the C-terminal fragment of Clostridium perfringens enterotoxin. Biochem. Pharmacol. 73, 206–214 [DOI] [PubMed] [Google Scholar]
- 18. Van Itallie C. M., Betts L., Smedley J. G., 3rd, McClane B. A., Anderson J. M. (2008) Structure of the claudin-binding domain of Clostridium perfringens enterotoxin. J. Biol. Chem. 283, 268–274 [DOI] [PubMed] [Google Scholar]
- 19. Kitadokoro K., Nishimura K., Kamitani S., Fukui-Miyazaki A., Toshima H., Abe H., Kamata Y., Sugita-Konishi Y., Yamamoto S., Karatani H., Horiguchi Y. (2011) Crystal structure of Clostridium perfringens enterotoxin displays features of β-pore-forming toxins. J. Biol. Chem. 286, 19549–19555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Briggs D. C., Naylor C. E., Smedley J. G., 3rd, Lukoyanova N., Robertson S., Moss D. S., McClane B. A., Basak A. K. (2011) Structure of the food-poisoning Clostridium perfringens enterotoxin reveals similarity to the aerolysin-like pore-forming toxins. J. Mol. Biol. 413, 138–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fujita K., Katahira J., Horiguchi Y., Sonoda N., Furuse M., Tsukita S. (2000) Clostridium perfringens enterotoxin binds to the second extracellular loop of claudin-3, a tight junction integral membrane protein. FEBS Lett. 476, 258–261 [DOI] [PubMed] [Google Scholar]
- 22. Kimura J., Abe H., Kamitani S., Toshima H., Fukui A., Miyake M., Kamata Y., Sugita-Konishi Y., Yamamoto S., Horiguchi Y. (2010) Clostridium perfringens enterotoxin interacts with claudins via electrostatic attraction. J. Biol. Chem. 285, 401–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Winkler L., Gehring C., Wenzel A., Müller S. L., Piehl C., Krause G., Blasig I. E., Piontek J. (2009) Molecular determinants of the interaction between Clostridium perfringens enterotoxin fragments and claudin-3. J. Biol. Chem. 284, 18863–18872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Escudero-Esparza A., Jiang W. G., Martin T. A. (2011) The claudin family and its role in cancer and metastasis. Front. Biosci. 16, 1069–1083 [DOI] [PubMed] [Google Scholar]
- 25. Ouban A., Ahmed A. A. (2010) Claudins in human cancer: a review. Histol. Histopathol. 25, 83–90 [DOI] [PubMed] [Google Scholar]
- 26. Blasig I. E., Winkler L., Lassowski B., Mueller S. L., Zuleger N., Krause E., Krause G., Gast K., Kolbe M., Piontek J. (2006) On the self-association potential of transmembrane tight junction proteins. Cell. Mol. Life Sci. 63, 505–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Piontek J., Fritzsche S., Cording J., Richter S., Hartwig J., Walter M., Yu D., Turner J. R., Gehring C., Rahn H. P., Wolburg H., Blasig I. E. (2011) Elucidating the principles of the molecular organization of heteropolymeric tight junction strands. Cell. Mol. Life Sci. 68, 3903–3918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Piontek J., Winkler L., Wolburg H., Müller S. L., Zuleger N., Piehl C., Wiesner B., Krause G., Blasig I. E. (2008) Formation of tight junction: determinants of homophilic interaction between classic claudins. FASEB J. 22, 146–158 [DOI] [PubMed] [Google Scholar]
- 29. Krause G., Winkler L., Piehl C., Blasig I., Piontek J., Müller S. L. (2009) Structure and function of extracellular claudin domains. Ann. N.Y. Acad. Sci. 1165, 34–43 [DOI] [PubMed] [Google Scholar]
- 30. Robertson S. L., Smedley J. G., 3rd, McClane B. A. (2010) Identification of a claudin-4 residue important for mediating the host cell binding and action of Clostridium perfringens enterotoxin. Infect. Immun. 78, 505–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kakutani H., Takahashi A., Kondoh M., Saito Y., Yamaura T., Sakihama T., Hamakubo T., Yagi K. (2011) A novel screening system for claudin binder using baculoviral display. PLoS One 6, e16611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yu A. S., Cheng M. H., Angelow S., Günzel D., Kanzawa S. A., Schneeberger E. E., Fromm M., Coalson R. D. (2009) Molecular basis for cation selectivity in claudin-2-based paracellular pores: identification of an electrostatic interaction site. J. Gen. Physiol. 133, 111–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Piehl C., Piontek J., Cording J., Wolburg H., Blasig I. E. (2010) Participation of the second extracellular loop of claudin-5 in paracellular tightening against ions, small and large molecules. Cell. Mol. Life Sci. 67, 2131–2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang J., Piontek J., Wolburg H., Piehl C., Liss M., Otten C., Christ A., Willnow T. E., Blasig I. E., Abdelilah-Seyfried S. (2010) Establishment of a neuroepithelial barrier by claudin-5a is essential for zebrafish brain ventricular lumen expansion. Proc. Natl. Acad. Sci. U.S.A. 107, 1425–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ebihara C., Kondoh M., Hasuike N., Harada M., Mizuguchi H., Horiguchi Y., Fujii M., Watanabe Y. (2006) Preparation of a claudin-targeting molecule using a C-terminal fragment of Clostridium perfringens enterotoxin. J. Pharmacol. Exp. Ther. 316, 255–260 [DOI] [PubMed] [Google Scholar]
- 36. Fofana I., Krieger S. E., Grunert F., Glauben S., Xiao F., Fafi-Kremer S., Soulier E., Royer C., Thumann C., Mee C. J., McKeating J. A., Dragic T., Pessaux P., Stoll-Keller F., Schuster C., Thompson J., Baumert T. F. (2010) Monoclonal anti-claudin 1 antibodies prevent hepatitis C virus infection of primary human hepatocytes. Gastroenterology 139, 953-964 [DOI] [PubMed] [Google Scholar]
- 37. Nitta T., Hata M., Gotoh S., Seo Y., Sasaki H., Hashimoto N., Furuse M., Tsukita S. (2003) Size-selective loosening of the blood-brain barrier in claudin-5-deficient mice. J. Cell Biol. 161, 653–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.