Background: Tumor differentiation factor (TDF) is a newly identified pituitary protein with no known receptor.
Results: Heat shock 70-kDa proteins are potential TDF receptor candidates.
Conclusion: TDF acts on breast cells through a novel pathway.
Significance: These data may help to elucidate the role of TDF.
Keywords: Analytical Biochemistry, Cell Differentiation, Mass Spectrometry (MS), Protein Chemistry, Receptor, Hormone, Signaling
Abstract
Tumor differentiation factor (TDF) is a recently discovered protein, produced by the pituitary gland and secreted into the bloodstream. TDF and TDF-P1, a 20-amino acid peptide selected from the open reading frame of TDF, induce differentiation in human breast and prostate cancer cells but not in other cells. TDF protein has no identified site of action or receptor, and its mechanism of action is unknown. Here, we used TDF-P1 to purify and identify potential TDF receptor (TDF-R) candidates from MCF7 steroid-responsive breast cancer cells and non-breast HeLa cancerous cells using affinity purification chromatography (AP), and mass spectrometry (MS). We identified four candidate proteins from the 70-kDa heat shock protein (HSP70) family in MCF7 cells. Experiments in non-breast HeLa cancerous cells did not identify any TDF-R candidates. AP and MS experiments were validated by AP and Western blotting (WB). We additionally looked for TDF-R in steroid-resistant BT-549 cells and human dermal fibroblasts (HDF-a) using AP and WB. TDF-P1 interacts with potential TDF-R candidates from MCF7 and BT-549 breast cells but not from HeLa or HDF-a cells. Immunofluorescence (IF) experiments identified GRP78, a TDF-R candidate, at the cell surface of MCF7, BT-549 breast cells, and HeLa cells but not HDF-a cells. IF of other HSP70 proteins demonstrated labeling on all four cell types. These results point toward GRP78 and HSP70 proteins as strong TDF-R candidates and suggest that TDF interacts with its receptor, exclusively on breast cells, through a steroid-independent pathway.
Introduction
TDF2 is a protein produced by the pituitary gland and secreted into the bloodstream, with no identified receptor and no known mechanism of action. TDF and TDF-P1, a 20-amino acid peptide selected from the open reading frame of TDF, induce morphological and biochemical changes in vitro and in vivo that suggest that TDF is involved in the differentiation of human breast and prostate cancer cells (1, 2) Specifically, TDF induces markers of differentiation, such as the polarization and formation of cell junctions and basement membrane, and furthermore induces milk protein synthesis and the overexpression of E-cadherin (3–10). However, TDF has no known morphological differentiation effect on fibroblasts or on kidney, hepatoma, and leukemic lymphocytic cell lines (1, 2). The differentiation activity of TDF has not been reproduced by any of the known pituitary hormones or growth factors (1, 2). TDF is secreted by the pituitary directly into the blood, suggesting that this protein has an endocrine role (1, 2). However, TDF protein is very understudied. It is not yet clear where this protein acts and to what receptor it binds. It is also not clear how TDF protein promotes cell differentiation.
MCF7 human breast cancer cells express the estrogen receptors and are responsive to steroid hormones, manifested through activation of transcription of some genes, leading to increased cell proliferation (11–14). MCF7 human breast cancer cells are also responsive to TDF protein in vitro and in vivo through induction of cell differentiation (1, 2). Therefore, it is of interest to understand whether TDF protein induces differentiation of MCF7 breast cancer cells through a steroid-dependent or steroid-independent pathway. It is of additional interest as to whether the TDF pathway is similar to the estrogen pathway or is a novel pathway.
The first step in understanding the TDF pathway is through the isolation and characterization of the TDF-R. The standard procedure for isolation and characterization of most membrane-bound or intracellular receptors for hormones or growth factors is through AP and Edman sequencing or MS (15, 16). Due to its higher accuracy, sensitivity, cost, and speed, MS has become the method of choice for identifying and sequencing proteins (15, 17–20). Validation of these findings is typically performed using AP, followed by WB using antibodies against TDF-R candidates identified by MS. If validation is positive, then the potential receptor (or receptors) warrants further investigation.
Here, we used TDF-P1 to purify potential TDF-R candidates from MCF7 steroid-responsive breast cancer cells and non-breast HeLa cancerous cells using AP and MS. We used TDF-P1 because we reasoned that if TDF-P1 mimics the effect of full-length TDF protein and induces cell differentiation, then TDF-P1 must interact with the receptor of full-length TDF, and therefore, TDF-P1 could be used to purify the potential TDF receptor candidates. We further investigated the potential TDF-R in these two cell types and additionally in steroid-resistant BT-549 cells (these cells do not express estrogen receptors) (21) and HDF-a by AP, WB, and IF. Our results suggest that TDF-R candidates are members of the HSP70 protein family, are present on breast and cancerous cells but not other cells, and act through a novel steroid-independent pathway. The possibility that TDF-R is a multisubunit protein complex is also discussed.
EXPERIMENTAL PROCEDURES
Peptide Synthesis and Coupling to Agarose Beads
TDF-P1 peptide, with the amino acid sequence NH2-RESQGTRVGQALSFLCKGTA-COOH, was synthesized by standard peptide synthesis (Creative Biolabs), and its correct sequence was confirmed by MS (data not shown). The TDF-P1 peptide was then coupled to CNBr-activated beads (Sigma) according to the manufacturer's instructions.
Cell Culture and Cell Lysis
All cell lines were grown in RPMI 1650 medium supplemented with 10% (v/v) fetal bovine serum (FBS) and 100 units/ml penicillin/streptomycin (all from Invitrogen) and were grown to confluence under standard cell culture conditions (5% CO2, 37 °C). The cells were then washed with cold PBS, pH 7.4, and then lysed using lysis buffer (150 mm NaCl, 20 mm Tris-HCl (pH 8.0), 0.2 mm EDTA, 1% (v/v) Triton-X-100, and SIGMAFASTTM protease inhibitor tablets (Sigma-Aldrich; 1 tablet/100 ml of buffer). The cell lysate was pooled to have a uniform concentration of proteins and then split into 1-ml aliquots in Eppendorf tubes. These tubes were then incubated on ice for 30 min and centrifuged for 20 min at 14,000 rpm at 4 °C in an Eppendorf centrifuge (Epifuge). The pellet contained DNA and unsolubilized material and was discarded. The supernatant was collected and combined and then stored at −80 °C and used as starting material for SDS-PAGE and MS or for validation by SDS-PAGE and WB.
AP of the Potential TDF-R Candidate(s)
The cell lysates were incubated for 4 h at 4 °C with 0.1 mg of agarose-bound TDF-P1 peptide, 10 ml of lysate, washed three times with lysis buffer, and eluted with ImmunoPure® IgG elution buffer (Pierce). The eluates were collected by centrifugation in an Epifuge microcentrifuge for 3 min at 5000 rpm (twice) and then stored at −80 °C for further experiments. A control experiment in which the beads alone were used as bait for the AP experiments was also performed. The eluates from the control AP experiments were also analyzed by MS and WB.
Immunoaffinity Precipitation (IAP)
Recombinant human GRP78 protein was obtained from ProSpec-Tany TechnoGene Ltd. (Rehovolt, Israel). GRP78 protein at a concentration of 1 μg/100 μl was incubated with TDF-P1 (at a concentration of 10 μg/100 μl) in lysis buffer (150 mm NaCl, 20 mm Tris-HCl (pH 8.0), 0.2 mm EDTA, 1% (v/v) Triton X-100, and SIGMAFASTTM protease inhibitor tablets (Sigma-Aldrich; 1 tablet/100 ml of buffer) for 2 h at 4 °C. The mixture of GRP78 protein and TDF-P1 was then incubated for 2 h with 1 μg of mouse monoclonal anti-GRP78 antibodies (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) and protein A/G UltraLink resin (Thermo Scientific, Waltham, MA). Samples were then centrifuged for 30 s at 3000 rpm, and the supernatant was saved for analysis. Pellet was washed three times with ice-cold lysis buffer and boiled for 10 min in Laemmli sample buffer. A negative control with protein A/G and no antibodies was processed in the same manner. All samples were loaded on 16% Tris-HCl gels and separated by SDS-PAGE. Because the molecular mass of the TDF-P1 is small (about 2 kDa), the gels were stopped before the front electrophoresis ran out of the gel. The gels were then electroblotted and incubated with relevant primary antibody (produced in a different host than the antibodies used for immunoprecipitation): rat monoclonal GRP78 (Santa Cruz Biotechnology, Inc.) and rabbit polyclonal TDF generated against TDF-P1 (Creative Biolabs, Shirley, NY). Secondary antibodies were HRP-conjugated: goat-anti-rat IgG-HRP and goat-anti-rabbit IgG-HRP (Santa Cruz Biotechnology, Inc.). The signal was visualized by an enhanced chemiluminescent (ECL) reaction kit (Pierce).
SDS-PAGE and WB
The eluates obtained from AP experiments were solubilized for 5 min in Laemmli sample buffer at 95 °C, loaded on 8% Tris-HCl gels, and separated by SDS-PAGE. In some experiments, the APs were first concentrated five times on a low molecular weight Amicon concentrator (Millipore, Bedford, MA). The gels were then stained by Coomassie or electroblotted on BiotraceTM NT nitrocellulose membrane (Pall Life Sciences, Port Washington, NY), incubated with different commercial primary GRP78, HSP70 (Santa Cruz Biotechnology, Inc.), and HRP-conjugated secondary antibodies (goat-anti-rat IgG-HRP and goat-anti-mouse IgG-HRP (Santa Cruz Biotechnology, Inc.)) and then visualized by an ECL reaction kit (Pierce). The Coomassie-stained gels were the starting material for MS-based analysis.
Protein Digestion and Peptide Extraction
Proteins that were separated by SDS-PAGE and stained by Coomassie dye were cut in 4–6 gel pieces and washed in high purity, high performance liquid chromatography (HPLC) grade water, cut into small pieces, and destained by incubating in 50 mm ammonium bicarbonate; 50 mm ammonium bicarbonate, 50% acetonitrile; and 100% acetonitrile under moderate shaking, followed by drying in a SpeedVac concentrator (22). The gel bands were then rehydrated with 50 mm ammonium bicarbonate. The procedure was repeated twice. The gel bands were then rehydrated in 50 mm ammonium bicarbonate containing 10 mm DTT and incubated at 56 °C for 45 min. The DTT solution was then replaced by 50 mm ammonium bicarbonate containing 100 mm iodoacetamide for 45 min in the dark with occasional vortexing. The gel pieces were then reincubated in 50 mm ammonium bicarbonate, 50% acetonitrile, and 100% acetonitrile under moderate shaking, followed by drying in a SpeedVac concentrator. The dry gel pieces were then rehydrated using 50 mm ammonium bicarbonate containing 10 ng/μl trypsin and incubated overnight at 37 °C under low shaking. The resulting peptides were extracted twice with 5% formic acid, 50 mm ammonium bicarbonate, 50% acetonitrile and once with 100% acetonitrile under moderate shaking. The peptide mixture was then dried in a SpeedVac, solubilized in 20 μl of 0.1% formic acid, 2% acetonitrile.
MS and Protein Identification
The resulting peptide mixture was analyzed by reverse phase liquid chromatography (LC) and MS (LC-MS/MS) using an Alliance 2695 HPLC (Waters Corp., Milford, MA) coupled to a Q-Tof Micro MS (Micromass/Waters, Milford, MA). The peptides were loaded onto an XBridgeTM C18 3.5-μm, 2.1 × 100-mm column (Waters Corp.) and eluted over a 60-min gradient of 2–100% acetonitrile in 0.1% formic acid at a flow rate of 200 μl/min. MS data acquisition involved survey MS scans and automatic data-dependent MS/MS of 2+, 3+, or 4+ ions. The MS/MS was triggered when the MS signal intensity exceeded 10 counts/s. In survey MS scans, the three most intense peaks were selected for collision-induced dissociation and fragmented until the total MS/MS ion counts reached 5000 or for up to 6 s each. Additional experiments were performed using a nanoLC-MS/MS that contained a Micromass-QTOF hybrid mass spectrometer (Waters Corp.) with a nanoelectrospray source. A fused silica tip mounting adaptor, fitted with a 75-μm (inner diameter) fused silica tip (New Objective), was connected through 50-μm (inner diameter) fused silica tubing to the LC detector outlet. An LC Packings system (Dionex Corp., Sunnyvale, CA), equipped with an Ultimate micropump and solvent organizer and a Switchos loading pump and Famos autosampler, was used for LC-MS. Separation was carried out on a 75 μm × 15-cm column (LC Packings C18 PepMap; 5-μl injection volume) at a flow rate of 200 nl/min, using a gradient of 2–80% acetonitrile in 0.1% formic acid. The mass spectrometer was operated in the data-dependent mode and automatically switched between MS and MS/MS. In survey MS scans, the seven most intense peaks were selected for collision-induced dissociation and fragmented. A full description of the nanoLC-MS/MS analysis and data processing can be found elsewhere (23–25). The raw data were processed using ProteinLynx Global Server (PLGS, version 2.4) software with the following parameters: background subtraction of polynomial order 5 adaptive with a threshold of 35%, two smoothings with a window of three channels in Savitzky-Golay mode, and centroid calculation of the top 80% of peaks based on a minimum peak width of 4 channels at half-height. The resulting pkl files were submitted for database search and protein identification to the public Mascot database search (see the Web site for Matrix Science (London, UK)) using the following parameters: human databases from NCBI and SwissProt; parent mass error, 1.2 Da; product ion error, 0.6 Da; enzyme used, trypsin; one missed cleavage; and carbamidomethyl-cysteine as fixed modification and methionine oxidized as variable modification. To identify the false negative results, we varied our database search parameters and compared the results with each other. To eliminate false positive results, we manually checked the MS/MS spectra that led to identification of a protein. The pkl files were also searched against an in-house PLGS database (see the Waters Web site) using searching parameters similar to the ones used for Mascot search. The Mascot and PLGS database search provided a list of proteins for each gel band. Only proteins identified by two or more peptides and with a Mascot score of >50 were considered. For the proteins identified by a mascot score lower than 50, the MS/MS spectra of these peptides were manually inspected to confirm the identity of the peptide.
IF
Cells were grown for 24 h on coverslides in 6-well plates and washed with PBS three times before staining using 3 μm CM-Dil (Invitrogen) for 5 min at 37 °C followed by 15 min at 4 °C. Afterward, cells were fixed in 4% paraformaldehyde in PBS for 15 min at room temperature. They were then incubated in 0.1% Triton, 50 mm NH4Cl, PBS for 5 min at room temperature. Cells were blocked for 60 min at room temperature in blocking buffer (10% normal donkey serum in Tris-buffered saline, pH 7.4 (TBS)) and then incubated with relevant primary antibody (GRP78 or HSP70 (Santa Cruz Biotechnology, Inc.)) in blocking buffer for 60 min at room temperature, washed with TBS, and followed by secondary antibody (AlexaFluor 488-conjugated antibody, 1:500 (Invitrogen)) for 30 min at room temperature in the dark. Cells were washed with TBS, nuclei were counterstained with 0.5 μm DAPI (Sigma) for 4 min at room temperature, and coverslides were mounted using VectaShield HardSet mounting medium. All images were obtained using a Nikon Eclipse TE200 inverted microscope and were processed using IPLab 4.0.
Peptide Docking to HSP Proteins
The tentative peptide (TDF-P1) docking sites were computationally predicted using Protein Data Bank crystal structures 1YUW (26), 3LDN (27), 3N8E (http://www.rcsb.org), and 2E88 (29) as receptors. 1YUW is bovine heat shock 70-kDa protein 8, composed of a nucleotide binding domain and substrate binding domain β. 1YUW shares 68, 88, and 55% sequence identity with GRP78, HSP1, and HSPA9, respectively (30). 3LDN corresponds to the human GRP78 ATPase domain (apo form). 3N8E is a substrate binding domain of human heat shock protein. It is heat shock 70-kDa protein 9. 2E88 is the apo form of the ATPase domain of human heat shock 70-kDa protein 1.
Following the previously reported framework of “blind docking,” we have used the whole protein chain as a model receptor to identify the potential peptide binding regions (31). The docking operation was carried out using the GRAMM-X Protein-Protein Docking Web Server version 1.2.0 (32, 33). The simulation generated several protein receptor/peptide conformations with an output in the form of a Protein Data Bank file. Graphic views of the proposed docked model simulations were generated using Discovery Studio Visualizer version 3.1. To substantiate these results further, PatchDock and FireDock simulation servers were used to develop another set of models. To do this, preliminary models were developed using PatchDock and refined again by using FireDock (34–37).
RESULTS
Protein Isolation and Identification of TDF-R Candidates by AP and MS
Isolation of the TDF-R was performed by AP, followed by MS. We synthesized TDF-P1 peptide, coupled the peptide to agarose beads, and incubated the peptide beads with the cell lysate from estrogen-responsive MCF7 human breast cancer cells. After incubation, we washed the beads, eluted the TDF-R candidates, and separated them by SDS-PAGE. The gel bands were excised and digested with trypsin, and the peptide mixture was then analyzed by LC-MS/MS. We analyzed all gel bands from each lane.
Isolation and Identification of TDF-R Candidates from Steroid-responsive MCF7 Breast Cancer Cells
In LC-MS/MS experiments performed with AP eluate isolated from estrogen-responsive MCF7 cells, we identified four proteins with high confidence: glucose-regulated protein (GRP78 precursor, gi386758), heat shock 70-kDa protein 8 isoform 1 (HSP8, gi5729877), heat shock 70-kDa protein 1 (HSP1, gi4529893), and heat shock 70-kDa protein 9 (HSPA9, gi12653415). Glucose-regulated protein (GRP78, gi386758) is a 78-kDa protein and a member of the HSP family, also named heat shock 70-kDa protein 5 or immunoglobulin heavy chain-binding protein (BiP). The other three proteins identified in our experiments (HSP8 (gi5729877), HSP1 (gi4529893), and HSPA9 (gi12653415)) were all part of the HSP70 protein family. Examples of total ion current (TIC), MS, and MS/MS of peptides identified in our LC-MS/MS experiments, which were part of GRP78, HSP1, HSP8, and HSPA9, are shown in Fig. 1 (GRP78), Fig. 2 (HSP1), Fig. 3 (HSP8), and Fig. 4 (HSPA9).
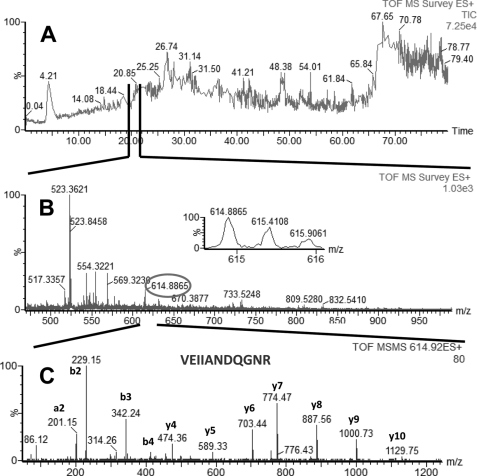
FIGURE 1.
LC-MS/MS analysis of a peptide mixture for identification of GRP78 protein as a potential TDF-R. The TDF-P1 AP protein sample was separated on SDS-PAGE, and the gel bands were excised and digested by trypsin. The resulting peptide mixture was separated on a C18 reverse phase column over an acetonitrile gradient. A, TIC of the chromatogram; B, MS survey mass spectrum, in which one double charged peak at m/z of 614.88 (expanded in the inset) was fragmented by MS/MS and produced a MS/MS spectrum (C). The resulting peaks in the MS/MS spectrum correspond to a series of b and y ions that resulted from fragmentation of a peptide that was part of GRP78. Data analysis of these MS/MS peaks led to identification of the sequence shown in C. Data analysis of this MS/MS spectrum, combined with data analysis of other MS/MS spectra (data not shown), led to identification of GRP78 as a potential TDF-R.
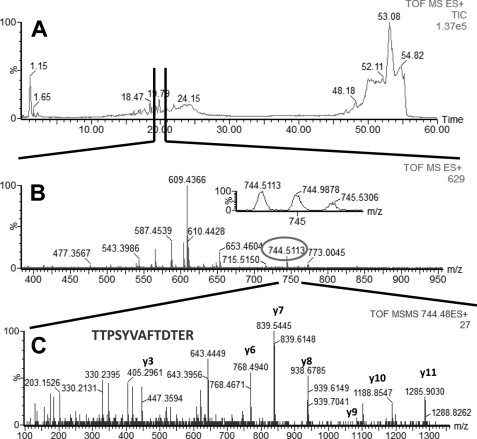
FIGURE 2.
LC-MS/MS analysis of a peptide mixture for identification of HSP1 protein as a potential TDF-R. The experimental procedure was performed as described in the legend to Fig. 1. The TDF-P1 AP protein sample was separated on SDS-PAGE, and the gel bands were excised and digested by trypsin. The resulting peptide mixture was separated on a C18 reverse phase column over an acetonitrile gradient. A, TIC of the chromatogram. B, MS survey mass spectrum, in which one double charged peak at m/z of 744.51 (expanded in the inset) was fragmented by MS/MS and produced an MS/MS spectrum (C). The resulting peaks in the MS/MS spectrum correspond to a series of b and y ions that resulted from fragmentation of a peptide that was part of HSP1. Data analysis of these peaks led to identification of the sequence shown in C. Data analysis of this MS/MS spectrum, combined with data analysis of other MS/MS spectra (data not shown) led to identification of HSP1 as a potential TDF-R.
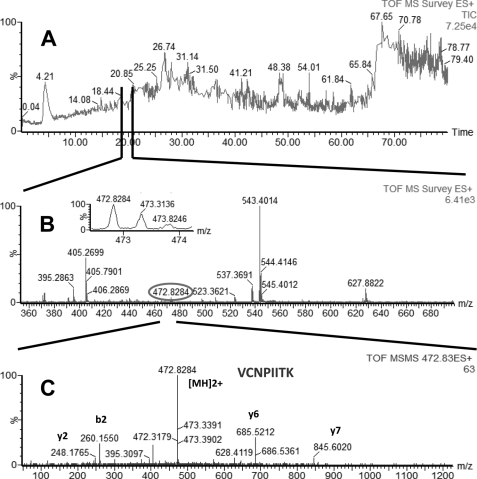
FIGURE 3.
LC-MS/MS analysis of a peptide mixture for identification of HSP8 protein as a potential TDF-R. The TDF-P1 AP protein sample was separated on SDS-PAGE, and the gel bands were excised and digested by trypsin. The resulting peptide mixture was separated on a C18 reverse phase column over an acetonitrile gradient. A, TIC of the chromatogram. B, MS survey mass spectrum, in which one double charged peak at m/z of 472.83 (expanded in the inset) was fragmented by MS/MS and produced an MS/MS spectrum (C). The resulting peaks in the MS/MS spectrum correspond to a series of b and y ions that resulted from fragmentation of a peptide that was part of HSP8. Data analysis of these peaks led to identification of the sequence shown in C. Data analysis of this MS/MS spectrum and other additional MS/MS spectra (data not shown) led to identification of HSP8 as a potential TDF-R.
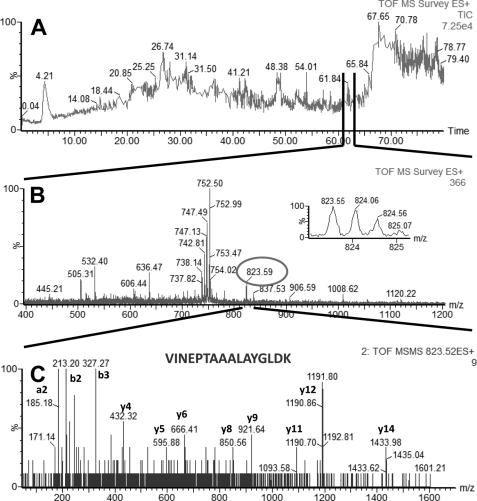
FIGURE 4.
LC-MS/MS analysis of a peptide mixture for identification of HSPA9 protein as a potential TDF-R. The experimental procedure was performed as described in the legend to Fig. 1. The TDF-P1 AP protein sample was separated on SDS-PAGE, and the gel bands were excised and digested by trypsin. The resulting peptide mixture was separated on a C18 reverse phase column over an acetonitrile gradient. A, TIC of the chromatogram. B, MS survey mass spectrum, in which one double charged peak at m/z of 823.59 (expanded in the inset) was fragmented by MS/MS and produced a MS/MS spectrum (C). The resulting peaks in the MS/MS spectrum correspond to a series of b and y ions that resulted from fragmentation of a peptide that was part of HSPA9. Data analysis of these peaks led to identification of the sequence shown in C. Data analysis of these peaks and other MSMS spectra (data not shown) led to identification of HSPA9 as a potential TDF-R.
Additional structural proteins, such as actin, keratin, cytokeratin, and tubulin, were also identified. In control experiments, where we used only agarose beads without TDF-P1 peptide, we identified cytokeratins (8 and 18), actin, and tubulin, suggesting that these proteins are false positive identifications in our experiments. However, in these control experiments, we did not identify any protein that was part of the HSP family, suggesting that these proteins are true TDF-R candidates. Examples of TIC, MS, and MS/MS of peptides that were part of cytokeratin 8 and 18 are shown in supplemental Figs. 1 and 2. A summary of the peptides and proteins identified in our AP and MS experiments using MCF7 cells is shown in Table 1. Taken together, these experiments suggest that GRP78, HSP8, HSP1, and HSPA9 proteins are potential TDF-R candidates.
TABLE 1.
Summary of the proteins and peptides identified in our AP and MS experiments from MCF7 steroid-responsive breast cancer cells
In the control AP and MS experiments, where we used agarose beads without TDF-P1 peptide for AP, we identified the same cytokeratins 8 and 18, known to be overexpressed in MCF7 cells.
| Identified protein | Identified peptides | m/z (charge state) |
|---|---|---|
| GRP78 precursor | ITITNDQNR | 537.85 (2+) |
| VEIIANDQGNR | 614.88 (2+) | |
| Heat shock 70-kDa protein 9 (HSPA9) | LLGQFTLIGIPPAPR | 797.09 (2+) |
| VINEPTAAALAYGLDK | 823.59 (2+) | |
| Heat shock 70-kDa protein 8 isoform 1 (HSP8) | VCNPIITK | 472.83 (2+) |
| VEIIANDQGNR | 614.88 (2+) | |
| TTPSYVAFTDTER | 744.51 (2+) | |
| ITITNDKGR | 509.38 (2+) | |
| Heat shock 70-kDa protein 1 (HSP1) | LSKEEIER | 502.35 (2+) |
| TTPSYVAFTDTER | 744.51 (2+) | |
| ITITNDKGR | 509.38 (2+) | |
| VEIIANDQGNR | 614.88 (2+) | |
| α-Tubulin | VGINYQPPTVVGGDLAK | 913.18 (2+) |
| TIGGGDDSFNTFFSETGAGK | 1004.62 (2+) | |
| Cytokeratin 18 | IMADIR | 359.29 (2+) |
| LASYLDR | 419.32 (2+) | |
| AQYDELAR | 483.33 (2+) | |
| VVSETNDTK | 496.84 (2+) | |
| YETELAMR | 506.84 (2+) | |
| VIDDTNITR | 521.35 (2+) | |
| LEAEIATYR | 523.87 (2+) | |
| VKYETELAMR | 533.38 (2+) | |
| KVIDDTNITR | 413.96 (3+) 620.44 (2+) | |
| QSVENDIHGLR | 392.29 (3+) | |
| AQIFANTVDNAR | 423.29 (3+) 634.44 (2+) | |
| GLQAQIASSGLTVEVDAPK | 440.65 (3+) 660.47 (2+) | |
| IVLQIDNAR | 942.70 (2+) | |
| Cytokeratin 8 | AQYEDIANR | 540.32 (2+) |
| LALDIEIATYR | 639.46 (2+) | |
| LESGMQNMSIHTK | 738.48 (2+) | |
| VGSSNFR | 383.77 (2+) | |
| ISSSSFSR | 435.80 (2+) | |
| QLETLGQEK | 523.38 (2+) | |
| LVSESSDVLPK | 587.43 (2+) | |
| ASLEAAIADAEQR | 672.97 (2+) | |
| LEGLTDEINFLR | 710.49 (2+) |
Isolation and Identification of TDF-R Candidates from Non-breast HeLa Cancerous Cells
To investigate whether the TDF-R candidates can also be purified by AP and identified by MS, we analyzed, in addition to MCF7 cells, non-breast HeLa cancerous cells. In our AP and MS experiments using TDF-P1 peptide to purify the TDF-R candidates from HeLa cancer cells, we did not identify any protein. In control experiments, in which we used only agarose beads for AP from the lysate of HeLa cells, followed by MS, we did not identify any protein. These experiments suggest that non-breast HeLa cells either do not have TDF-R at the cell surface or do not have the receptor at all or that TDF-P1 peptide does not interact with the potential TDF-R candidates.
Validation of AP and MS Experiments
To validate our experiments, we performed a similar AP experiment followed by WB using antibodies against GRP78 and against HSP70 protein. Whereas anti-GRP78 antibodies were specific against human GRP78 protein, the anti-HSP70 protein antibodies were made against whole HSP70 protein, and part of its amino acid sequence was also common to the amino acid sequences of HSPA9, HSP8, and HSP1 (all of them part of HSP70 protein family). Due to the small differences in the molecular mass, we could not differentiate between HSP8 (70 kDa), HSP1 (70 kDa), and HSPA9 (72 kDa) proteins, and, because they were already identified by MS, we considered that the anti-HSP70 antibodies identified all HSP8, HSP1, and HSPA9 proteins.
HSP70 Proteins Are Potential TDF-Rs in Steroid-responsive MCF7 Cells
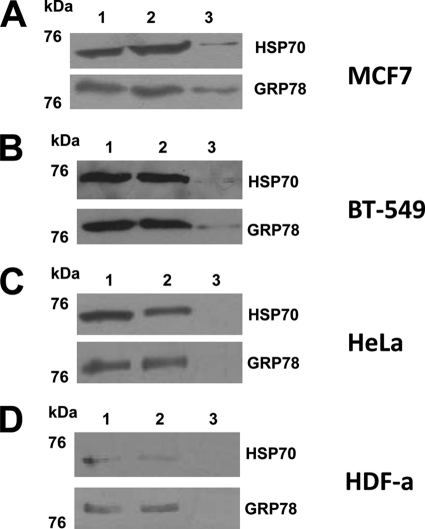
When we analyzed the MCF7 cells by AP and WB using anti-GRP78 and anti-HSP70 antibodies, we could observe an immune reaction at around 73 kDa with anti-HSP70 antibodies and around 78 kDa with anti-GRP78 antibodies (Fig. 5A). In an additional WB experiment, when a more concentrated eluate was used, a stronger reaction was observed (supplemental Fig. 3). In control experiments of AP using beads alone, followed by WB with anti-GRP78 and anti-HSP70 antibodies, no immune reaction was observed (data not shown). Therefore, the interaction of TDF-P1 peptide with GRP78 and HSP70 proteins, previously found by AP and MS, are true interactions, and GRP78 and HSP70 proteins are true potential TDF-R candidates.
FIGURE 5.
WB of the affinity-purified TDF-R candidates. The TDF-R candidates were purified by AP using TDF-P1 peptide, and the eluate was investigated by WB using anti-GRP78 and anti-HSP70 antibodies. The cell lysate was prepared from MCF7 steroid-responsive cells (A), BT-549 steroid-resistant cells (B), HeLa cancer cells (C), and HDF-a cells (D). The molecular mass marker is shown (in kDa) on the left. Each WB contains input cell lysate (lane 1), flow-through (lane 2), and eluate (lane 3) of the AP experiments.
HSP70 Proteins Are Potential TDF-Rs in both Steroid-responsive MCF7 Cells and Steroid-resistant BT-549 Cells
To investigate whether TDF-P1 interacts with GRP78 and HSP70 proteins in steroid-resistant human breast cancer cells (BT-549 cell line), we performed an AP and WB using anti-GRP78 and anti-HSP70 antibodies. In BT-549 cells, we observed the same immune reaction as observed previously in MCF7 cells with antibodies against both GRP78 and HSP70 (Fig. 5B). In an additional WB experiment, when a more concentrated eluate was used, a stronger reaction was observed (data not shown). These results suggest that GRP78 and/or HSP70 proteins are receptors for TDF protein in both MCF7 steroid-responsive and BT-549 steroid-resistant cells. These results also suggest that TDF protein activates, through GRP78 and/or HSP70 proteins, a pathway that is different from the steroid pathway.
HSP70 Proteins Are Potential TDF-Rs in Breast Cancer Cells but Not in HeLa Cancer Cells
To further investigate whether TDF-P1 interacts with GRP78 and HSP70 proteins in other, non-breast cells, we chose HeLa cancer cells for further investigation. These cells are not steroid-regulated cells and do not contain steroid receptors; therefore, we can eliminate any possible interference of the TDF-R pathway with the steroid receptor pathway. We performed an AP experiment from HeLa cell lysate using TDF-P1 as bait, followed by WB using the same anti-GRP78 and anti-HSP70 antibodies. As observed in Fig. 5C, no reaction was observed in the AP and WB experiments when anti-GRP78 or anti-HSP70 antibodies were used. In an additional WB experiment, when a more concentrated eluate was used, again, no reaction was observed (supplemental Fig. 3). These results suggest that GRP78 and HSP70 proteins are potential TDF-R candidates in breast cancer cells but not in other cancer cells (HeLa).
HSP70 Proteins Are Potential TDF-Rs in Breast Cancer Cells but Not in Non-breast, Normal HDF-a
To investigate whether GRP78 and/or HSP70 proteins are TDF-R candidates in non-breast, non-cancer cells, we investigated HDF-a using AP and WB experiments that utilized anti-GRP78 or anti-HSP70 antibodies (Fig. 5D). As observed, no reaction was observed in the AP and WB experiments when anti-GRP78 or anti-HSP70 antibodies were used. We would, however, like to mention that HDF-a cells grew slower, and the cell lysate that was accumulated and used for AP and WB experiments was about 8 times lower compared with the other cells. However, in an additional WB experiment, when a more concentrated eluate was used, no reaction was observed (data not shown). Therefore, GRP78 and HSP70 proteins are potential TDF-R candidates in breast cells but not in other cells (HeLa and HDF-a). These data suggest that the TDF protein has its own pathway that is independent of the steroid pathway and is restricted to breast cells.
Characterization of Expression of GRP78 Protein in Steroid-responsive MCF7 Cells, Steroid-resistant BT-549 Cells, Non-breast Cancerous HeLa Cells, and Normal HDF-a
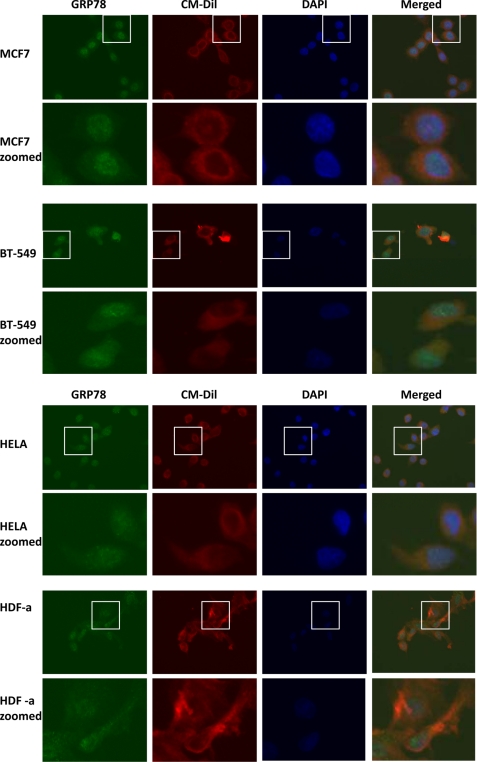
Identification of GRP78 and HSP70 family members of proteins as potential TDF-R candidates in breast cancer cells warranted further IF investigation. Therefore, to investigate whether HSP70 proteins that were identified as potential TDF-R candidates are expressed on the cell surface of breast cells but not on HeLa cancerous cells or on HDF-a fibroblasts, we analyzed these cell lines by IF. We used antibodies against GRP78 and HSP70 proteins to track expression of both proteins in all investigated cell lines. The outcome of this experiment is presented in Figs. 6 and 7. As expected, GRP78 protein was expressed in the plasma membrane fractions of both steroid-responsive MCF7 and steroid-resistant BT-549 cells (Fig. 6). Staining can be observed not only in the cytosol but also in the membrane of the cells. Membrane was also stained using selective membrane tracker CM-Dil, and it can be clearly seen that GRP78 is detected outside the cells. This experiment is consistent with the AP and MS experiments and with AP and WB experiments and further suggests a possible role of GRP78 as a receptor for TDF protein. A similar expression pattern of GRP78 protein was observed when non-breast HeLa cancer cells were used (Fig. 6). However, when normal HDF-a cells were used, no staining outside the cells was observed (Fig. 6). Both HeLa and HDF-a exhibited much weaker GRP78 membrane staining, further confirming that GRP78 might be a TDF-R candidate selective only for breast cell lines.
FIGURE 6.
IF detection of GRP78 protein. Expression of GRP78 was investigated in steroid-responsive MCF7 and steroid-resistant BT-549 breast cancer cells, non-breast HeLa cancer cells and normal HDF-a. The cells were incubated with anti-GRP78 antibodies and then AlexaFluor 488 antibodies for detection of GRP78 protein (green). Plasma membrane was stained with CM-Dil (red), and nuclei were stained with DAPI (blue). The merged images are also shown. Enlarged images for each cell line are also shown.
FIGURE 7.
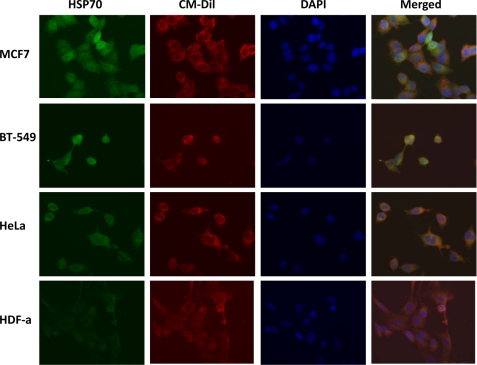
IF detection of HSP70 proteins. Expression of HSP70 proteins was investigated in steroid-responsive MCF7 and steroid-resistant BT-549 breast cancer cells, non-breast HeLa cancer cells, and normal HDF-a. The cells were incubated with anti-HSP70 antibodies and then AlexaFluor 488 antibodies for detection of HSP70 proteins (green). Plasma membrane was stained with CM-Dil (red), and nuclei were stained with DAPI (blue). The merged images are also shown.
Characterization of Expression of HSP70 Proteins in Steroid-responsive MCF7 Cells, Steroid-resistant BT-549 Cells, Non-breast Cancerous HeLa Cells, and Normal HDF-a
When IF experiments using anti-HSP70 antibodies were performed, the staining was similar in all four cell lines studied (MCF7, BT-549 breast cancer cell lines, HeLa cancer cells, and HDF-a cells; Fig. 7). Staining was observed both in the cytosol and in the membrane of the cells. Membrane was also stained using selective membrane tracker CM-Dil, and it can be clearly seen that HSP70 is detected outside the membrane of all cells studied. Taken together, the IF experiments suggest that GRP78 protein is a true potential TDF-R candidate. These results, corroborated with AP and MS and with AP and WB experiments, suggest that GRP78 protein is the main receptor of TDF protein and that HSP70 proteins are contaminants co-purified together with GRP78 protein. Alternatively, TDF-R may have GRP78 as a main receptor and HSP70 proteins as co-receptors.
Investigation of Interaction between TDF-P1 and HSP70 Family Proteins
To further investigate whether TDF-P1 and HSP70 proteins interact with each other and to further confirm the AP and MS experiments (Figs. 1–4) and the AP and WB experiments (Fig. 5), we used two additional approaches: 1) direct infusion followed by electrospray ionization mass spectrometry (ESI-MS) and 2) IAP of GRP78/BiP preincubated with TDF-P1, followed by WB using anti-GRP78 and anti-TDF antibodies (IAP and WB). Using the first approach, we monitored the levels of TDF-P1 in the ESI-MS, either alone or when the TDF-P1 peptide was preincubated with myoglobin, bovine serum albumin (BSA), or recombinant GRP78/BiP. We reasoned that if TDF-P1 interacts with one of these proteins, then we should observe the disappearance of TDF-P1 from the ESI-MS spectrum, mostly due to its interaction with one of the three proteins analyzed. The outcome of this experiment is shown in supplemental Fig. 4. TDF-P1 was observed mostly as a (2+) double-charged peak with m/z of 1106.59 and as a triple-charged (3+) peak with m/z of 738.09 (supplemental Fig. 4A), but variation in the m/z of the measured triple-charged peak was also observed (e.g. m/z of 738.075 and 738.1144; supplemental Fig. 4, B and C). When TDF-P1 was preincubated with myoglobin or BSA, a large amount of TDF-P1 was observed in its monomeric form, and its relative intensity in these two spectra was not different from its intensity of the TDF-P1 peptide measured alone, suggesting that TDF-P1 does not interact with myoglobin or with BSA. However, when TDF-P1 was preincubated with recombinant GRP78/BiP, almost no TDF-P1 could be identified in its monomeric form, suggesting that TDF-P1 interacts naturally with GRP78/BiP. We also investigated the interaction of TDF-P1 with these three proteins by monitoring the shift in m/z of these proteins before and after incubation with TDF-P1. Although we detected no mass shift between myoglobin and myoglobin preincubated with TDF-P1 or between BSA and BSA preincubated with TDF-P1, the results using GRP78/BiP and GRP78/BiP preincubated with TDF-P1 were inconclusive (data not shown).
To further investigate whether TDF-P1 interacts with the members of the HSP70 family of proteins, we preincubated GRP78 with TDF-P1 and then purified them by IAP using anti-GRP78 antibodies. The IAP eluate was then tested by WB using antibodies against TDF-P1 and GRP78. We reasoned that WB of the IAP using antibodies against TDF-P1 and GRP78 should demonstrate a direct interaction between GRP78 and TDF-P1. The outcome of the experiment is shown in Fig. 8. When the GRP78/TDF-P1 mixture was analyzed by anti-GRP78 or anti-TDF-P1 antibodies, both the protein and the peptide were detected (Fig. 8, lane 1). When the IAP was performed using anti-GRP78 antibodies, no protein was detected in the IAP flow-through (Fig. 8, lane 2). Protein A/G-bound beads used as a negative control also showed no reaction (Fig. 8, lane 3). However, when the IAP eluate was tested by WB, it reacted with both GRP78 and TDF-P1 antibodies (Fig. 8, lane 4). Taken together, these data suggest that TDF-P1 indeed interacts naturally with GRP78/BiP but not with other proteins, such as myoglobin or BSA, consistent with our previous finding.
FIGURE 8.
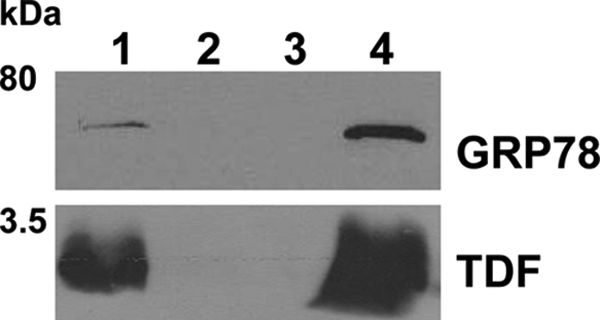
IAP of GRP78 and TDF-P1 mixture. GRP78 protein was incubated with TDF-P1 for 2 h, and then the mixture was precipitated by IAP using anti-GRP78 antibodies (mouse monoclonal). The input, flow-through, control, and eluate were then investigated by WB using anti-GRP78 (rat monoclonal) and anti-TDF (rabbit polyclonal) antibodies. The molecular mass marker is shown (in kDa) on the left. Each WB contains input mixture of GRP78 protein and TDF-P1 (lane 1), flow-through after IP (lane 2), negative control (lane 3), and eluate from IAP (lane 4).
Investigation of Interaction between TDF-P1 and HSP70 Family Proteins Using Structural Biology (Peptide Docking)
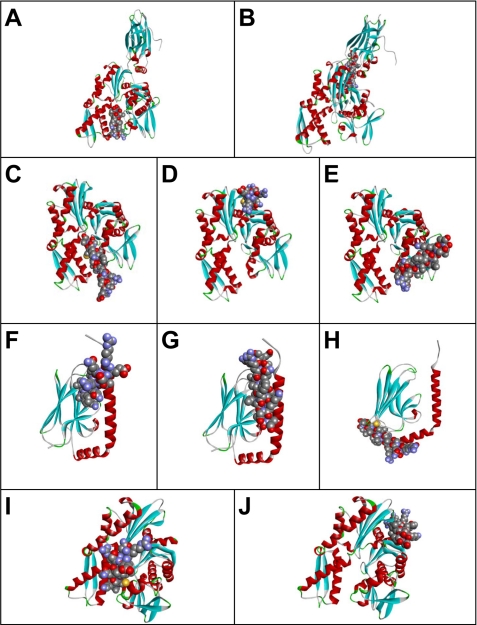
We also investigated whether TDF-P1 interacts with native GRP78 and with members of the HSP70 family of proteins using structural biology. Specifically, we investigated docking of TDF-P1 on binding sites of GRP78 or HSP70 proteins, using the available crystal structure of these proteins. The outcome of this experiment is presented in Fig. 9. The TDF-P1 peptide binding pockets were predicted using Protein Data Bank structures 1YUW (Fig. 9, A and B), 3LDN (Fig. 9, C–E), 3N8E (Fig. 9, F–H), and 2E88 (Fig. 9, I and J).
FIGURE 9.
Possible peptide binding pockets tentatively identified by GRAMM-X Protein-Protein Docking Web Server version 1.2.0. The TDF-P1 peptide (P1) binding pockets were predicted using Protein Data Bank structures 1YUW (A and B), 3LDN (C–E), 3N8E (F–H), and 2E88 (I and J). Receptor proteins are shown in the form of a ribbon diagram colored by secondary structure. P1 peptide is shown in space-filling mode colored by atom type.
The first 10 most optimal scoring structures that resulted from the GRAMM-X simulation were considered for detailed scrutiny. The frequencies with which the locations of the docking pockets appeared among these models were used as the primary criteria for determining the most probable structure. The order of sequencing of the results was also used as a second condition for selection. Based on these considerations, we selected two docked models from this list. Fig. 9, A and B, shows these two docked models in the case of 1YUW, indicating the most probable (tentative) binding pockets. Six of the top 10 simulations had almost the same docking site as Fig. 9A. Two of the 10 simulations had almost the same binding pocket as in Fig. 9B. The results using PatchDock and FireDock simulation servers were almost identical to those shown in Fig. 9, A and B, with the relative frequencies of the different probable binding pockets slightly differing between the different sets of runs.
Fig. 9, C–E, shows tentative models of TDF-P1 binding pockets in 3LDN (chain A). After carrying out the docking operation using GRAMM-X, four of the 10 simulations had almost the same binding region as that shown in Fig. 9C. In three of these 10 simulations, the TDF-P1 peptide docked at the binding site identified in Fig. 9D, and two of the results had almost the same conformation as the one shown in Fig. 9E. Results similar to those of Fig. 9, C and D, were also obtained using PatchDock and FireDock.
The TDF-P1 binding pocket was predicted using Protein Data Bank structure 3N8E (chain A). Fig. 9, F–H, shows the tentative models of peptide binding sites in the substrate binding domain of human heat shock 70-kDa protein 9. The docking operation was carried out using GRAMM-X. Six of the 10 simulations had nearly the same binding pockets as Fig. 9F. One of the 10 simulations essentially mimicked the binding configuration of Fig. 9G, and in two cases, the docking occurred approximately in the same region as that shown in Fig. 9H. Results very similar to that of Fig. 9G were also obtained using PatchDock and FireDock. In the last two cases, however, only a single binding location, similar to the one mentioned above, was observed.
Fig. 9, I and J, shows tentative models of TDF-P1 binding regions in the ATPase domain of human heat shock 70-kDa protein 1 (Protein Data Bank entry 2E88). The docking operation was carried out using GRAMM-X. One of the 10 simulations yielded the same binding region as that shown in Fig. 9I. In six of these 10 simulations, the peptide docked in the binding site identified in Fig. 9J. Additional results similar to those of Fig. 9I were also obtained using PatchDock and FireDock. In the latter case, however, the configuration considered in Fig. 9J remained excluded. It is worth noting that the simulations reported here did not account for possible structural changes of the receptor protein upon peptide binding.
DISCUSSION
TDF is a relatively new protein secreted by the pituitary into the bloodstream, with no definitive function and no known receptor. It is an underinvestigated protein, yet to be universally recognized as a new hormone. The first step that we took in unveiling the function of TDF was to isolate its receptor. In our experiments using AP and MS, we identified four proteins that are potential TDF-R candidates: GRP78, HSP8, HSP1, and HSPA9. All of these proteins are members of the HSP70 family of proteins and are involved in the folding and assembly of proteins in the endoplasmic reticulum, but they may also be identified in the cytosol or cell membrane (38–48).
HSPs are highly expressed in cancerous cells and are essential to the survival of these cells (49–58); therefore, HSP inhibitors show promise as anticancer agents (27, 59–66). HSPs (HSP70 and HSP90) have been associated with both estrogen and androgen receptors (67–71). HSPs have a role in cell proliferation. For example, inhibition of HSP90 led to the dysregulation of HPS70 and inhibition of cell proliferation (61). HSPs also have a role in apoptosis and cell differentiation, especially HSP70 and HSP90. These two proteins interact with apoptotic proteins and block the apoptotic pathways, thus promoting cell differentiation (61). HSPs may even determine whether cells undergo apoptosis or differentiation (44). Recently, it was demonstrated that GRP78 forms a cell surface complex with Cripto, an oncoprotein that signals via MAPK/ERK, PI3K/Akt, and Smad2/3 pathways, and mediates signaling in human tumor, mammary epithelial, and embryonic stem cells (43). Active Cripto from the Cripto-GRP78 complex promotes cellular proliferation, decrease of cell adhesion, and down-regulation of E-cadherin. However, Cripto alone is not able to signal and promote the above mentioned cellular events. Therefore, GRP78 (when in complex with Cripto), is an oncogene (43). When it is not in a protein complex with Cripto, it may promote cell differentiation (44).
In our AP experiments using estrogen-responsive MCF7 cells, we expected to identify estrogen receptors, but instead we identified HSP70 proteins. Therefore, TDF may have a natural receptor that is different from estrogen receptors and that promotes differentiation of steroid-responsive cells by a non-steroid signal transduction pathway. We wanted to investigate whether TDF protein has HSP70 proteins as TDF-Rs only on MCF7 estrogen-responsive breast cancer cells or in both estrogen-responsive and -resistant breast cancer cells. We therefore investigated BT-549 estrogen-resistant breast cancer cells in AP and WB experiments using antibodies against GRP78 and HSP70. In BT-549 cells, we identified the same HSP70 proteins as receptors for TDF protein as we had previously found in MCF7 cells, suggesting that the TDF acts on both steroid-responsive and steroid-resistant breast cancer cells through the same TDF-R, but through a pathway independent of estrogen receptors and independent of the estrogen pathway.
TDF-R was identified in MCF7 cells and BT-549 cells, both of which are breast cancer cells. This indicates that HSP70 proteins may act as TDF-R specifically in cancerous cells, in breast cells, or in both types of cells. To investigate whether other cancerous, non-breast cells have the same TDF-R candidates, we analyzed non-breast HeLa cancerous cells. We found that the AP did not identify any HSP70 proteins in either AP and MS or AP and WB. Therefore, it seems that TDF-R is linked somehow to breast cells specifically and not globally to cancer cells. To further investigate whether TDF has a TDF-R (or interacts with it) only in breast cells or in any (non-cancerous) cells, we investigated HDF-a fibroblasts by AP and WB. As in HeLa cells, we did not see any reaction, again suggesting that TDF-R is somehow linked to breast cells and is not present (or it does not interact with its ligand) in normal or cancerous cells.
It is it possible that we identified GRP78 as a potential TDF-R candidate, because GRP78, when present at the cell surface, has many roles, acting as a hub for signaling to the nucleus (28, 42, 50, 66, 72–78). We should also remember that TDF-P1 peptide produces a differentiation effect on breast and prostate cancer cells (2). However, it does not make sense that GRP78 and the other HSP70 proteins are specific only to breast cells and not to cancer or non-cancer cells, unless 1) a specific number of receptors act as co-receptors for TDF and 2) only when all co-receptors are in the same place and at the same time as a protein complex receptor, TDF binds to its receptor and promotes cell differentiation.
To further investigate whether the HSP70 proteins are localized at the cell membrane, we investigated MCF7, BT-549, HeLa, and HDF-a cell lines by IF using anti-GRP78 and anti-HSP70 antibodies. Whereas GRP78 was identified at the cell membrane of MCF7, BT-549, and HeLa cells, the other HSP70 proteins were identified in all cell lines investigated. It should also be kept in mind that GRP78 and other HSP70 proteins have been identified at the cell surface of all cancer cell lines investigated in the current study and in other studies (49–58), but they were not identified by AP and MS or by AP and WB in any non-breast cancer cell line that we investigated. Therefore, comparison of AP and MS with AP and WB and further using IF suggest that TDF-R is restricted to breast cells. These data also suggest that GRP78 is the main receptor for TDF-R and that the other HSP70 proteins are either co-receptors for TDF protein or isolation artifacts.
In conclusion, we used AP and MS experiments to investigate different cell lysates to identify TDF-R candidates and AP and WB, ESI-MS, IAP and WB, and structural biology experiments to further validate our findings. We found that all TDF-R candidates identified are members of the HSP70 family (possibly as a protein complex) and were detected only in breast cancer cells and not in non-breast HeLa cancerous or normal non-breast HDF-a cells. The HSP70 proteins were found in both MCF7 steroid-responsive cells and BT-549 steroid-resistant cells. This suggests that TDF protein acts in both cell types through a steroid-independent receptor and activates a TDF pathway that is specific to breast cells. All four HSP70 proteins are being further investigated as potential TDF-R candidates.
Supplementary Material
Acknowledgments
We thank Dr. Linda Hendershot (St. Jude Children's Research Hospital, Memphis, TN) for providing the GRP78 clone and for anti-GRP78 antibodies. We also thank Dr. Erasmus Schneider (Wadsworth Center, Albany, NY) for sharing MCF7 cells, Dr. Craig Woodworth (Clarkson University) for providing HeLa and HDF-a cells, and Dr. Kenneth Wallace (Clarkson University) for support with fluorescence microscopy. We also thank Laura Mulderig and colleagues (Waters Corp.) for generous support in setting up the Proteomics Center at Clarkson University. C. C. D. thanks Dr. Micsunica Platica for strong moral and scientific support during his work at Mount Sinai School of Medicine (New York).
This work was supported in part by a start-up grant provided by Clarkson University (to C. C. D.) and by support from the United States Army Research Office through the Defense University Research Instrumentation Program (DURIP Grant W911NF-11-1-0304).

This article contains supplemental Figs. 1–4.
- TDF
- tumor differentiation factor
- TDF-R
- TDF receptor
- AP
- affinity chromatography
- IAP
- immunoaffinity precipitation
- TIC
- total ion current
- WB
- Western blotting
- IF
- immunofluorescence
- GRP78
- glucose-regulated protein (accession number gi386758)
- HSP
- heat shock protein.
REFERENCES
- 1. Platica M., Chen H. Z., Ciurea D., Gil J., Mandeli J., Hollander V. P. (1992) Pituitary extract causes aggregation and differentiation of rat mammary tumor MTW9/Pl cells. Endocrinology 131, 2573–2580 [DOI] [PubMed] [Google Scholar]
- 2. Platica M., Ivan E., Holland J. F., Ionescu A., Chen S., Mandeli J., Unger P. D., Platica O. (2004) A pituitary gene encodes a protein that produces differentiation of breast and prostate cancer cells. Proc. Natl. Acad. Sci. U.S.A. 101, 1560–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brabant G., Hoang-Vu C., Cetin Y., Dralle H., Scheumann G., Mölne J., Hansson G., Jansson S., Ericson L. E., Nilsson M. (1993) E-cadherin. A differentiation marker in thyroid malignancies. Cancer Res. 53, 4987–4993 [PubMed] [Google Scholar]
- 4. De Leeuw W. J., Berx G., Vos C. B., Peterse J. L., Van de Vijver M. J., Litvinov S., Van Roy F., Cornelisse C. J., Cleton-Jansen A. M. (1997) Simultaneous loss of E-cadherin and catenins in invasive lobular breast cancer and lobular carcinoma in situ. J. Pathol. 183, 404–411 [DOI] [PubMed] [Google Scholar]
- 5. Edelman G. M., Crossin K. L. (1991) Cell adhesion molecules. Implications for a molecular histology. Annu. Rev. Biochem. 60, 155–190 [DOI] [PubMed] [Google Scholar]
- 6. Frixen U. H., Behrens J., Sachs M., Eberle G., Voss B., Warda A., Löchner D., Birchmeier W. (1991) E-cadherin-mediated cell-cell adhesion prevents invasiveness of human carcinoma cells. J. Cell Biol. 113, 173–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ray D. B., Horst I. A., Jansen R. W., Kowal J. (1981) Normal mammary cells in long term culture. I. Development of hormone-dependent functional monolayer cultures and assay of α-lactalbumin production. Endocrinology 108, 573–583 [DOI] [PubMed] [Google Scholar]
- 8. Sommers C. L., Thompson E. W., Torri J. A., Kemler R., Gelmann E. P., Byers S. W. (1991) Cell adhesion molecule uvomorulin expression in human breast cancer cell lines. Relationship to morphology and invasive capacities. Cell Growth Differ. 2, 365–372 [PubMed] [Google Scholar]
- 9. Thean E. T., Toh B. H. (1990) Serum human α-lactalbumin as a marker for breast cancer. Br. J. Cancer 61, 773–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vleminckx K., Vakaet L., Jr., Mareel M., Fiers W., van Roy F. (1991) Genetic manipulation of E-cadherin expression by epithelial tumor cells reveals an invasion suppressor role. Cell 66, 107–119 [DOI] [PubMed] [Google Scholar]
- 11. Levin E. R. (2005) Integration of the extranuclear and nuclear actions of estrogen. Mol. Endocrinol. 19, 1951–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nilsson S., Gustafsson J. A. (2002) Estrogen receptor action. Crit. Rev. Eukaryot. Gene Expr. 12, 237–257 [DOI] [PubMed] [Google Scholar]
- 13. Nilsson S., Mäkelä S., Treuter E., Tujague M., Thomsen J., Andersson G., Enmark E., Pettersson K., Warner M., Gustafsson J. A. (2001) Mechanisms of estrogen action. Physiol. Rev. 81, 1535–1565 [DOI] [PubMed] [Google Scholar]
- 14. Pfeffer U., Fecarotta E., Vidali G. (1995) Coexpression of multiple estrogen receptor variant messenger RNAs in normal and neoplastic breast tissues and in MCF-7 cells. Cancer Res. 55, 2158–2165 [PubMed] [Google Scholar]
- 15. Darie C. C., Shetty V., Spellman D. S., Zhang G., Xu C., Cardasis H. L., Blais S., Fenyo D., Neubert T. A. (2008) Blue native PAGE and mass spectrometry analysis of the ephrin stimulation-dependent protein-protein interactions in NG108-EphB2 cells. in Applications of Mass Spectrometry in Life Safety (Popescu C., Zamfir A. D., Dinca N., eds) pp. 3–22, NATO Science for Peace and Security Series, Springer-Verlag, Düsseldorf, Germany [Google Scholar]
- 16. Darie C. C., Deinhardt K., Zhang G., Cardasis H. S., Chao M. V., Neubert T. A. (2011) Identifying transient protein-protein interactions in EphB2 signaling by blue native PAGE and mass spectrometry. Proteomics 11, 4514–4528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Aebersold R., Mann M. (2003) Mass spectrometry-based proteomics. Nature 422, 198–207 [DOI] [PubMed] [Google Scholar]
- 18. Darie C. C., Biniossek M. L., Winter V., Mutschler B., Haehnel W. (2005) Isolation and structural characterization of the Ndh complex from mesophyll and bundle sheath chloroplasts of Zea mays. FEBS J. 272, 2705–2716 [DOI] [PubMed] [Google Scholar]
- 19. Spellman D. S., Deinhardt K., Darie C. C., Chao M. V., Neubert T. A. (2008) Stable isotopic labeling by amino acids in cultured primary neurons. Application to brain-derived neurotrophic factor-dependent phosphotyrosine-associated signaling. Mol. Cell Proteomics 7, 1067–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Darie C. C., Litscher E. S., Wassarman P. M. (2008) Structure, processing, and polymerization of rainbow trout egg vitelline envelope proteins. in Applications of Mass Spectrometry in Life Safety (Popescu C., Zamfir A. D., Dinca N., eds) pp. 23–36, NATO Science for Peace and Security Series, Springer-Verlag, Düsseldorf, Germany [Google Scholar]
- 21. Vladusic E. A., Hornby A. E., Guerra-Vladusic F. K., Lakins J., Lupu R. (2000) Expression and regulation of estrogen receptor β in human breast tumors and cell lines. Oncol. Rep. 7, 157–167 [DOI] [PubMed] [Google Scholar]
- 22. Shevchenko A., Wilm M., Vorm O., Mann M. (1996) Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 23. Darie C. C., Biniossek M. L., Gawinowicz M. A., Milgrom Y., Thumfart J. O., Jovine L., Litscher E. S., Wassarman P. M. (2005) Mass spectrometric evidence that proteolytic processing of rainbow trout egg vitelline envelope proteins takes place on the egg. J. Biol. Chem. 280, 37585–37598 [DOI] [PubMed] [Google Scholar]
- 24. Darie C. C., Biniossek M. L., Jovine L., Litscher E. S., Wassarman P. M. (2004) Structural characterization of fish egg vitelline envelope proteins by mass spectrometry. Biochemistry 43, 7459–7478 [DOI] [PubMed] [Google Scholar]
- 25. Darie C. C., Janssen W. G., Litscher E. S., Wassarman P. M. (2008) Purified trout egg vitelline envelope proteins VEβ and VEγ polymerize into homomeric fibrils from dimers in vitro. Biochim. Biophys. Acta 1784, 385–392 [DOI] [PubMed] [Google Scholar]
- 26. Jiang J., Prasad K., Lafer E. M., Sousa R. (2005) Structural basis of interdomain communication in the Hsc70 chaperone. Mol. Cell 20, 513–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Macias A. T., Williamson D. S., Allen N., Borgognoni J., Clay A., Daniels Z., Dokurno P., Drysdale M. J., Francis G. L., Graham C. J., Howes R., Matassova N., Murray J. B., Parsons R., Shaw T., Surgenor A. E., Terry L., Wang Y., Wood M., Massey A. J. (2011) Adenosine-derived inhibitors of 78-kDa glucose regulated protein (Grp78) ATPase. Insights into isoform selectivity. J. Med. Chem. 54, 4034–4041 [DOI] [PubMed] [Google Scholar]
- 28. Saba J. A., McComb M. E., Potts D. L., Costello C. E., Amar S. (2007) Proteomic mapping of stimulus-specific signaling pathways involved in THP-1 cells exposed to Porphyromonas gingivalis or its purified components. J. Proteome Res. 6, 2211–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shida M., Arakawa A., Ishii R., Kishishita S., Takagi T., Kukimoto-Niino M., Sugano S., Tanaka A., Shirouzu M., Yokoyama S. (2010) Direct intersubdomain interactions switch between the closed and open forms of the Hsp70 nucleotide-binding domain in the nucleotide-free state. Acta Crystallogr. D Biol. Crystallogr. 66, 223–232 [DOI] [PubMed] [Google Scholar]
- 30. Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Gapped BLAST and PSI-BLAST. A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hetényi C., van der Spoel D. (2002) Efficient docking of peptides to proteins without prior knowledge of the binding site. Protein Sci. 11, 1729–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tovchigrechko A., Vakser I. A. (2005) Development and testing of an automated approach to protein docking. Proteins 60, 296–301 [DOI] [PubMed] [Google Scholar]
- 33. Tovchigrechko A., Vakser I. A. (2006) GRAMM-X public web server for protein-protein docking. Nucleic Acids Res. 34, W310–W314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Andrusier N., Nussinov R., Wolfson H. J. (2007) FireDock. Fast interaction refinement in molecular docking. Proteins 69, 139–159 [DOI] [PubMed] [Google Scholar]
- 35. Duhovny D., Nussinov R., Wolfson H. J. (2002) Efficient unbound docking of rigid molecules. in Proceedings of the 2nd Workshop on Algorithms in Bioinformatics (WABI) Lecture Notes in Computer Science (Gussfield D., Guigo R., eds) Springer Verlag, Rome [Google Scholar]
- 36. Mashiach E., Schneidman-Duhovny D., Andrusier N., Nussinov R., Wolfson H. J. (2008) FireDock. A web server for fast interaction refinement in molecular docking. Nucleic Acids Res. 36, W229–W232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schneidman-Duhovny D., Inbar Y., Nussinov R., Wolfson H. J. (2005) PatchDock and SymmDock. Servers for rigid and symmetric docking. Nucleic Acids Res. 33, W363–W367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Al-Hashimi A. A., Caldwell J., Gonzalez-Gronow M., Pizzo S. V., Aboumrad D., Pozza L., Al-Bayati H., Weitz J. I., Stafford A., Chan H., Kapoor A., Jacobsen D. W., Dickhout J. G., Austin R. C. (2010) Binding of anti-GRP78 autoantibodies to cell surface GRP78 increases tissue factor procoagulant activity via the release of calcium from endoplasmic reticulum stores. J. Biol. Chem. 285, 28912–28923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Arap M. A., Lahdenranta J., Mintz P. J., Hajitou A., Sarkis A. S., Arap W., Pasqualini R. (2004) Cell surface expression of the stress response chaperone GRP78 enables tumor targeting by circulating ligands. Cancer Cell 6, 275–284 [DOI] [PubMed] [Google Scholar]
- 40. Delpino A., Castelli M. (2002) The 78-kDa glucose-regulated protein (GRP78/BIP) is expressed on the cell membrane, is released into cell culture medium, and is also present in human peripheral circulation. Biosci. Rep. 22, 407–420 [DOI] [PubMed] [Google Scholar]
- 41. Delpino A., Piselli P., Vismara D., Vendetti S., Colizzi V. (1998) Cell surface localization of the 78-kDa glucose regulated protein (GRP 78) induced by thapsigargin. Mol. Membr. Biol. 15, 21–26 [DOI] [PubMed] [Google Scholar]
- 42. Graner M. W., Raynes D. A., Bigner D. D., Guerriero V. (2009) Heat shock protein 70-binding protein 1 is highly expressed in high-grade gliomas, interacts with multiple heat shock protein 70 family members, and specifically binds brain tumor cell surfaces. Cancer Sci. 100, 1870–1879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kelber J. A., Panopoulos A. D., Shani G., Booker E. C., Belmonte J. C., Vale W. W., Gray P. C. (2009) Blockade of Cripto binding to cell surface GRP78 inhibits oncogenic Cripto signaling via MAPK/PI3K and Smad2/3 pathways. Oncogene 28, 2324–2336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lanneau D., de Thonel A., Maurel S., Didelot C., Garrido C. (2007) Apoptosis versus cell differentiation. Role of heat shock proteins HSP90, HSP70, and HSP27. Prion 1, 53–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Misra U. K., Pizzo S. V. (2010) Ligation of cell surface GRP78 with antibody directed against the COOH-terminal domain of GRP78 suppresses Ras/MAPK and PI 3-kinase/AKT signaling while promoting caspase activation in human prostate cancer cells. Cancer Biol. Ther. 9, 142–152 [DOI] [PubMed] [Google Scholar]
- 46. Ni M., Zhang Y., Lee A. S. (2011) Beyond the endoplasmic reticulum. Atypical GRP78 in cell viability, signaling, and therapeutic targeting. Biochem. J. 434, 181–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ni M., Zhou H., Wey S., Baumeister P., Lee A. S. (2009) Regulation of PERK signaling and leukemic cell survival by a novel cytosolic isoform of the UPR regulator GRP78/BiP. PLoS One 4, e6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang Y., Liu R., Ni M., Gill P., Lee A. S. (2010) Cell surface relocalization of the endoplasmic reticulum chaperone and unfolded protein response regulator GRP78/BiP. J. Biol. Chem. 285, 15065–15075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Daneshmand S., Quek M. L., Lin E., Lee C., Cote R. J., Hawes D., Cai J., Groshen S., Lieskovsky G., Skinner D. G., Lee A. S., Pinski J. (2007) Glucose-regulated protein GRP78 is up-regulated in prostate cancer and correlates with recurrence and survival. Hum. Pathol. 38, 1547–1552 [DOI] [PubMed] [Google Scholar]
- 50. Fu Y., Lee A. S. (2006) Glucose regulated proteins in cancer progression, drug resistance, and immunotherapy. Cancer Biol. Ther. 5, 741–744 [DOI] [PubMed] [Google Scholar]
- 51. Katanasaka Y., Ishii T., Asai T., Naitou H., Maeda N., Koizumi F., Miyagawa S., Ohashi N., Oku N. (2010) Cancer antineovascular therapy with liposome drug delivery systems targeted to BiP/GRP78. Int. J. Cancer 127, 2685–2698 [DOI] [PubMed] [Google Scholar]
- 52. Lee A. S. (2007) GRP78 induction in cancer. Therapeutic and prognostic implications. Cancer Res. 67, 3496–3499 [DOI] [PubMed] [Google Scholar]
- 53. Li J., Lee A. S. (2006) Stress induction of GRP78/BiP and its role in cancer. Curr. Mol. Med. 6, 45–54 [DOI] [PubMed] [Google Scholar]
- 54. Misra U. K., Payne S., Pizzo S. V. (2011) Ligation of prostate cancer cell surface GRP78 activates a proproliferative and antiapoptotic feedback loop. A role for secreted prostate-specific antigen. J. Biol. Chem. 286, 1248–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pfaffenbach K. T., Lee A. S. (2011) The critical role of GRP78 in physiologic and pathologic stress. Curr. Opin. Cell Biol. 23, 150–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Quinones Q. J., de Ridder G. G., Pizzo S. V. (2008) GRP78. A chaperone with diverse roles beyond the endoplasmic reticulum. Histol. Histopathol. 23, 1409–1416 [DOI] [PubMed] [Google Scholar]
- 57. Watson L. M., Chan A. K., Berry L. R., Li J., Sood S. K., Dickhout J. G., Xu L., Werstuck G. H., Bajzar L., Klamut H. J., Austin R. C. (2003) Overexpression of the 78-kDa glucose-regulated protein/immunoglobulin-binding protein (GRP78/BiP) inhibits tissue factor procoagulant activity. J. Biol. Chem. 278, 17438–17447 [DOI] [PubMed] [Google Scholar]
- 58. Zhang L. H., Zhang X. (2010) Roles of GRP78 in physiology and cancer. J. Cell Biochem. 110, 1299–1305 [DOI] [PubMed] [Google Scholar]
- 59. Didelot C., Lanneau D., Brunet M., Joly A. L., De Thonel A., Chiosis G., Garrido C. (2007) Anti-cancer therapeutic approaches based on intracellular and extracellular heat shock proteins. Curr. Med. Chem. 14, 2839–2847 [DOI] [PubMed] [Google Scholar]
- 60. Solit D. B., Rosen N. (2006) Hsp90. A novel target for cancer therapy. Curr. Top. Med. Chem. 6, 1205–1214 [DOI] [PubMed] [Google Scholar]
- 61. Jensen M. R., Schoepfer J., Radimerski T., Massey A., Guy C. T., Brueggen J., Quadt C., Buckler A., Cozens R., Drysdale M. J., Garcia-Echeverria C., Chène P. (2008) NVP-AUY922. A small molecule HSP90 inhibitor with potent antitumor activity in preclinical breast cancer models. Breast Cancer Res. 10, R33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Liu Y., Steiniger S. C., Kim Y., Kaufmann G. F., Felding-Habermann B., Janda K. D. (2007) Mechanistic studies of a peptidic GRP78 ligand for cancer cell-specific drug delivery. Mol. Pharm. 4, 435–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Rauschert N., Brändlein S., Holzinger E., Hensel F., Müller-Hermelink H. K., Vollmers H. P. (2008) A new tumor-specific variant of GRP78 as target for antibody-based therapy. Lab. Invest. 88, 375–386 [DOI] [PubMed] [Google Scholar]
- 64. Sato M., Yao V. J., Arap W., Pasqualini R. (2010) GRP78 signaling hub a receptor for targeted tumor therapy. Adv. Genet. 69, 97–114 [DOI] [PubMed] [Google Scholar]
- 65. Schwarze S., Rangnekar V. M. (2010) Targeting plasma membrane GRP78 for cancer growth inhibition. Cancer Biol. Ther. 9, 153–155 [DOI] [PubMed] [Google Scholar]
- 66. Zhang L. H., Yang X. L., Zhang X., Cheng J. X., Zhang W. (2011) Association of elevated GRP78 expression with increased astrocytoma malignancy via Akt and ERK pathways. Brain Res. 1371, 23–31 [DOI] [PubMed] [Google Scholar]
- 67. Nair S. C., Toran E. J., Rimerman R. A., Hjermstad S., Smithgall T. E., Smith D. F. (1996) A pathway of multichaperone interactions common to diverse regulatory proteins. Estrogen receptor, Fes tyrosine kinase, heat shock transcription factor Hsf1, and the aryl hydrocarbon receptor. Cell Stress Chaperones 1, 237–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Nemoto T., Ohara-Nemoto Y., Ota M. (1992) Association of the 90-kDa heat shock protein does not affect the ligand-binding ability of androgen receptor. J. Steroid Biochem. Mol. Biol. 42, 803–812 [DOI] [PubMed] [Google Scholar]
- 69. Sanchez E. R., Faber L. E., Henzel W. J., Pratt W. B. (1990) The 56–59-kilodalton protein identified in untransformed steroid receptor complexes is a unique protein that exists in cytosol in a complex with both the 70- and 90-kilodalton heat shock proteins. Biochemistry 29, 5145–5152 [DOI] [PubMed] [Google Scholar]
- 70. Veldscholte J., Berrevoets C. A., Brinkmann A. O., Grootegoed J. A., Mulder E. (1992) Anti-androgens and the mutated androgen receptor of LNCaP cells. Differential effects on binding affinity, heat-shock protein interaction, and transcription activation. Biochemistry 31, 2393–2399 [DOI] [PubMed] [Google Scholar]
- 71. Veldscholte J., Berrevoets C. A., Zegers N. D., van der Kwast T. H., Grootegoed J. A., Mulder E. (1992) Hormone-induced dissociation of the androgen receptor-heat-shock protein complex. Use of a new monoclonal antibody to distinguish transformed from nontransformed receptors. Biochemistry 31, 7422–7430 [DOI] [PubMed] [Google Scholar]
- 72. Awad W., Estrada I., Shen Y., Hendershot L. M. (2008) BiP mutants that are unable to interact with endoplasmic reticulum DnaJ proteins provide insights into interdomain interactions in BiP. Proc. Natl. Acad. Sci. U.S.A. 105, 1164–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Calvert M. E., Digilio L. C., Herr J. C., Coonrod S. A. (2003) Oolemmal proteomics. Identification of highly abundant heat shock proteins and molecular chaperones in the mature mouse egg and their localization on the plasma membrane. Reprod. Biol. Endocrinol 1, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Dul J. L., Davis D. P., Williamson E. K., Stevens F. J., Argon Y. (2001) Hsp70 and antifibrillogenic peptides promote degradation and inhibit intracellular aggregation of amyloidogenic light chains. J. Cell Biol. 152, 705–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Häfker T., Techel D., Steier G., Rensing L. (1998) Differential expression of glucose-regulated (grp78) and heat-shock-inducible (hsp70) genes during asexual development of Neurospora crassa. Microbiology 144, 37–43 [DOI] [PubMed] [Google Scholar]
- 76. Jang J. H., Hanash S. (2003) Profiling of the cell surface proteome. Proteomics 3, 1947–1954 [DOI] [PubMed] [Google Scholar]
- 77. Jiang H., He J., Pu S., Tang C., Xu G. (2007) Heat shock protein 70 is translocated to lipid droplets in rat adipocytes upon heat stimulation. Biochim. Biophys. Acta 1771, 66–74 [DOI] [PubMed] [Google Scholar]
- 78. Jørgensen M. M., Jensen O. N., Holst H. U., Hansen J. J., Corydon T. J., Bross P., Bolund L., Gregersen N. (2000) Grp78 is involved in retention of mutant low density lipoprotein receptor protein in the endoplasmic reticulum. J. Biol. Chem. 275, 33861–33868 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.