Background: Myelin basic protein (MBP) synthesis and Fyn kinase activity are important for CNS myelination.
Results: Oligodendroglial hnRNP F is associated with RNA granules, a target of Fyn, and modulates MBP translation.
Conclusion: hnRNP F is a crucial regulator of MBP synthesis.
Significance: Dysregulation of hnRNP F levels as occurs in some white matter diseases will compromise myelination.
Keywords: Neuroscience, Protein Synthesis, RNA-binding Protein, Translation Control, Protein-tyrosine Kinase (Tyrosine kinase), Fyn Kinase, Myelin Basic Protein, hnRNP A2, hnRNP F, Oligodendrocyte
Abstract
Myelin basic protein (MBP) is a major component of central nervous system (CNS) myelin. The absence of MBP results in the loss of almost all compact myelin in the CNS. MBP mRNA is sorted into RNA granules that are transported to the periphery of oligodendrocytes in a translationally inactive state. A central mediator of this transport process is the trans-acting factor heterogeneous nuclear ribonucleoprotein (hnRNP) A2 that binds to the cis-acting A2-response element in the 3′UTR of MBP mRNA. Recently, we found that activation of the Src family nonreceptor tyrosine kinase Fyn in oligodendrocytes leads to phosphorylation of hnRNP A2 and to increased translation of MBP mRNA. Here, we identify the RNA-binding protein hnRNP F as a novel component of MBP mRNA transport granules. It is associated with hnRNP A2 and MBP mRNA in cytoplasmic granular structures and is involved in post-transcriptional regulation of MBP expression. Fyn kinase activity results in phosphorylation of hnRNP F in the cytoplasm and its release from MBP mRNA and RNA granules. Our results define hnRNP F as a regulatory element of MBP expression in oligodendrocytes and imply an important function of hnRNP F in the control of myelin synthesis.
Introduction
Myelination of neuronal axons by oligodendrocytes allows rapid and energetically efficient propagation of action potentials over long distances and is essential for maintenance of axonal integrity (1). At the onset of myelination, oligodendrocytes extend cell processes to axonal segments that they enwrap with myelin membrane. The generation of large amounts of myelin necessitates high biosynthetic activity and tightly controlled transport of proteins and lipids toward the axon-glial contact site (2–4). Proteolipid protein (PLP)4 and myelin basic protein (MBP) are the most abundant proteins in CNS myelin. Whereas knock-out mice for the tetraspan protein PLP show only subtle myelination deficits but late onset axonal degeneration (5), a lack of the cytosolic protein MBP results in almost complete failure to form compact myelin leading to shivering symptoms and premature death (6, 7). MBP compacts the forming myelin sheaths but may indeed have additional roles in the oligodendrocyte (8–10). Activation of the Src family nonreceptor tyrosine kinase Fyn is instrumental for oligodendrocyte differentiation and myelination (11–14); Fyn mutant mice are hypomyelinated, and MBP levels are reduced (15–17).
MBP mRNA is transported to the most distal regions of oligodendrocyte processes in ribonucleoprotein complexes referred to as RNA (transport) granules (18–20). The trans-acting factor heterogeneous nuclear ribonucleoprotein (hnRNP) A2 recruits MBP mRNA by binding to its cis-acting A2-response element (A2RE) in the 3′UTR. During granule transport from the nucleus to the periphery of the cell, MBP mRNA is maintained in a translationally silenced state. We recently demonstrated that activation of oligodendroglial Fyn kinase results in phosphorylation of hnRNP A2 and its release from the granules resulting in translation of MBP mRNA (21). Here, we identify the RNA-binding protein hnRNP F, a known regulator of PLP/DM20 splicing (22, 23), as a novel target of the Fyn pathway and a new component of RNA granules associating with hnRNP A2 and MBP mRNA in oligodendrocytes. Furthermore, we demonstrate that altered hnRNP F levels result in reduced MBP synthesis. Strikingly, in patients with the leukodystrophy vanishing white matter disease/childhood ataxia with central hypomyelination (VWM/CACH), mutations in the ubiquitous translational initiation factor eIF2B result in demyelination (24, 25). In cells of these patients, transcription of hnRNP F is reduced.5 Our results thus identify hnRNP F as a critical component of the MBP synthesis pathway. This novel role of hnRNP F in MBP synthesis in addition to its known function in the regulation of PLP splicing underscores its importance for myelination and has important implications for leukodystrophies.
EXPERIMENTAL PROCEDURES
Materials and Antibodies
General chemicals were purchased from Roth and Sigma. ECL reagents were from Thermo Scientific; protein A- and G-Sepharose were from GE Healthcare, and the BCA protein assay kit was from Novagen. Monoclonal antibodies were used against phosphotyrosine (mouse; clone 4G10; Millipore); 1:500 on Western blot (WB), hnRNP A2 (mouse; clone EF67; provided by W. Rigby, Dartmouth Medical School, Lebanon, NH); 1:500 on WB, 1:100 in immunoprecipitations (IP) or 1:200 in immunocytochemistry (ICC), the Myc epitope (mouse; Sigma); 1:100 in IP, PLP (rat; clone AA3; M. B. Lees, Waltham, MA); 1:10 in ICC, 2′,3′-cyclic nucleotide 3′-phosphodiesterase (mouse; Sigma); 1:500 on WB, MOG (mouse; clone 8-18C5; provided by C. Linington, University of Glasgow); 1:500 on WB, NG2 (rat; Trotter laboratory, University of Mainz); 1:20 in ICC and MBP (rat; Serotec); 1:1000 on WB or 1:500 in ICC. Polyclonal antibodies were used against hnRNP F (rabbit; Abcam); 1:1500 on WB or 1:250 in ICC, GAPDH (rabbit; Bethyl Laboratories); 1:3000 on WB, Fyn kinase (rabbit; Santa Cruz Biotechnology); 1:500 on WB, activated Fyn (rabbit; Src-pY418; Invitrogen); 1:1000 on WB and the Myc epitope (rabbit; Cell Signaling); 1:1000 on WB or 1:100 in IP. Secondary antibodies were from Dianova (HRP-coupled antibodies and Cy dyes) or Invitrogen (Alexa dyes).
Cell Culture
Primary oligodendrocytes were prepared from embryonic day 14–16 C57/BL6 mice, as described previously (27), and cultured in B27 medium containing 1% (v/v) horse serum on poly-l-lysine-coated dishes. Immediately after plating, 10 ng/ml platelet-derived growth factor (AA) and 5 ng/ml basic fibroblast growth factor were added to the medium. Cells were kept in the presence of growth factors for 2 days. To inhibit proteasomal degradation, cells were treated with the peptide N-acetyl-l-leucyl-l-leucyl-leucyl-l-norleucinal (ALLN) with a concentration of 100 μm in dimethyl sulfoxide (DMSO) for 4 h prior to lysis. Oli-neu cells (28) were cultured in Sato medium containing 1% (v/v) horse serum on poly-l-lysine-coated dishes. In some experiments, cells were differentiated by daily addition of 1 mm (final concentration) N6,2′-O-dibutyryl-adenosine-3′,5′-cyclic monophosphate.
Plasmids and RNAi
For hnRNP F expression vectors, full-length cDNA of hnRNP F was generated by RT-PCR on total RNA from Oli-neu cells, and the coding sequence was cloned into the XhoI-PstI sites of the pEGFP C3 vector (Clontech). By RT-PCR on total RNA from primary mouse oligodendrocytes, cDNA of hnRNP F lacking the stop codon was amplified and cloned into the BamHI-XhoI site of the pcDNA 4 TO/myc-His vector (Invitrogen) to obtain an hnRNP F-myc (F-myc) expression vector. Construction of the luciferase MBP reporter, including the A2RE (+A2RE) and the wild type and constitutive active Fyn constructs, was described before (21). Additionally, a luciferase MBP reporter was constructed (−A2RE) that is identical to the +A2RE plasmid but lacks 20 nucleotides at the 3′ end containing the A2RE of the MBP 3′UTR. Kinase-inactive Fyn was obtained by site-directed mutagenesis (QuickChange II; Stratagene) by replacing lysine 299 with methionine (29).
For knockdown experiments, Smartpool SiGenome siRNA (Thermo Fisher Scientific) against hnRNP F (M-051363-00-0005) and nonsilencing siRNA (target sequence 5′-AATTCTCCGAACGTGTCACGT-3′, Qiagen) were used.
qPCR
Total RNA was extracted using the RNeasy mini kit (Qiagen). RNA was reverse-transcribed using the Transcriptor High Fidelity cDNA synthesis kit (Roche Applied Science) according to the manufacturer's protocol using random hexameric primers. cDNA was amplified with the LightCycler TaqMan master kit (Roche Applied Science) and analyzed with a LightCycler 1.5 capillary-based system (Roche Applied Science) using Universal ProbeLibrary (Roche Applied Science) for detection. The primers and probes for hnRNP F, MBP, hnRNP A2, and Firefly and Renilla luciferase were designed using the Roche Applied Science website-based Universal ProbeLibrary Assay Design Center.
Transfection
Plasmids were transfected with a Gene Pulser Xcell (Bio-Rad). 10–15 μg of plasmid DNA were mixed with 1.8–2 million Oli-neu cells in Sato 1% horse serum and electroporated at 220 V and 950 microfarads (exponential decay program). After 4 or 16 h, the medium was changed completely, and 1 mm N6,2′-O-dibutyryl-adenosine-3′,5′-cyclic monophosphate was applied if desired. Alternatively Oli-neu cells were transfected using FuGENE HD (Roche Applied Science) according to the manufacturer's instructions. siRNA was introduced into primary oligodendrocytes by nucleofection (basic nucleofector kit for primary mammalian neurons; Lonza) using 160 pmol of siRNA for 4 million cells.
Immunofluorescence
Cells were fixed for 15 min at room temperature in 4% (w/v) paraformaldehyde and permeabilized with 0.1% Triton X-100 in PBS for 2 min. After blocking with 10% horse serum in PBS for 1 h, primary antibodies were applied for 1 h at room temperature in blocking medium. Detection was performed with secondary antibodies conjugated with Cy2 (1:50–1:200), Cy3 (1:1000), Cy5 (1:100), and Alexa488 or -546 (both 1:400) in blocking medium for 20 min at room temperature. In some cases, nuclei were stained with DAPI for 2 min. Cells were mounted in Mowiol, and images were acquired with a microscope (DMLB) with a 40×/0.7 NA objective lens or a 100×/1.3 NA oil objective lens connected to a digital camera (DFC 350F) using Application Suite 2.5.0 software or with a DM 6000 B microscope with a 63×/1.32 NA oil objective lens connected to a digital camera (DFC 360) using LASAF software (all from Leica). Stacked images were processed by blind deconvolution with five iterations, and single planes were shown. Images were adjusted using ImageJ and Photoshop (Adobe).
Cell Lysates and Immunoprecipitation
Cells were scraped off in cold lysis buffer (50 mm Tris, pH 7.4; 150 mm NaCl; 1 mm EDTA, pH 7.4; 1% (v/v) Nonidet P-40; in some cases plus 0.25% (v/v) sodium deoxycholate) containing protease and phosphatase inhibitor mixtures (Thermo Scientific) and incubated on a rotating wheel for 45 min. Postnuclear supernatants were obtained by pelleting the nuclei for 10 min at 300 × g at 4 °C. For simultaneous analysis of nuclear and cytoplasmic fractions, cells were lysed using the NE-PER nuclear and cytoplasmic fractionation kit (Pierce) according to the manufacturer's instructions. To deplete from RNA, lysates were treated with 50 μg/ml RNase A (Invitrogen) or with 50 units/ml RNasin (Promega; control condition) for 12 min at 37 °C. Granule-free lysates were obtained by ultracentrifugation of post-nuclear supernatants for 30 min at 136,000 × g at 4 °C.
For IP, cell lysates were incubated with 20 μl packed protein A- or G-Sepharose for 45 min (preclear). Precleared lysates were incubated with the appropriate antibody bound to 20 μl of packed protein A- or G-Sepharose for 4 h or overnight at 4 °C. Beads were washed four times with 1 ml of lysis buffer and once with 1 ml of PBS. Tyrosine-phosphorylated proteins were purified by IP using 4G10 antibody-coupled agarose beads according to the manufacturer's instructions (Millipore).
For protein-RNA coIPs, buffers were prepared with diethyl pyrocarbonate-treated H2O, and 50 units/ml RNasin (Promega) were added. Proteins and RNA were eluted from the beads by incubation with 0.2% (w/v) SDS and 2% (v/v) β-mercaptoethanol in nuclease-free H2O for 5 min at 70 °C. cDNA from total RNA (lysate) and RNA eluted from IP and control-IP was generated with the Transcriptor High Fidelity cDNA synthesis kit (Roche Applied Science) according to the manufacturer's protocol using random hexameric primers. Relative cDNA levels were quantified by real time-PCR using a one-step real time-PCR system and the TaqMan assays Mm01225301_m1, Mm00607939_s1, and Mm01266402_m1 (all Applied Biosystems). The relative enrichment of mRNAs in the hnRNP F-myc and control IPs was calculated as described before (30). In individual experiments the enrichment factor of MBP and β-actin mRNAs was normalized to the enrichment factor of phosphoglycerate kinase 1 mRNA.
Proteins were separated by SDS-PAGE (Mini PROTEAN 3 system; Bio-Rad) and analyzed on Western blots (Mini Trans-Blot Electrophoretic Transfer Cell; Bio-Rad). Prior to antibody incubation, the PVDF membranes (Immobilon-P; Millipore) were blocked for 30 min with 4% (w/v) milk in TBST (0.05 m Tris, 0.15 m NaCl, pH 7.2, 1% (v/v) Tween 20). Primary antibodies were incubated overnight at 4 °C and secondary antibodies for 30 min at room temperature, all in blocking medium. For the phosphotyrosine antibody, 3% (w/v) bovine serum albumin in TBST was used as blocking medium.
In Vitro Fyn Kinase Assay
Immunoprecipitations of F-myc were washed twice with lysis buffer, followed by two washing steps with kinase buffer (50 mm PIPES, pH 7.02; 10 mm MgCl2) and kinase buffer containing 50 μm ATP, respectively. 30 ng of GST-tagged active recombinant human FynB (SignalChem) were added to the beads in 40 μl of kinase buffer with 50 μm ATP. As a control condition, the kinase was replaced with an equal amount of distilled H2O. The assay was carried out incubating the beads for 20 min at 30 °C. The reaction was stopped, and the proteins were eluted by the addition of sample buffer and heating of the beads for 5 min at 90 °C. Phosphorylated proteins were analyzed by SDS-PAGE and Western blotting.
Luciferase Assay
After siRNA treatment, Oli-neu cells were transfected with 250 ng +A2RE or −A2RE Firefly luciferase reporter (see above), 100 ng of Renilla luciferase, and 1650 ng of pEGFP C3 plasmids. All luciferase constructs are driven by the same promoter (CMV). In hnRNP F overexpression experiments, 1650 ng of pEGFP was replaced with 1650 ng of hnRNP F plasmid. After 2 days, cells were scraped off in PBS, and a luciferase assay was performed according the manufacturer's instructions (DualGlo; Promega). Firefly luciferase activities were normalized with Renilla luciferase activities. Total RNA was isolated from remaining cells and analyzed by qRT-PCR for Firefly and Renilla luciferase mRNA.
RESULTS
hnRNP F Is Associated with hnRNP A2 in Cytoplasmic Granular Structures in Oligodendrocytes
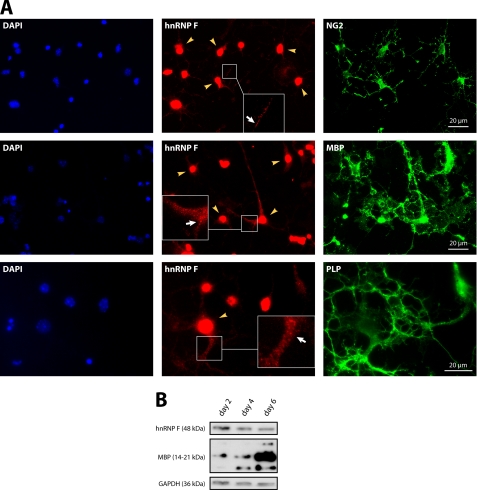
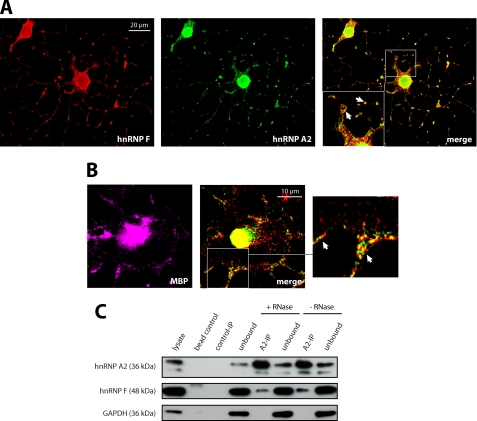
To identify downstream target proteins of the nonreceptor tyrosine kinase Fyn in oligodendrocytes, we introduced a constitutively active mutant of Fyn into the oligodendroglial precursor cell line Oli-neu (28) by transfection. We then purified tyrosine-phosphorylated proteins by immunoprecipitation and identified these by subsequent SDS-PAGE and mass spectrometry (data not shown). The RNA-binding protein hnRNP F was one of these tyrosine-phosphorylated proteins, and we analyzed the role of this protein in oligodendrocytes. We confirmed the expression of hnRNP F in NG2-positive oligodendrocyte precursor cells as well as in more mature MBP- and PLP-positive cells by immunocytochemistry and Western blotting (Fig. 1). hnRNP F showed a dominant nuclear localization in agreement with the previously reported nuclear role in the alternative splicing of PLP and DM20 in oligodendrocytes (22, 23). Interestingly, however, the hnRNP F staining revealed granular structures in the cytoplasm and processes of oligodendrocytes at all differentiation stages (see insets, Fig. 1A). The homologue of hnRNP F in Drosophila, Glorund, has been shown to regulate the translation of specific mRNAs in the cytoplasm of oocytes (31). As the granular pattern of hnRNP F staining resembled the cytoplasmic distribution of hnRNP A2 and E1, proteins that play a role in the localization and translation of MBP mRNA in oligodendrocytes (21, 32), we further evaluated a potential cytoplasmic function of hnRNP F. A substantial fraction of hnRNP F colocalizes with hnRNP A2 in cytoplasmic granular structures of differentiated Oli-neu cells and primary oligodendrocytes (Fig. 2, A and B). Moreover, hnRNP F coimmunoprecipitates with hnRNP A2 from postnuclear supernatants, further endorsing the interaction of hnRNP A2 and hnRNP F in the cytoplasm (Fig. 2C). As this coimmunoprecipitation is RNase-resistant (Fig. 2C), we conclude that the interaction between hnRNP F and A2 does not require the presence of RNA. Together, these results indicate that hnRNP F is associated with hnRNP A2 in cytoplasmic granules in oligodendrocytes.
FIGURE 1.
hnRNP F is present in the nucleus and the cytoplasm of oligodendrocytes. A, in addition to its prominent nuclear localization (yellow arrowheads), hnRNP F is present in granular structures throughout the cytoplasm (white arrows). Primary mouse oligodendrocytes after 2 or 4 days in culture were stained with antibodies to hnRNP F and markers for oligodendrocyte precursor cells (NG2; 2 days in vitro) or more mature oligodendrocytes (MBP, PLP; 4 days in vitro). Cell nuclei were stained with DAPI. Insets show enlarged areas. B, Western blots confirming the expression of hnRNP F in immature and mature oligodendrocytes (2, 4, and 6 days in vitro). MBP serves as marker for the ongoing differentiation of the cells, and GAPDH is shown as a loading control.
FIGURE 2.
hnRNP F and hnRNP A2 associate in cytoplasmic granular structures in oligodendroglial cells. A, partial colocalization of hnRNP F and hnRNP A2 in granular structures (white arrows) in the cytoplasm of Oli-neu cells that were allowed to differentiate for 3 days and immunostained for hnRNP F and hnRNP A2. Single deconvoluted planes are depicted; the inset shows an enlarged area. B, partial colocalization of hnRNP F and hnRNP A2 in granular structures (enlargement: white arrows) in the cytoplasm of primary mouse oligodendrocytes that were immunostained for hnRNP F and hnRNP A2 after 3 days in culture. MBP is shown as oligodendroglial marker. Single deconvoluted planes are depicted. C, hnRNP F coimmunoprecipitates with hnRNP A2. Immunoprecipitations with hnRNP A2 or isotype-matched control antibodies were performed from RNase- or untreated Oli-neu postnuclear lysates and analyzed on Western blots. The absence of GAPDH in the hnRNP A2-IP confirms the specificity of hnRNP F binding. Note that the hnRNP A2 Western blots show an additional lower band at ∼32 kDa, which is likely to be the hnRNP A2/B1 splice variant hnRNP A2b.
hnRNP F Associates with MBP mRNA in RNA Transport Granules
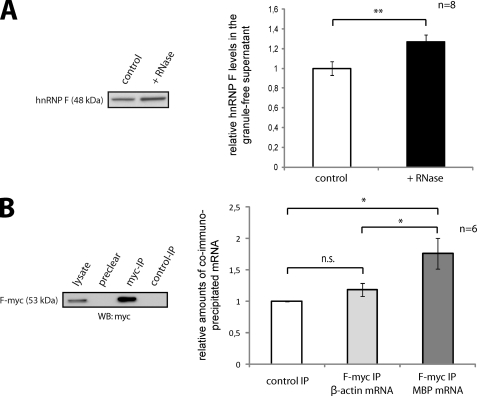
It is well documented that hnRNP A2 is a major constituent of RNA transport granules containing mRNAs with an A2-response element in the 3′UTR (19, 33). The association of the RNA-binding protein hnRNP F with hnRNP A2 in granular cytoplasmic structures alludes to a function of hnRNP F in mRNA localization. To verify that the granular distribution of hnRNP F is RNA-dependent, we treated granule-containing oligodendroglial post-nuclear supernatants with RNase A to disrupt RNA granules. We subsequently created a granule-free supernatant by ultracentrifugation as described previously (21). Western blot analysis of this granule-free supernatant containing soluble cytoplasmic proteins showed an enrichment of hnRNP F in RNase A-treated compared with untreated samples (Fig. 3A). The disruption of RNA granules thus leads to an increase of free hnRNP F in the cytoplasmic fraction and supports the concept that hnRNP F is a component of RNA transport granules in oligodendrocytes. The cytoplasmic association of hnRNP F and hnRNP A2 (Fig. 2) suggests that hnRNP F is involved in the assembly and transport of MBP mRNA granules. We performed experiments to further investigate the potential interaction between hnRNP F and MBP mRNA. Oli-neu cells were cotransfected with expression constructs coding for recombinant Myc-tagged hnRNP F (F-myc) and MBP14 (including the 3′UTR). F-myc was immunoprecipitated, and copurifying mRNAs were quantified by reverse transcription and qPCR. To examine whether MBP14 mRNA was specifically enriched, it was related to the nongranule-associated phosphoglycerate kinase 1 mRNA as a control. A significant enrichment of MBP mRNA in F-myc immunoprecipitated samples compared with immunoprecipitations with isotype-matched control antibodies (Fig. 3B) was found, supporting an association of MBP mRNA with hnRNP F. Interestingly, β-actin mRNA, which is transported in ZBP1-dependent granules (34), is not enriched in F-myc immunoprecipitated samples (Fig. 3B).
FIGURE 3.
hnRNP F is associated with MBP mRNA and RNA granules. A, disruption of RNA granules increases the amount of soluble cytoplasmic hnRNP F. Postnuclear lysates of Oli-neu cells were treated with RNase A, and after depletion of RNA granules by ultracentrifugation, soluble cytoplasmic proteins were analyzed on Western blots. The change in soluble hnRNP F protein was densitometrically quantified and normalized to GAPDH. **, p < 0.01 (Student's t test). B, MBP mRNA coimmunoprecipitates with hnRNP F-myc. Oli-neu cells were cotransfected with hnRNP F-myc (F-myc) and MBP14 including its 3′UTR. After differentiation for 3 days, immunoprecipitations with antibodies against the Myc tag or isotype-matched control antibodies were performed from postnuclear lysates, and associated mRNAs were analyzed by qRT-PCR. MBP and β-actin mRNA levels were normalized to phosphoglycerate kinase 1 mRNA. n.s., not significant. *, p < 0.05 (Wilcoxon signed-rank test).
hnRNP F Is Involved in Post-transcriptional Regulation of MBP Expression
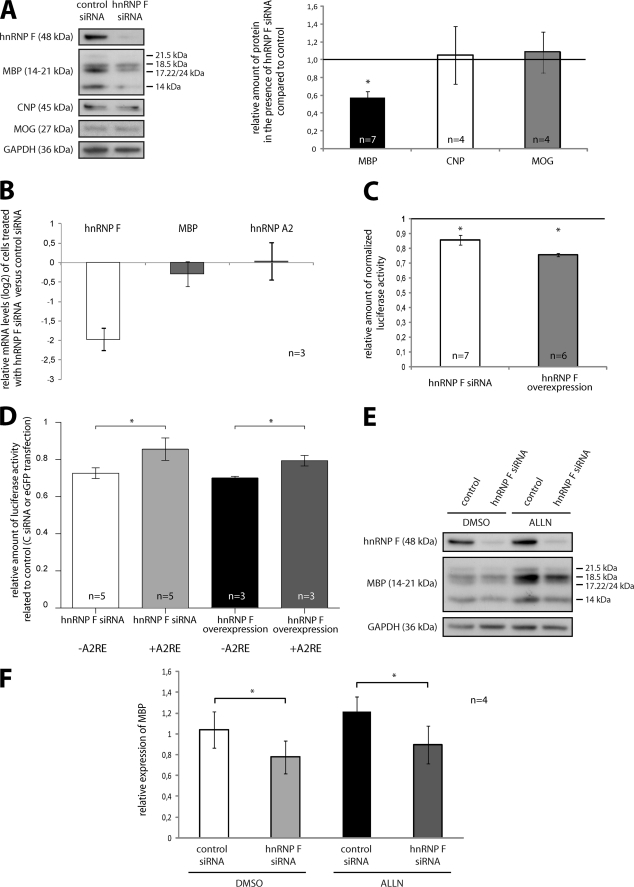
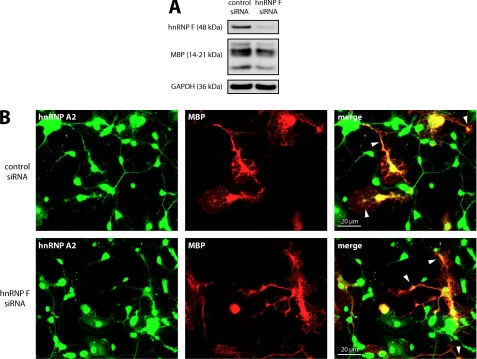
Having shown an association of hnRNP F with hnRNP A2-dependent MBP RNA granules and a binding to MBP mRNA, we investigated a role of hnRNP F in the regulation of MBP expression. hnRNP F protein levels were reduced in primary oligodendrocytes by siRNA, and we analyzed by Western blotting if this influenced the expression of MBP. As shown in Fig. 4A, hnRNP F knockdown leads to a significant reduction of MBP levels, whereas the amount of the other myelin proteins CNP and MOG are unaltered. In addition, qPCR experiments showed that MBP mRNA levels are not altered by the reduction of hnRNP F, leading to the conclusion that hnRNP F functions at the translational rather than the transcriptional level (Fig. 4B). The expression of hnRNP A2 was not affected by hnRNP F silencing demonstrating the specificity of hnRNP F-directed siRNA. We confirmed a potential role of hnRNP F on MBP translation in a second approach by using a luciferase reporter-based translational assay as described previously. In this assay, a construct is used in which Firefly luciferase expression is driven by a cytomegalovirus (CMV) promoter and is fused to a 378-nucleotide-long region of the MBP 3′UTR upstream of and including the A2RE sequence, which was reported to control the translation of the Firefly luciferase reporter in a Fyn-dependent manner (21). To normalize the readouts, this vector was cotransfected into Oli-neu cells with a Renilla luciferase that is driven by the same promoter but lacks a downstream regulatory element. These experiments hence focus on the role of the 3′UTR of MBP on translation and exclude general transcriptional or translational effects. We observed a significant reduction of normalized Firefly luciferase activity in response to reduction of hnRNP F by transfection of hnRNP F-directed siRNA. Intriguingly, overexpression of hnRNP F also led to reduction of normalized reporter activity (Fig. 4C). To exclude the possibility that mRNA stability is affected in these assays, we analyzed the Firefly and Renilla luciferase mRNA levels by qPCR, and they were unaltered (data not shown). The discrepancy between the modest reduction in reporter activity and the stronger reduction in MBP may be due to the overexpression of Firefly luciferase and an escape of the highly abundant mRNA from incorporation into RNA granules and thus from translation repression or from additional regulatory effects of hnRNP F on other regions of MBP mRNA (35). To further analyze if the hnRNP F-mediated effect on MBP mRNA translation requires the binding site of hnRNP A2, we used an additional luciferase reporter construct lacking the A2RE resulting in a 358-nucleotide fragment of the 3′UTR. As shown in Fig. 4D, both in the presence and absence of the hnRNP A2-binding site, overexpression and siRNA-mediated reduction of hnRNP F shows a reduction of normalized luciferase activity compared with enhanced GFP or control siRNA-transfected cells, respectively. The absence of an A2RE significantly reduces the amount of reporter activity in both experimental approaches suggesting that binding of hnRNP A2 to the mRNA does not seem to be required for the action of hnRNP F. Interestingly, when the luciferase assay was performed with a reporter construct, including the entire 3′UTR of MBP mRNA, the knockdown of hnRNP F led to comparable results indicating that the interaction of hnRNP F with MBP mRNA occurs within the first 358 nucleotides of the 3′UTR (data not shown). To entirely exclude that the reduction of MBP levels is not a result of increased degradation specific for MBP but not affecting other proteins such as GAPDH, MOG, or CNP, we treated primary oligodendrocytes with control or hnRNP F-directed siRNA in the presence of the proteasomal inhibitor N-acetyl-l-leucyl-l-leucyl-leucyl-l-norleucinal (ALLN) as described (36). As depicted in Fig. 4, E and F, proteasomal inhibition does not alter the hnRNP F siRNA-mediated effect leading to the conclusion that decreased synthesis rather than increased degradation are responsible for the reduction in MBP levels. Taken together, the results shown in Fig. 4, strongly suggest a function of hnRNP F in the synthesis pathway of MBP. Synthesis of protein from localized mRNAs is precluded by their transport to their specific destination where translation ensues. hnRNP A2 binding to MBP mRNA is required to transport these RNA granules to the periphery of the cell (33). We thus performed experiments in which we reduced hnRNP F levels in primary oligodendrocytes by siRNA resulting in a reduction of MBP measured by Western blot (Fig. 5A) and then visualized hnRNP A2 in cells from the same experiment by immunocytochemistry. We could not detect any differences in the cytoplasmic localization of hnRNP A2 and MBP between hnRNP F and control siRNA-transfected primary oligodendrocytes (Fig. 5B). These results make it very unlikely that the reduction of MBP levels is caused by a defect in RNA granule transport to the periphery of the cell.
FIGURE 4.
MBP synthesis is influenced by hnRNP F. A, knockdown of hnRNP F specifically affects MBP levels. Primary mouse oligodendrocytes were treated with hnRNP F or control siRNA and allowed to differentiate for 3 days. Protein levels were quantified by densitometric analysis of Western blots, and the expression of MBP, CNP, and MOG was normalized to GAPDH. B, knockdown of hnRNP F has no impact on levels of MBP mRNA. Primary mouse oligodendrocytes were treated as in A, but here mRNA levels were analyzed by qRT-PCR and normalized to β-actin mRNA. C, altered hnRNP F levels impair the translation of MBP luciferase reporters. hnRNP F levels in Oli-neu cells were reduced by transfection of hnRNP F-directed siRNA or increased by transfection of hnRNP F expression vectors. Subsequently, luciferase-based translational reporters containing parts of the 3′UTR of MBP as regulatory elements were used to measure translational activity in DualGlo assays (see “Experimental Procedures” for details). Relative luciferase activity is reduced in cells treated with hnRNP F siRNA as well as in hnRNP F-overexpressing cells that were related to control siRNA- or GFP-transfected cells, respectively. D, effect of hnRNP F levels on the translation of MBP reporters, including or lacking the A2RE. Cells were treated as in C, but here a luciferase reporter construct lacking the A2RE was included. E, reduced MBP levels in response to hnRNP F knockdown are independent of proteasomal activity in primary oligodendrocytes. The experiment was performed according to A. Before lysis, the siRNA-treated cells were incubated with the proteasomal inhibitor ALLN or as control with DMSO. F, statistical evaluation of four independent experiments as shown in E. Note the persistence in MBP reduction despite inhibition of the proteasome. *, p < 0.05 (Student's t test (D and F) or Wilcoxon signed-rank test (A and C)).
FIGURE 5.
Cytoplasmic localization of hnRNP A2 and MBP distribution appear unaffected by hnRNP F knockdown. A, primary mouse oligodendrocytes were treated with hnRNP F or control siRNA and allowed to differentiate for 3 days. The knockdown of hnRNP F was assessed by Western analysis. GAPDH serves as loading control. B, cells from A were immunostained for hnRNP A2 and MBP. Note the characteristic distribution of MBP and the cytoplasmic localization of hnRNP A2, in particular its concentration at foci where MBP is highly abundant (white arrowheads).
Cytoplasmic hnRNP F Is a Downstream Target of Fyn
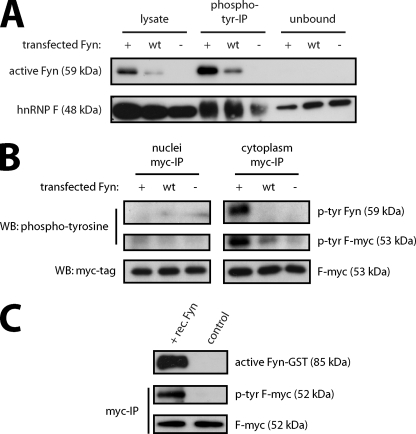
We previously reported a role of Fyn kinase in the translational control of MBP expression by phosphorylation of hnRNP A2 (21). As described above, we identified hnRNP F as a tyrosine-phosphorylated protein in response to Fyn activity in oligodendrocytes. To confirm the role of Fyn kinase in the phosphorylation of endogenous hnRNP F, we transfected Oli-neu cells with constructs expressing constitutively active (Fyn+), wild type (Fyn WT), or kinase-inactive Fyn (Fyn−). We then isolated tyrosine-phosphorylated proteins by immunoprecipitation and visualized hnRNP F by Western blotting (Fig. 6A). The amount of tyrosine-phosphorylated hnRNP F is strongly increased in the samples from cells transfected with wild type and constitutively active Fyn compared with kinase-inactive Fyn. Furthermore, the remaining unphosphorylated hnRNP F, which is not precipitated by phosphotyrosine antibodies, is reduced in Fyn+ and Fyn WT (unbound in Fig. 6A) compared with Fyn-transfected cells. These results thus confirm that hnRNP F is a downstream target in the Fyn kinase pathway. To exclude that hnRNP F is detected in the phosphotyrosine precipitations solely due to coprecipitation with tyrosine-phosphorylated hnRNP A2 (21), we analyzed Fyn-dependent phosphorylation of hnRNP F by precipitation of hnRNP F followed by phosphotyrosine detection. Moreover, hnRNP F shows a dominant nuclear localization in addition to its presence in cytoplasmic granules (Figs. 1A and 2A), and it was thus important to analyze whether the phosphorylation of hnRNP F takes place in the cytoplasm where it would be accessible to Fyn kinase. We cotransfected Myc-tagged hnRNP F (F-myc) with constitutively active (Fyn+), wild type (Fyn WT), or kinase-inactive Fyn (Fyn−) and purified F-myc from cytoplasmic and nuclear fractions using antibodies directed against the Myc tag. As shown by Western analysis in Fig. 6B, F-myc was purified from nuclear and cytoplasmic fractions in equal amounts regardless of which of the Fyn plasmids was cotransfected. A phosphotyrosine Western blot illustrates that F-myc is tyrosine-phosphorylated in the cytoplasmic fraction of cells transfected with Fyn+ and Fyn WT. Moreover, active Fyn is coimmunoprecipitated with hnRNP F only in the cytoplasmic fraction. This provides further evidence for Fyn-dependent phosphorylation of hnRNP F in the cytoplasm. We then tested whether hnRNP F is a direct target of Fyn by incubating immunoprecipitated F-myc with recombinant Fyn kinase in vitro. Fig. 6C shows that F-myc is tyrosine-phosphorylated in the presence of recombinant Fyn demonstrating the ability of Fyn to phosphorylate hnRNP F directly.
FIGURE 6.
Cytoplasmic hnRNP F is a target of Fyn kinase. A, hnRNP F is tyrosine-phosphorylated in response to Fyn activity. Oli-neu cells were transfected with constitutive active (Fyn+), wild type (Fyn WT), or kinase-inactive (Fyn−) Fyn constructs. After 2 days, tyrosine-phosphorylated proteins were immunoprecipitated and analyzed on Western blots together with total lysates and proteins that were not precipitated (unbound). B, Fyn-dependent tyrosine phosphorylation of hnRNP F occurs in the cytoplasm of oligodendrocytes. Oli-neu cells were cotransfected with Fyn+, Fyn WT, or Fyn− together with Myc-tagged hnRNP F (F-myc). 2 days later, separate nuclear and cytoplasmic fractions were prepared, and immunoprecipitations with antibodies against the Myc tag were performed. The immunoprecipitations were analyzed on Western blots for tyrosine-phosphorylated proteins and total amounts of F-myc. C, Fyn phosphorylates hnRNP F directly. Myc-tagged hnRNP F was immunoprecipitated and incubated in the presence or absence of recombinant Fyn. Western blot analysis with phosphotyrosine- and Myc-specific antibodies shows that the addition of Fyn results in tyrosine phosphorylation of hnRNP F. An activity-dependent antibody (Src-pY418) demonstrates that recombinant Fyn is active in the experiment.
hnRNP F Dissociates from RNA Granules and MBP mRNA in a Fyn-dependent Manner
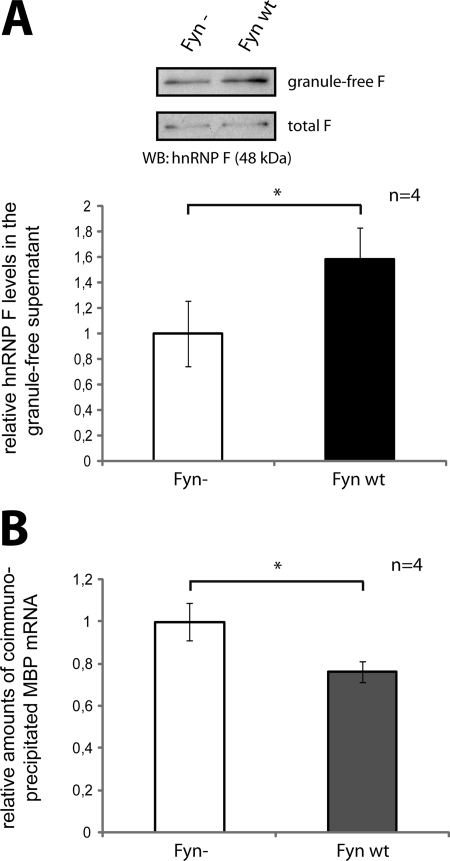
We further analyzed if the association of hnRNP F with RNA granules and MBP mRNA was altered by Fyn activity. Oli-neu cells were transfected with Fyn− or Fyn WT, and postnuclear supernatants were subjected to ultracentrifugation to isolate granule-free supernatants containing cytoplasmic proteins. As shown by Western and densitometric analysis, the overexpression of Fyn WT leads to a significant increase of cytosolic hnRNP F that is not associated with RNA granules (Fig. 7A). This suggests that Fyn induces the dissociation of hnRNP F from RNA transport granules. To see if the Fyn-induced dissociation of hnRNP F from the RNA granules also affects the association of hnRNP F with MBP mRNA, we cotransfected F-myc with either Fyn− or Fyn WT. We then analyzed the levels of associated endogenous MBP mRNA copurifying with F-myc in Fyn WT versus Fyn-transfected cells. Interestingly, Fyn WT leads to a significant reduction in MBP mRNA associated with F-myc (Fig. 7B). This suggests that MBP mRNA dissociates from hnRNP F in a Fyn-dependent manner. Together, these results imply that Fyn activity induces a dissociation of hnRNP F from MBP mRNA and the granule, thus facilitating translation.
FIGURE 7.
Fyn activity leads to a release of hnRNP F from MBP mRNA. A, Fyn activity releases hnRNP F from the granule fraction into the cytosol. Oli-neu cells were transfected with Fyn WT or Fyn− constructs. After 2 days, postnuclear total lysates and granule-free lysates, obtained by ultracentrifugation, were analyzed on Western blots. The levels of hnRNP F in the nongranule fraction were quantified densitometrically and normalized to total hnRNP F levels. B, Fyn activity releases hnRNP F from MBP mRNA. Oli-neu cells were cotransfected with F-myc and Fyn WT or Fyn−, respectively. After differentiation for 3 days, immunoprecipitations with antibodies against the Myc tag or control antibodies were performed from postnuclear lysates, and associated mRNAs were analyzed by qRT-PCR. MBP mRNA levels were normalized to levels of phosphoglycerate kinase 1 mRNA. *, p < 0.05 (Student's t test).
DISCUSSION
Oligodendroglial synthesis of MBP is essential for myelination. Here, we define hnRNP F as a novel protein component of MBP RNA transport granules in oligodendrocytes that binds to MBP mRNA and is important for the post-transcriptional regulation of MBP synthesis. Distinct levels of hnRNP F appear to be required for normal MBP synthesis, as this process is affected by the knockdown as well as the overexpression of hnRNP F. Cytoplasmic hnRNP F is phosphorylated in response to Fyn activation resulting in its release from MBP mRNA and the granule.
Regulated Localization and Translation of MBP mRNA in Oligodendrocytes
Regulated translation of localized mRNAs is widespread in biology and includes cell types of the CNS. In neurons, mRNAs encoding postsynaptic proteins and β-actin are transported in RNA granules to dendrites and the axonal growth cone, respectively (34, 37). The isolation of ribosomes and MBP mRNA from biochemically purified myelin fractions (38) led to the postulation of localized regulated translation of MBP mRNA in myelinating oligodendrocytes. A transport pathway was subsequently identified in which MBP mRNA is transported from the nucleus to the most distal regions of oligodendrocyte processes in ribonucleoprotein complexes mediated by hnRNP A2 (19). During granule transport, MBP mRNA is maintained in a translationally silenced state, which has been proposed to be regulated by hnRNP E1 that is recruited to RNA granules by hnRNP A2 (Fig. 8) (32). Src family kinases have been shown to regulate translation initiation of mRNAs transported in RNA granules in both neurons and erythrocytes by phosphorylating distinct RNA-binding proteins (34, 39). We showed previously that activation of Fyn kinase leads to a phosphorylation of hnRNP A2, dissociation of hnRNP A2 and E1 from RNA granules, and increased translation of an A2RE containing reporter construct (21). These results demonstrated that Fyn activation is a critical regulator of translation of MBP mRNA in oligodendrocytes, in addition to a published role in regulating transcription of the MBP gene (40). Fyn-deficient mice show reduced MBP levels and are strongly hypomyelinated in the forebrain (16, 17, 41). Fyn activity is controlled by several pathways, including the binding of neuronal L1-CAM to glial F3/Contactin (21), ligation of glial integrins (42–44), and binding to FcRγ (common γ chain of immunoglobulin Fc receptors) (45) on the surface of oligodendrocytes (12).
FIGURE 8.
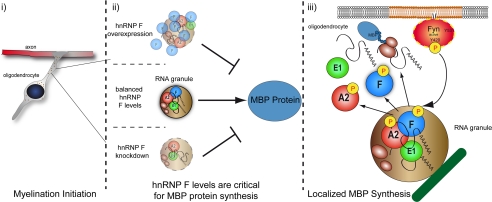
Model. MBP mRNA is transported in RNA granules toward the periphery of the cell where Fyn-mediated translational initiation occurs by phosphorylation of RNA-binding proteins and their liberation from the granule and a dissociation of MBP mRNA. Distinct levels of hnRNP F seem to be required to form fully functional MBP mRNA granules thus facilitating efficient protein synthesis.
hnRNP F Is a Novel Structural Component of Oligodendrocyte MBP mRNA Granules
The RNA-binding protein hnRNP F contains three RNA recognition motifs and is expressed by many cell types (46, 47). We show here that hnRNP F is expressed at all stages of the oligodendrocyte lineage from NG2+ precursor cells to mature PLP+ and MBP+ oligodendrocytes. In Oli-neu cells, which represent a precursor stage of the oligodendroglial lineage, hnRNP F together with hnRNP H have been reported to regulate differential splicing of PLP/DM20 in the nucleus during oligodendrocyte differentiation (23). Our results confirm that in primary cultured oligodendrocytes, endogenous hnRNP F is strongly expressed in the nucleus. Additionally, we could detect both endogenous and recombinantly expressed hnRNP F in the cytoplasm, which implies an additional and novel function of hnRNP F outside the nucleus. Cytoplasmic hnRNP F localizes in granular structures in the oligodendroglial cytoplasm and processes. We show that hnRNP F is present in RNA granules, associates with hnRNP A2 in an RNA-independent manner, and coimmunoprecipitates with MBP mRNA, demonstrating that hnRNP F is a structural component of MBP mRNA granules in oligodendrocytes. Interestingly, our observation of cytoplasmic granules containing both hnRNP A2 and hnRNP F, or only one of these proteins, suggests heterogeneity among oligodendroglial RNA granules. It remains unclear if the granules containing only hnRNP F (or hnRNP A2) are intermediates in the maturation process or if they represent distinct types of granules that may potentially differ in their cargo. As reported recently for neurons, different types of A2RE-containing mRNAs can be transported within the same hnRNP A2-containing granule (37), which may represent an energy-efficient localization of mRNA within the cell or, more specifically, a pre-selection of molecules that interact functionally at a specific place or time. There are four splice variants of the hnRNP A2/B1 gene resulting in four different proteins (48). Most former studies have focused on hnRNP A2, which is the most abundant isoform in the brain. It was recently proposed that the hnRNP A2b splice variant rather than hnRNP A2 is primarily localized in the cytoplasm (49). The antibody used in this study does not discriminate between the different isoforms, and it remains unclear if the binding of hnRNP F to hnRNP A2 is isoform-specific. hnRNP A2 binds to a defined linear sequence in the mRNA. In contrast, in Drosophila, the binding of nanos mRNA to the Drosophila homologue of hnRNP F, Glorund, is thought to be mediated by conformational epitopes in the mRNA that are hard to define. A 358-nucleotide region of the 3′UTR upstream of the A2RE appears sufficient to mediate the interaction of hnRNP F with MBP mRNA (Fig. 4D). Together with our observation that hnRNP A2 and F colocalize only partially in cytoplasmic granules, it is possible that hnRNP F binds to additional mRNAs in oligodendrocytes independently of hnRNP A2 and the A2RE. However, our results demonstrate a degree of specificity for hnRNP F as β-actin mRNA, which has been shown to be present in ZBP1-dependent granules (34), does not associate with hnRNP F (Fig. 3B).
hnRNP F Is a Novel Functional Component of Oligodendrocyte MBP mRNA Granules
Intriguingly, Glorund has been shown to regulate the translation of nanos mRNA during Drosophila oogenesis (31). The cytoplasmic distribution in nurse cells and oocytes resembles the hnRNP F-containing RNA granules we observed in oligodendrocytes. We show a role of hnRNP F in post-transcriptional regulation of MBP as down-regulation of hnRNP F by siRNA reduces the translation of an A2RE-containing mRNA reporter and furthermore leads to a significant decrease of MBP levels in oligodendrocytes, although MBP mRNA remains unchanged. The hnRNP F siRNA-mediated reduction of MBP is independent of proteasomal activity as the ALLN application does not interfere with this effect. Hence, it is very likely that MBP synthesis rather than its degradation pathway is affected by hnRNP F manipulation. Given the fact that other myelin proteins are unaffected by hnRNP F manipulation, this regulation of MBP seems to be specific and not a general effect on myelin protein synthesis. Interestingly, the down-regulation of other MBP mRNA granule components such as tumor overexpressed gene, hnRNP A2, and hnRNP K also results in decreased MBP levels (26, 50). Importantly, in our experiments hnRNP A2 expression levels remain unaltered, which emphasizes a direct effect of hnRNP F and excludes a secondary effect by hnRNP A2. Moreover, overexpression and knockdown of hnRNP F decreases translational activity of luciferase reporters containing or lacking the A2RE, suggesting an hnRNP A2-independent potential of F in regulating MBP synthesis. Similar to results with knockdown of tumor overexpressed gene, transport of MBP mRNA-containing granules appears unaffected in the absence of hnRNP F as hnRNP A2 and MBP distribution throughout the cytoplasm of oligodendrocytes appears normal. In contrast, the knockdown of hnRNP K leads to an accumulation of transport granules at oligodendroglial process branch points, although reduced hnRNP A2 levels result in a cytoplasmic retention of granules (26). Our work, where overexpression or reduction of hnRNP F shows similar effects in the luciferase assays, suggests that balanced levels of hnRNP F are required to form fully functional MBP mRNA transport granules, thus facilitating protein synthesis (Fig. 8).
Cytoplasmic Phosphorylation and Release of hnRNP F from RNA Granules by Fyn Activity
In neurons, the nonreceptor tyrosine kinase Src binds to the Src homology 3 binding domain and phosphorylates the RNA-binding protein ZBP-1, which relieves the translational inhibition of β-actin mRNA, allowing actin protein synthesis at the tip of growing processes (34). We identified hnRNP F as a potential oligodendroglial phosphorylation target of the Src family nonreceptor tyrosine kinase Fyn. We confirmed the role of Fyn kinase activity in hnRNP F phosphorylation and demonstrated that hnRNP F is phosphorylated in the cytoplasm that is in agreement with the subcellular localization of Fyn kinase that is associated with the plasma membrane by acylation. By motif prediction analysis, we determined that amino acids 311–316 (PLNPVR) within the hnRNP F sequence represent a potential binding site for Src homology 3 domains such as is present in Fyn. An in vitro kinase assay strongly suggests that Fyn phosphorylates hnRNP F in a direct manner (Fig. 6C).
Translation of an A2RE-containing luciferase construct is stimulated by active Fyn (21). Strikingly, overexpression of active Fyn in Oli-neu cells resulted in an increase of cytosolic hnRNP F that is not granule-associated and a decrease of associated MBP mRNA, suggesting that upon Fyn activation hnRNP F dissociates from the mRNA and leaves the granule, allowing translation to ensue. Based on our experiments, it cannot be excluded that the dissociation of hnRNP F from the granule in response to Fyn activity is (partly) mediated by the phosphorylation of hnRNP A2. Our results indicate, however, that hnRNP F influences MBP reporter translation independent of hnRNP A2. Abnormal hnRNP F levels lead to a significant reduction of MBP reporter levels lacking the A2RE when related to A2RE-containing reporters. This can be the result of decreased translational initiation at the target site as hnRNP A2 is required for transport of MBP mRNA to the periphery of the cell (33). Taken together, our results suggest that Fyn activation mediates translation initiation of MBP mRNA by several means, including phosphorylation of hnRNP A2 and F and release of hnRNP F, A2, and E1 from the RNA granules. It seems that specific levels of hnRNP F are required to allow normal MBP synthesis as knockdown and overexpression of hnRNP F both reduce MBP reporter levels. We suggest that one has to distinguish between the action of Fyn on “normal” granules and the malfunction of granules that contain unbalanced levels of hnRNP F (Fig. 8).
Diminished Levels of hnRNP F in Leukodystrophies May Contribute to Demyelination
A subgroup of leukodystrophies known as VWM/CACH result from mutations in the ubiquitous translation initiation factor eIF2B (24, 25). The patients suffer from demyelination with subsequent neuronal degeneration that is worsened by a variety of stress conditions. The eIF2B factor is involved in translation initiation regulation by activating the eIF2 complex via guanine nucleotide exchange factor activity. EIF2B mutations strikingly affect the white matter of the CNS, which is thought to result from an enhanced sensitivity of glial cells to eIF2B dysfunction. In transcriptome analyses of fibroblasts or fetal white matter from these patients, several mRNAs are substantially down-regulated, including hnRNP F.5 Our results presented here demonstrate that in addition to the recently discovered role in splicing of PLP/DM20 (22, 23), hnRNP F has an important function in post-transcriptional control of MBP mRNA. Thus, hnRNP F regulates expression of the two main myelin proteins of the CNS. As MBP is known to be critical for myelination, insufficient levels of hnRNP F in the patients with eIF2B mutations could lead to myelin instability and demyelination due to misregulated MBP translation.
Acknowledgments
We thank U. Stapf and L. Niedens for technical assistance, Dr. Rigby for providing hnRNP A2 antibodies (clone EF67), and Dr. Linington for providing MOG antibodies (clone 8-18C5).
This work was supported by Deutsche Forschungsgemeinschaft SPP Glia-Synapse, European Union FP6 Signalling and Traffic, and Deutsche Forschungsgemeinschaft Grant GRK 1044.
L. Horzinski, A. Huyghe, E. Bertini, R. Schiffmann, D. Rodriguez, Y. Dantal, O. Boespflug-Tanguy, and A. Fogli, poster presented at the 9th European Meeting on Glial Cells in Health and Disease, Paris, France, September 8–12, 2009.
- PLP
- proteolipid protein
- MBP
- myelin basic protein
- hnRNP
- heterogeneous nuclear ribonucleoprotein
- A2RE
- A2-response element
- VWM/CACH
- vanishing white matter disease/childhood ataxia with central hypomyelination
- WB
- Western blot
- IP
- immunoprecipitation
- ICC
- immunocytochemistry
- qPCR
- quantitative PCR
- MOG
- myelin oligodendrocyte glycoprotein
- CNP
- 2′,3′-cyclic nucleotide 3′-phosphodiesterase
- ALLN
- N-acetyl-l-leucyl-l-leucyl-leucyl-l-norleucinal.
REFERENCES
- 1. Nave K. A. (2010) Myelination and the trophic support of long axons. Nat. Rev. Neurosci 11, 275–283 [DOI] [PubMed] [Google Scholar]
- 2. Aggarwal S., Yurlova L., Simons M. (2011) Central nervous system myelin. Structure, synthesis, and assembly. Trends Cell Biol. 21, 585–593 [DOI] [PubMed] [Google Scholar]
- 3. Emery B. (2010) Regulation of oligodendrocyte differentiation and myelination. Science 330, 779–782 [DOI] [PubMed] [Google Scholar]
- 4. Simons M., Trotter J. (2007) Wrapping it up. The cell biology of myelination. Curr. Opin. Neurobiol. 17, 533–540 [DOI] [PubMed] [Google Scholar]
- 5. Griffiths I., Klugmann M., Anderson T., Yool D., Thomson C., Schwab M. H., Schneider A., Zimmermann F., McCulloch M., Nadon N., Nave K. A. (1998) Axonal swellings and degeneration in mice lacking the major proteolipid of myelin. Science 280, 1610–1613 [DOI] [PubMed] [Google Scholar]
- 6. Readhead C., Hood L. (1990) The dysmyelinating mouse mutations shiverer (shi) and myelin-deficient (shimld). Behav. Genet. 20, 213–234 [DOI] [PubMed] [Google Scholar]
- 7. Popko B., Puckett C., Lai E., Shine H. D., Readhead C., Takahashi N., Hunt S. W., 3rd, Sidman R. L., Hood L. (1987) Myelin-deficient mice. Expression of myelin basic protein and generation of mice with varying levels of myelin. Cell 48, 713–721 [DOI] [PubMed] [Google Scholar]
- 8. Aggarwal S., Yurlova L., Snaidero N., Reetz C., Frey S., Zimmermann J., Pähler G., Janshoff A., Friedrichs J., Müller D. J., Goebel C., Simons M. (2011) A size barrier limits protein diffusion at the cell surface to generate lipid-rich myelin-membrane sheets. Dev. Cell 21, 445–456 [DOI] [PubMed] [Google Scholar]
- 9. Fitzner D., Schneider A., Kippert A., Möbius W., Willig K. I., Hell S. W., Bunt G., Gaus K., Simons M. (2006) Myelin basic protein-dependent plasma membrane reorganization in the formation of myelin. EMBO J. 25, 5037–5048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Homchaudhuri L., Polverini E., Gao W., Harauz G., Boggs J. M. (2009) Influence of membrane surface charge and post-translational modifications to myelin basic protein on its ability to tether the Fyn-SH3 domain to a membrane in vitro. Biochemistry 48, 2385–2393 [DOI] [PubMed] [Google Scholar]
- 11. Klein C., Kramer E. M., Cardine A. M., Schraven B., Brandt R., Trotter J. (2002) Process outgrowth of oligodendrocytes is promoted by interaction of fyn kinase with the cytoskeletal protein tau. J. Neurosci. 22, 698–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Krämer-Albers E. M., White R. (2011) From axon-glial signaling to myelination. The integrating role of oligodendroglial Fyn kinase. Cell. Mol. Life Sci. 68, 2003–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Osterhout D. J., Wolven A., Wolf R. M., Resh M. D., Chao M. V. (1999) Morphological differentiation of oligodendrocytes requires activation of Fyn tyrosine kinase. J. Cell Biol. 145, 1209–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wake H., Lee P. R., Fields R. D. (2011) Control of local protein synthesis and initial events in myelination by action potentials. Science 333, 1647–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Goto J., Tezuka T., Nakazawa T., Sagara H., Yamamoto T. (2008) Loss of Fyn tyrosine kinase on the C57BL/6 genetic background causes hydrocephalus with defects in oligodendrocyte development. Mol. Cell. Neurosci. 38, 203–212 [DOI] [PubMed] [Google Scholar]
- 16. Sperber B. R., Boyle-Walsh E. A., Engleka M. J., Gadue P., Peterson A. C., Stein P. L., Scherer S. S., McMorris F. A. (2001) A unique role for Fyn in CNS myelination. J. Neurosci. 21, 2039–2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Umemori H., Sato S., Yagi T., Aizawa S., Yamamoto T. (1994) Initial events of myelination involve Fyn tyrosine kinase signaling. Nature 367, 572–576 [DOI] [PubMed] [Google Scholar]
- 18. Boccaccio G. L. (2000) Targeting of mRNAs within the glial cell cytoplasm. How to hide the message along the journey. J. Neurosci. Res. 62, 473–479 [DOI] [PubMed] [Google Scholar]
- 19. Carson J. H., Barbarese E. (2005) Systems analysis of RNA trafficking in neural cells. Biol. Cell 97, 51–62 [DOI] [PubMed] [Google Scholar]
- 20. Maggipinto M., Rabiner C., Kidd G. J., Hawkins A. J., Smith R., Barbarese E. (2004) Increased expression of the MBP mRNA-binding protein HnRNP A2 during oligodendrocyte differentiation. J. Neurosci. Res. 75, 614–623 [DOI] [PubMed] [Google Scholar]
- 21. White R., Gonsior C., Krämer-Albers E. M., Stöhr N., Hüttelmaier S., Trotter J. (2008) Activation of oligodendroglial Fyn kinase enhances translation of mRNAs transported in hnRNP A2-dependent RNA granules. J. Cell Biol. 181, 579–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang E., Cambi F. (2009) Heterogeneous nuclear ribonucleoproteins H and F regulate the proteolipid protein/DM20 ratio by recruiting U1 small nuclear ribonucleoprotein through a complex array of G runs. J. Biol. Chem. 284, 11194–11204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang E., Dimova N., Cambi F. (2007) PLP/DM20 ratio is regulated by hnRNPH and F and a novel G-rich enhancer in oligodendrocytes. Nucleic Acids Res. 35, 4164–4178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bugiani M., Boor I., Powers J. M., Scheper G. C., van der Knaap M. S. (2010) Leukoencephalopathy with vanishing white matter. A review. J. Neuropathol. Exp. Neurol. 69, 987–996 [DOI] [PubMed] [Google Scholar]
- 25. Labauge P., Fogli A., Niel F., Rodriguez D., Boespflug-Tanguy O. (2007) CACH/VWM syndrome and leucodystrophies related to EIF2B mutations. Rev. Neurol. 163, 793–799 [DOI] [PubMed] [Google Scholar]
- 26. Laursen L. S., Chan C. W., Ffrench-Constant C. (2011) Translation of myelin basic protein mRNA in oligodendrocytes is regulated by integrin activation and hnRNP-K. J. Cell Biol. 192, 797–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Trotter J., Bitter-Suermann D., Schachner M. (1989) Differentiation-regulated loss of the polysialylated embryonic form and expression of the different polypeptides of the neural cell adhesion molecule by cultured oligodendrocytes and myelin. J. Neurosci. Res. 22, 369–383 [DOI] [PubMed] [Google Scholar]
- 28. Jung M., Krämer E., Grzenkowski M., Tang K., Blakemore W., Aguzzi A., Khazaie K., Chlichlia K., von Blankenfeld G., Kettenmann H. (1995) Lines of murine oligodendroglial precursor cells immortalized by an activated neu tyrosine kinase show distinct degrees of interaction with axons in vitro and in vivo. Eur. J. Neurosci. 7, 1245–1265 [DOI] [PubMed] [Google Scholar]
- 29. Twamley-Stein G. M., Pepperkok R., Ansorge W., Courtneidge S. A. (1993) The Src family tyrosine kinases are required for platelet-derived growth factor-mediated signal transduction in NIH 3T3 cells. Proc. Natl. Acad. Sci. U.S.A. 90, 7696–7700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Arthur P. K., Claussen M., Koch S., Tarbashevich K., Jahn O., Pieler T. (2009) Participation of Xenopus Elr-type proteins in vegetal mRNA localization during oogenesis. J. Biol. Chem. 284, 19982–19992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kalifa Y., Huang T., Rosen L. N., Chatterjee S., Gavis E. R. (2006) Glorund, a Drosophila hnRNP F/H homolog, is an ovarian repressor of nanos translation. Dev. Cell 10, 291–301 [DOI] [PubMed] [Google Scholar]
- 32. Kosturko L. D., Maggipinto M. J., Korza G., Lee J. W., Carson J. H., Barbarese E. (2006) Heterogeneous nuclear ribonucleoprotein (hnRNP) E1 binds to hnRNP A2 and inhibits translation of A2-response element mRNAs. Mol. Biol. Cell 17, 3521–3533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Munro T. P., Magee R. J., Kidd G. J., Carson J. H., Barbarese E., Smith L. M., Smith R. (1999) Mutational analysis of a heterogeneous nuclear ribonucleoprotein A2-response element for RNA trafficking. J. Biol. Chem. 274, 34389–34395 [DOI] [PubMed] [Google Scholar]
- 34. Hüttelmaier S., Zenklusen D., Lederer M., Dictenberg J., Lorenz M., Meng X., Bassell G. J., Condeelis J., Singer R. H. (2005) Spatial regulation of β-actin translation by Src-dependent phosphorylation of ZBP1. Nature 438, 512–515 [DOI] [PubMed] [Google Scholar]
- 35. Song K. Y., Choi H. S., Law P. Y., Wei L. N., Loh H. H. (2011) Post-transcriptional regulation of μ-opioid receptor. Role of the RNA-binding protein heterogeneous nuclear ribonucleoprotein H1 and F. Cell. Mol. Life Sci., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Krämer-Albers E. M., Gehrig-Burger K., Thiele C., Trotter J., Nave K. A. (2006) Perturbed interactions of mutant proteolipid protein/DM20 with cholesterol and lipid rafts in oligodendroglia. Implications for dysmyelination in spastic paraplegia. J. Neurosci. 26, 11743–11752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gao Y., Tatavarty V., Korza G., Levin M. K., Carson J. H. (2008) Multiplexed dendritic targeting of α calcium calmodulin-dependent protein kinase II, neurogranin, and activity-regulated cytoskeleton-associated protein RNAs by the A2 pathway. Mol. Biol. Cell 19, 2311–2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Colman D. R., Kreibich G., Frey A. B., Sabatini D. D. (1982) Synthesis and incorporation of myelin polypeptides into CNS myelin. J. Cell Biol. 95, 598–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ostareck-Lederer A., Ostareck D. H., Cans C., Neubauer G., Bomsztyk K., Superti-Furga G., Hentze M. W. (2002) c-Src-mediated phosphorylation of hnRNP K drives translational activation of specifically silenced mRNAs. Mol. Cell. Biol. 22, 4535–4543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Umemori H., Kadowaki Y., Hirosawa K., Yoshida Y., Hironaka K., Okano H., Yamamoto T. (1999) Stimulation of myelin basic protein gene transcription by Fyn tyrosine kinase for myelination. J. Neurosci. 19, 1393–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Goto J., Tezuka T., Nakazawa T., Tsukamoto N., Nakamura T., Ajima R., Yokoyama K., Ohta T., Ohki M., Yamamoto T. (2004) Altered gene expression in the adult brain of fyn-deficient mice. Cell. Mol. Neurobiol. 24, 149–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Colognato H., Ramachandrappa S., Olsen I. M., ffrench-Constant C. (2004) Integrins direct Src family kinases to regulate distinct phases of oligodendrocyte development. J. Cell Biol. 167, 365–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Laursen L. S., Chan C. W., ffrench-Constant C. (2009) An integrin-contactin complex regulates CNS myelination by differential Fyn phosphorylation. J. Neurosci. 29, 9174–9185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liang X., Draghi N. A., Resh M. D. (2004) Signaling from integrins to Fyn to Rho family GTPases regulates morphologic differentiation of oligodendrocytes. J. Neurosci. 24, 7140–7149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nakahara J., Tan-Takeuchi K., Seiwa C., Gotoh M., Kaifu T., Ujike A., Inui M., Yagi T., Ogawa M., Aiso S., Takai T., Asou H. (2003) Signaling via immunoglobulin Fc receptors induces oligodendrocyte precursor cell differentiation. Dev. Cell 4, 841–852 [DOI] [PubMed] [Google Scholar]
- 46. Honoré B., Baandrup U., Vorum H. (2004) Heterogeneous nuclear ribonucleoproteins F and H/H′ show differential expression in normal and selected cancer tissues. Exp. Cell Res. 294, 199–209 [DOI] [PubMed] [Google Scholar]
- 47. Matunis M. J., Xing J., Dreyfuss G. (1994) The hnRNP F protein. Unique primary structure, nucleic acid-binding properties, and subcellular localization. Nucleic Acids Res. 22, 1059–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hatfield J. T., Rothnagel J. A., Smith R. (2002) Characterization of the mouse hnRNP A2/B1/B0 gene and identification of processed pseudogenes. Gene 295, 33–42 [DOI] [PubMed] [Google Scholar]
- 49. Han S. P., Friend L. R., Carson J. H., Korza G., Barbarese E., Maggipinto M., Hatfield J. T., Rothnagel J. A., Smith R. (2010) Differential subcellular distributions and trafficking functions of hnRNP A2/B1 spliceoforms. Traffic 11, 886–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Francone V. P., Maggipinto M. J., Kosturko L. D., Barbarese E. (2007) The microtubule-associated protein tumor overexpressed gene/cytoskeleton-associated protein 5 is necessary for myelin basic protein expression in oligodendrocytes. J. Neurosci. 27, 7654–7662 [DOI] [PMC free article] [PubMed] [Google Scholar]